Abstract
Glycopeptidolipids (GPLs) are major components present on the outer layers of the cell walls of several nontuberculous mycobacteria. GPLs are antigenic molecules and have variant oligosaccharides in mycobacteria such as Mycobacterium avium. In this study, we identified four genes (gtf1, gtf2, gtf3, and gtf4) in the genome of Mycobacterium smegmatis. These genes were independently inactivated by homologous recombination in M. smegmatis, and the structures of GPLs from each gene disruptant were analyzed. Thin-layer chromatography, gas chromatography-mass spectrometry, and matrix-assisted laser desorption ionization-time-of-flight mass spectrometry analyses revealed that the mutants Δgtf1 and Δgtf2 accumulated the fatty acyl-tetrapeptide core having O-methyl-rhamnose and 6-deoxy-talose as sugar residues, respectively. The mutant Δgtf4 possessed the same GPLs as the wild type, whereas the mutant Δgtf3 lacked two minor GPLs, consisting of 3-O-methyl-rhamnose attached to O-methyl-rhamnose of the fatty acyl-tetrapeptide core. These results indicate that the gtf1 and gtf2 genes are responsible for the early glycosylation steps of GPL biosynthesis and the gtf3 gene is involved in transferring a rhamnose residue not to 6-deoxy-talose but to an O-methyl-rhamnose residue. Moreover, a complementation experiment showed that M. avium gtfA and gtfB, which are deduced glycosyltransferase genes of GPL biosynthesis, restore complete GPL production in the mutants Δgtf1 and Δgtf2, respectively. Our findings propose that both M. smegmatis and M. avium have the common glycosylation pathway in the early steps of GPL biosynthesis but differ at the later stages.
The mycobacterial cell envelope has a unique structure that contains a complex of covalently linked peptidoglycan, arabinogalactan, and mycolic acids (7, 11). The outer layer of the cell envelope is composed of several types of glycolipids that affect the surface properties of mycobacterial cells (7, 11). Glycopeptidolipids (GPLs) are a major class of glycolipid present on the outer layer of several species of nontuberculous mycobacteria, such as Mycobacterium avium complex, M. scrofulaceum, M. chelonae, M. fortuitum, and M. smegmatis (31). GPLs have a common fatty acyl-tetrapeptide core consisting of tetrapeptide amino alcohol (d-Phe-d-allo-Thr-d-Ala-l-alaninol) and amide-linked long-chain fatty acid (C26-34). The fatty acyl-tetrapeptide core is glycosylated with 6-deoxy-talose (6-d-Tal) and variable O-methyl-rhamnose (O-Me-Rha) residues, termed non-serovar-specific GPLs (nsGPLs), which are also the main products of M. smegmatis GPLs (1, 4, 10). The GPLs of M. avium have a more complicated structure, in which an additional Rha residue is added to 6-d-Tal of nsGPLs to be extended with various haptenic oligosaccharides, which are important surface antigens, resulting in serovar-specific GPLs (ssGPLs) (1, 4, 31).
There are some evidences that GPLs may be responsible for pathogenicity. It has been shown that the some of the ssGPLs are immunosuppressive and are able to induce a variety of cytokines, which affect host responses to infection (3, 15, 18, 29). Also, ssGPLs are identified as the factors modulating the phagocytosis and phagosome-lysosome fusion (17, 21). The altered GPL structure is also known to affect the colony morphology relevant to variable virulence (14, 30).
The biosyntheses of GPLs, particularly nsGPLs, have been characterized for M. smegmatis. Several biosynthetic genes encoding enzymes such as O-methyltransferase, acetyltransferase, and peptide synthetase have been identified (5, 16, 25, 26), but less is known about the genes involved in the glycosylation steps of the GPLs. The only glycosyltransferase gene that has been characterized is rtfA from M. avium, which is responsible for transferring the Rha residue to 6-d-Tal of nsGPLs to form ssGPLs (12). However, the initial glycosylation steps for the formation of nsGPLs remain unknown. Recently, it was shown that GPLs from M. smegmatis has a unique structure in which nsGPLs are further glycosylated, unlike ssGPLs (23, 24, 32), but these unique GPLs are produced in a carbon-starved situation, which is not a normal growth condition.
In this study, to clarify the glycosylation step leading to the formation of nsGPLs and its further products, we focused on four of the M. smegmatis genes having high similarity to M. avium rtfA, whose functions remain uncharacterized. Here, we have undertaken the gene disruption approach for generating each mutant in M. smegmatis, characterized their biochemical phenotypes, and finally hypothesized new biosynthetic pathways associated with glycosylation of GPLs.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and DNA manipulation.
Bacterial strains and vectors used and constructed are listed in Table 1. Mycobacterial strains for DNA manipulation were grown in Middlebrook 7H9 broth (Difco) with 0.05% Tween 80 or Middlebrook 7H10 agar (Difco) with 0.5% glycerol, and each was supplemented with 10% albumin-dextrose-catalase enrichment (Difco). M. smegmatis strains for GPL production were cultured in Luria-Bertani (LB) broth with 0.05% Tween 80. DNA manipulation including isolation of DNA, transformation, and PCR was carried out as described previously (22). E. coli strain DH5α was used for routine manipulation and propagation of plasmid DNA. E. coli strain STBL2 was used for construction of phasmid vectors derived from phAE87. Antibiotics was added as required: kanamycin, 50 μg/ml for E. coli and 25 μg/ml for M. smegmatis; hygromycin B, 150 μg/ml for E. coli and 75 μg/ml for M. smegmatis.
TABLE 1.
Bacterial strains and vectors used in this study
| Strain or vector | Characteristic(s) | Source or reference |
|---|---|---|
| Bacteria | ||
| E. coli | ||
| DH5α | Cloning host | |
| STBL2 | Cloning host | |
| M. smegmatis | ||
| mc2155 | Wild type | 27 |
| Δgtf1 | gtf1 disruptant | This study |
| Δgtf2 | gtf2 disruptant | This study |
| Δgtf3 | gtf3 disruptant | This study |
| Δgtf4 | gtf4 disruptant | This study |
| M. avium | ||
| JATA51-01 (ATCC 25291) | Source of gtfA and gtfB | |
| Vectors | ||
| pYUB854 | Cosmid vector | 2 |
| phAE87 | Phasmid vector carrying full-length DNA of mycobacteriophage PH101 | 2 |
| pMV261 | E. coli-Mycobacterium shuttle vector carrying hsp60 promoter cassette | 28 |
| pYUBgtf1 | pYUB854 with gtf1-disrupted sequences for generating recombinant mycobacteriophage | This study |
| pYUBgtf2 | pYUB854 with gtf2-disrupted sequences for generating recombinant mycobacteriophage | This study |
| pYUBgtf3 | pYUB854 with gtf3-disrupted sequences for generating recombinant mycobacteriophage | This study |
| pYUBgtf4 | pYUB854 with gtf4-disrupted sequences for generating recombinant mycobacteriophage | This study |
| pMVgtf1 | pMV261 with gtf1 | This study |
| pMVgtf2 | pMV261 with gtf2 | This study |
| pMVgtf3 | pMV261 with gtf3 | This study |
| pMVgtf4 | pMV261 with gtf4 | This study |
| pMVgtfA | pMV261 with gtfA | This study |
| pMVgtfB | pMV261 with gtfB | This study |
Generation of the gene disruptants.
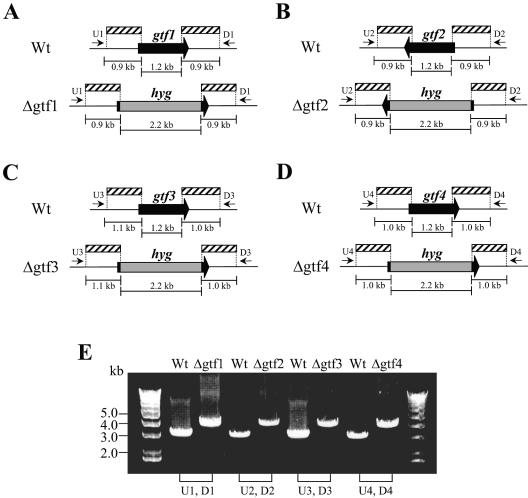
The targeted genes (gtf1, gtf2, gtf3, and gtf4) were selected by BLAST analysis of unfinished M. smegmatis genome sequences deposited in the database of The Institute for Genomic Research (TIGR) (http://www.tigr.org) with the rtfA gene of M. avium as the query nucleotide sequence. Each gene was inactivated by inserting a hygromycin-resistant cassette (hyg) using the specialized transducing phage system (2). To construct the disrupted sequences, around 1.0-kb fragments both upstream and downstream of each gene were amplified from M. smegmatis mc2155 genomic DNA using the following two pairs of primers: US1 and UA1 for upstream of gtf1 and DS1 and DA1 for downstream of gtf1; US2 and UA2 for upstream of gtf2 and DS2 and DA2 for downstream of gtf2; US3 and UA3 for upstream of gtf3 and DS3 and DA3 for downstream of gtf3; US4 and UA4 for upstream of gtf4 and DS4 and DA4 for downstream of gtf4. The PCR products were digested with each restriction enzyme and cloned into the corresponding sites flanking hyg of pYUB854 to give pYUBgtf1 (gtf1), pYUBgtf1 (gtf2), pYUBgtf3 (gtf3), and pYUBgtf4 (gtf4). These plasmids were used for packaging into the phasmid vector phAE87 to construct a specialized transducing mycobacteriophage for gene disruption as described previously (2). The M. smegmatis mc2155 strain infected with the above mycobacteriophage at a multiplicity of infection of 10 was incubated at 37°C for 3 h in 7H9 broth without Tween 80. Harvested bacterial cells were then plated and cultured on 7H10 agar containing 75 μg/ml hygromycin B for 1 week. The hygromycin B-resistant colonies were selected, and their genomic DNA was subjected to PCR analysis to confirm the disruption of each gene using the following primers: U1 and D1 for gtf1; U2 and D2 for gtf2; U3 and D3 for gtf3; and U4 and D4 for gtf4 (Fig. 1A to D).
FIG. 1.
Generation of gtf gene disruptants. (A to D) Schematic diagram of each gtf region on the chromosome of the wild-type M. smegmatis mc2155 strain (Wt) and its gene disruptants Δgtf1, Δgtf2, Δgtf3, and Δgtf4. The shaded boxes indicate the regions included in recombinant phage for gene disruption. The black arrows represent the coding region of each gtf gene. The gray boxes represent the hygromycin resistance cassette (hyg). The primers used for PCR analysis are indicated by small arrows. (E) PCR analyses of the wild type and each disruptant using the primers indicated above.
Construction of the gtf expression vectors.
The gtf genes of M. smegmatis and M. avium were amplified from each genomic DNA using the following primers: GTF1S and GTF1A for gtf1, GTF2S and GTF2A for gtf2, GTF3S and GTF3A for gtf3, GTF4S and GTF4A for gtf4, GTFAS and GTFAA for gtfA, and GTFBS and GTFBA for gtfB. The PCR products were digested with each restriction enzyme and cloned into the corresponding site of pMV261 to give pMVgtf1 (for the gtf1 gene), pMVgtf2 (for the gtf2 gene), pMVgtf3 (for the gtf3 gene), pMVgtf4 (for the gtf4 gene), pMVgtfA (for the gtfA gene), and pMVgtfB (for the gtfB gene). These vectors were used for complementation and overexpression experiment.
Isolation and purification of GPLs.
The total lipids were extracted from harvested bacterial cells with CHCl3/CH3OH (2:1, vol/vol) for several hours at room temperature. The extracts from the organic phase were separated from the aqueous phase and evaporated to dryness. For isolation of crude deacylated GPLs, total lipid fractions were subjected to mild alkaline hydrolysis as previously described (22, 25). For analytical thin-layer chromatography (TLC), the total lipid fraction after mild alkaline hydrolysis was spotted on silica gel 60 plates (Merck) and developed in CHCl3-CH3OH (9:1 [vol/vol]). Deacylated GPLs and other compounds were visualized by spraying with 10% H2SO4 and charring. Each total lipid fraction was extracted from an equal weight of harvested cells. Purified deacylated GPLs were separated from the total lipid fraction after mild alkaline hydrolysis by preparative TLC on the same plates and extracted from the bands corresponding to each GPLs. β-Elimination and perdeuteriomethylation treatment for determination of the linkage positions of sugar moieties were carried out as described previously (6, 9, 12).
GC/MS analysis.
For monosaccharide analysis, purified deacylated GPLs or total lipid fraction after mild alkaline hydrolysis was hydrolyzed in 2 M trifluoroacetic acid (2 h, 120°C), and released sugars from deacylated GPLs were reduced with NaBD4 (sodium borodeuteride) and then acetylated with pyridine-acetic anhydride (1:1 [vol/vol]) at room temperature overnight. Each total lipid fraction was extracted from an equal weight of harvested cells. The resulting alditol acetates were separated and analyzed by gas chromatography-mass spectrometry (GC/MS) on TRACE DSQ (Thermo electron) instrument equipped with an SP-2380 column (SUPELCO) using helium gas. The temperature program was from 52 to 172°C at 40°C/min and then 172 to 250°C at 3°C/min.
MALDI-TOF/MS analysis.
To determine the total mass of the purified deacylated GPLs, matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) mass spectra (in the positive mode) were acquired on a QSTAR XL (Applied Biosystems) with a pulse laser emitting at 337 nm. Samples mixed with 2,5-dihydroxybenzoic acid as the matrix were analyzed in the reflectron mode with an accelerating voltage operating in positive ion mode of 20 kV.
RESULTS
Disruption of gtf1, gtf2, gtf3, and gtf4 by allelic exchange.
Four genes showing high similarity to the rtfA gene, involved in GPL biosynthesis of M. avium, were identified for the M. smegmatis mc2155 strain (12). The homologies of their corresponding amino acid sequences with that of RtfA were around 60%. Three genes were found in the GPL biosynthetic gene cluster, namely, gtf1, gtf2, and gtf3 (GenBank accession no. AY138899.1) (16), whereas one gene, designated gtf4 (TIGR database no. 4839918 to 4841162), was located far from the other three genes. To examine whether these genes are responsible for GPL biosynthesis, we generated four gene disruptants, designated Δgtf1, Δgtf2, Δgtf3, and Δgtf4, using the specialized transducing mycobacteriophage containing the entire open reading frame, replacing with the hygromycin resistance cassette (2). For confirmation of the gene disruption, PCR analysis was performed on chromosomal DNA from each disruptant. To avoid the amplification of disrupted sequences derived from residual mycobacteriophage, we designed and used the primers located outside the sequences included in each mycobacteriophage as shown in Fig. 1A to D. As expected, around 3.0-kb fragments were amplified from mc2155 (wild type), whereas around 4.0-kb fragments were amplified from each disruptant, because most of the gtf coding region (1.2 kb) was replaced by the hygromycin resistance cassette (2.2 kb) (Fig. 1E). These results demonstrated that allelic exchanges involving replacement of the gtf genes with the disrupted constructs have been successful.
TLC analysis of gene disruptants.
To investigate the effects of the mutation in each gtf gene, we examined GPL production of four gene disruptants. TLC analyses of total lipid fraction after mild alkaline hydrolysis revealed that wild-type mc2155 mainly produced six components, designated GPL-1 to -6, whereas Δgtf1 and Δgtf2 lacked all six components and Δgtf3 lacked two minor ones (GPL-5 and GPL-6) found in the wild type (Fig. 2). In contrast, no differences in TLC profile were observed between Δgtf4 and the wild type (Fig. 2).
FIG. 2.
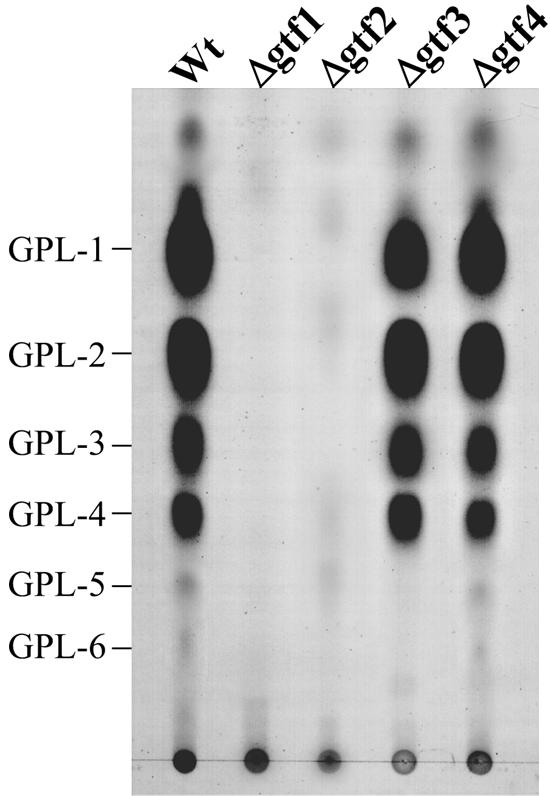
TLC analyses of crude GPL extracts from the M. smegmatis mc2155 strain (Wt) and its gene disruptants. The total lipid fraction after mild alkaline hydrolysis was spotted on plates and developed in CHCl3-CH3OH (9:1 [vol/vol]). GPLs were visualized by spraying with 10% H2SO4 and charring. Each total lipid fraction was extracted from an equal weight of harvested cells.
Characterization of Δgtf1 and Δgtf2.
In Δgtf1 and Δgtf2, the TLC analyses showed that six GPL components contained in the wild type had disappeared. On the other hand, there is the possibility that both disruptants contained GPL derivatives which are structurally incomplete and hard to be detected by TLC analyses. To characterize the sugars included in GPL derivatives from both disruptants and to compare with the wild type, each total lipid fraction after mild alkaline hydrolysis was hydrolyzed, and the released monosaccharides as their alditol acetates were examined by GC/MS. Figure 3 shows that the profiles of the wild type gave three peaks corresponding to 2,3,4-tri-O-Me-Rha, 3,4-di-O-Me-Rha, and 6-d-Tal (Fig. 3A), whereas Δgtf1 lacked 6-d-Tal (Fig. 3B) and Δgtf2 lacked 3,4-di-O-Me-Rha and 2,3,4-tri-O-Me-Rha (Fig. 3C). Complementation of both disruptants with each respective gene restored the TLC profile of GPLs to that observed for the wild type (not shown). Therefore, the gtf1 and gtf2 genes are found to be responsible for transferring the 6-d-Tal and Rha residues, respectively.
FIG. 3.
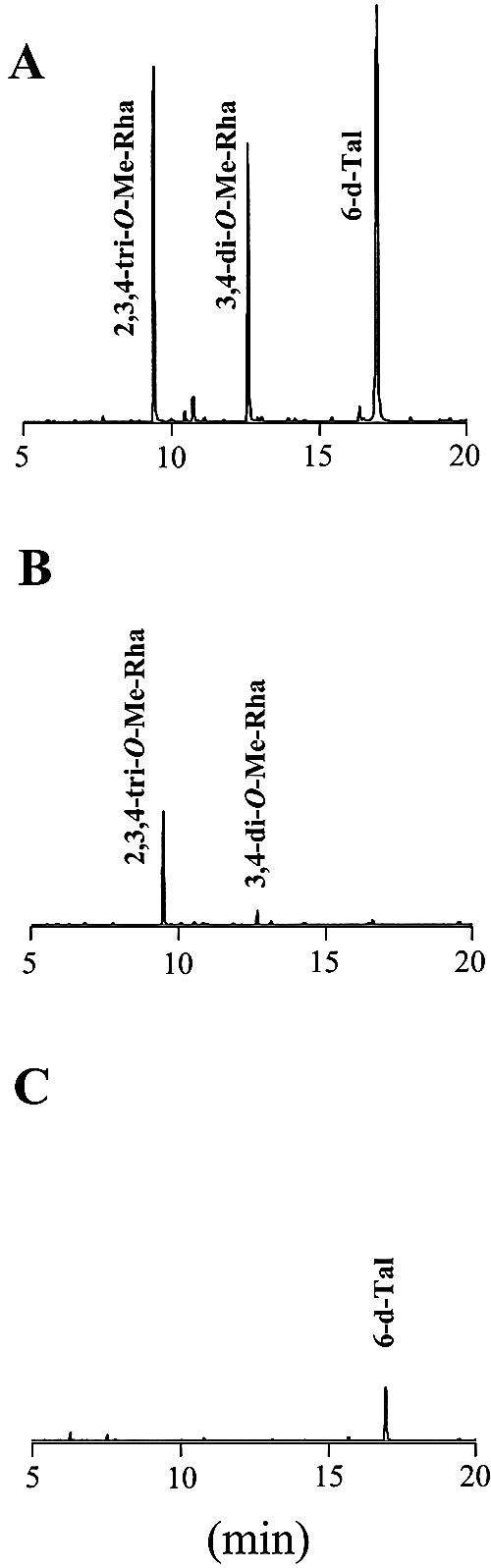
GC/MS analyses of alditol acetates of sugars released from crude GPLs. GPLs were extracted from M. smegmatis strains: (A) mc2155 strain, (B) Δgtf1, and (C) Δgtf2. Alditol acetate derivatives were prepared from the total lipid fraction after mild alkaline hydrolysis, which was extracted from an equal weight of harvested cells.
Structural determination of GPL-5 and GPL-6 for characterization of Δgtf3.
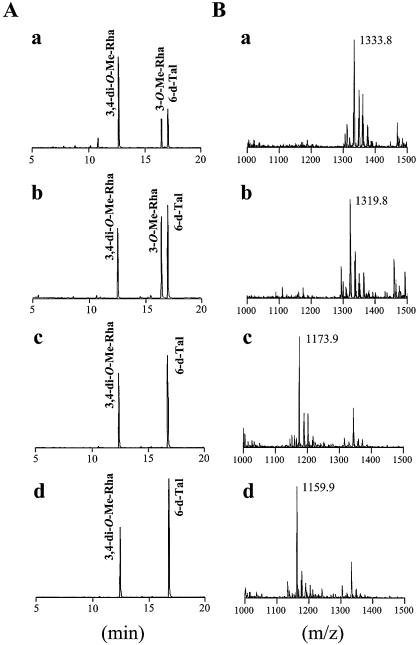
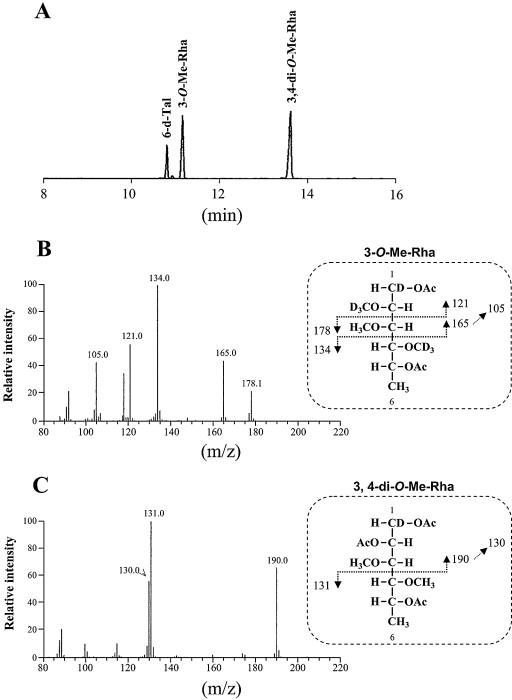
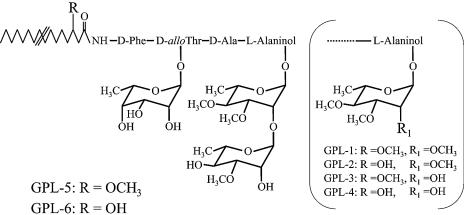
The TLC profile of Δgtf3 showed that two spots (GPL-5 and GPL-6) disappeared (Fig. 2). To reveal the biosynthetic role of the gtf3 gene, GPL-5 and GPL-6 were purified from mc2155 and their structures were determined. GC/MS analyses showed that GPL-5 and GPL-6 contained 6-d-Tal and 3,4-di-O-Me-Rha, which were identified as sugar moieties of GPL-3 and GPL-4 (Fig. 4A). However, an extra sugar, 3-O-Me-Rha, was also detected (Fig. 4A). MALDI-TOF/MS analyses revealed that the main molecular ions of GPL-5 (m/z 1,333.8) and GPL-6 (m/z 1,319.8) were 160 mass units higher than those of GPL-3 (m/z 1,173.9) and GPL-4 (m/z 1,159.9), respectively (Fig. 4B). These results confirmed the presence of 3-O-Me-Rha in GPL-5 and GPL-6 and also suggested that 3-O-Me-Rha was further added to GPL-3 and GPL-4. Although GPL-5 and GPL-6 contained same three sugars, the spectra showed that the main molecular ion of GPL-5 (m/z 1,333.8) was 14 mass units higher than that of GPL-6 (m/z 1,319.8) (Fig. 4Ba and 4Bb). These differences in total mass may be due to O methylation of fatty acid as observed in structures of GPL-1 and GPL-3, suggesting that fatty acid of GPL-5 was O methylated like GPL-1 and GPL-3 (16). To investigate the sugar linked to d-allo-Thr of the fatty acyl-tetrapeptide core, GPL-5 and GPL-6 were subjected to β-elimination treatment. The main ion peaks of treated GPL-5 and GPL-6 were m/z 1,171.7 and 1,157.7, respectively, which resulted in the loss of total mass of 162, suggesting that 6-d-Tal was linked to the position of d-allo-Thr (Fig. 4C). The linkage position of the sugars linked to the l-alaninol site of GPL-5 and GPL-6 was then determined by GC/MS analyses followed by perdeuteriomethylation. As shown in Fig. 5A, the GC profiles of alditol acetates from perdeuteriomethylated GPL-5 gave three peaks corresponding to 6-d-Tal, 3-O-Me-Rha, and 3,4-di-O-Me-Rha. The characteristic spectra of 3-O-Me-Rha and 3,4-di-O-Me-Rha, which are predicted to be linked to l-alaninol, are illustrated in Fig. 5B and C, respectively. The spectrum of 3-O-Me-Rha gave fragment ions at m/z 121, 134, and 165, which represent the presence of a deuteriomethyl group at positions C-2 and C-4. In contrast, no deuteriomethyl group was observed in 3,4-di-O-Me-Rha, whose C-2 position was acetylated, supported by the detection of fragment ions at m/z 131 and 190. The results from GC/MS analyses of perdeuteriomethylated GPL-6 were the same as those for GPL-5 (not shown). These observations demonstrated that GPL-5 and GPL-6 have the same sugar moieties, which are 6-d-Tal at D-allo-Thr and 3-O-Me-rhamnosyl-(1→2)-3,4-di-O-Me-Rha at l-alaninol, indicating that 3-O-Me-Rha was linked to GPL-3 and GPL-4 (Fig. 6).
FIG. 4.
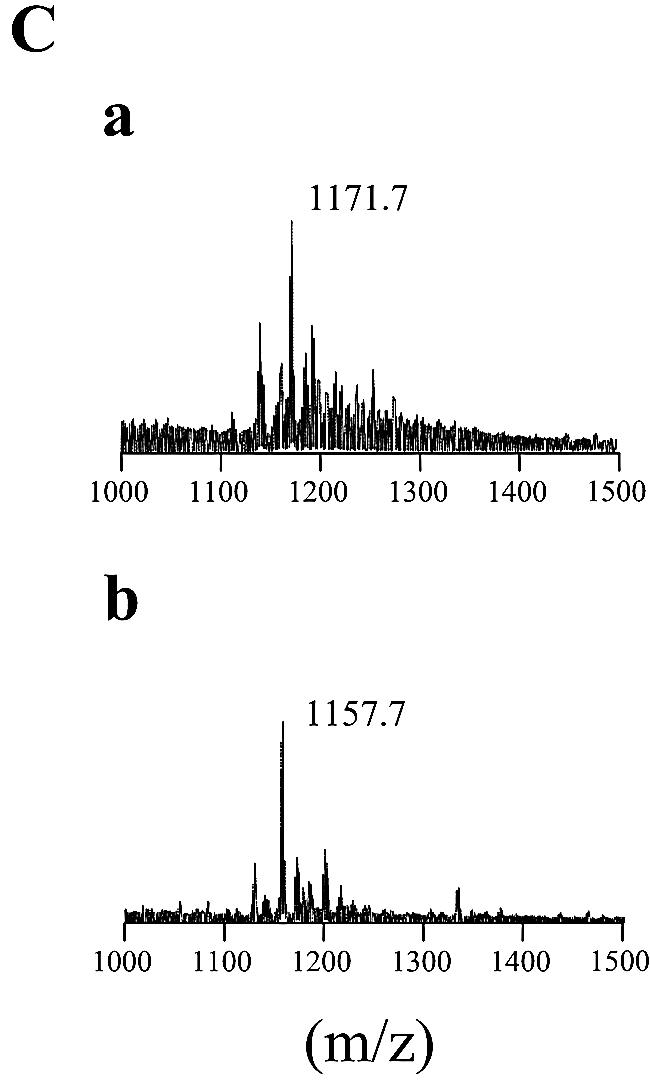
Biochemical characterization of GPL-5 (a), GPL-6 (b), GPL-3 (c), and GPL-4 (d). (A) GC/MS analysis of alditol acetates of sugars released from each purified GPL. (B) MALDI-TOF/MS analysis of total molecular mass of each purified GPLs. (C) MALDI-TOF/MS analysis of total molecular mass of purified GPL-5 (a) and GPL-6 (b), which were subjected to β-elimination.
FIG. 5.
GC/MS analysis of alditol acetates of sugars released from perdeuteriomethylated GPL-5. (A) GC profile. (B) Mass spectrum and fragment ion assignment corresponding to 3-O-Me-Rha. (C) Mass spectrum of fragment ion assignment corresponding to 3,4-di-O-Me-Rha.
FIG. 6.
Proposed structures of GPL-5 and GPL-6. Figure in parentheses shows the structure of GPL-1, GPL-2, GPL-3, and GPL-4, which were characterized in previous studies (10, 16, 25).
Overexpression of gtf1, gtf2, gtf3, and gtf4 in M. smegmatis mc2155.
To investigate the effects of overexpression of each gene on GPL biosynthesis, we constructed four gtf-overexpressed strains in wild-type mc2155 and compared the profile of total GPLs by TLC analyses. The results showed that the profiles of Wt/pMVgtf1, Wt/pMVgtf2, and Wt/pMVgtf4 were the same as that of Wt/pMV261, whereas Wt/pMVgtf3 produced two major compounds whose biochemical data corresponded to those of GPL-5 and GPL-6 (Fig. 7).
FIG. 7.
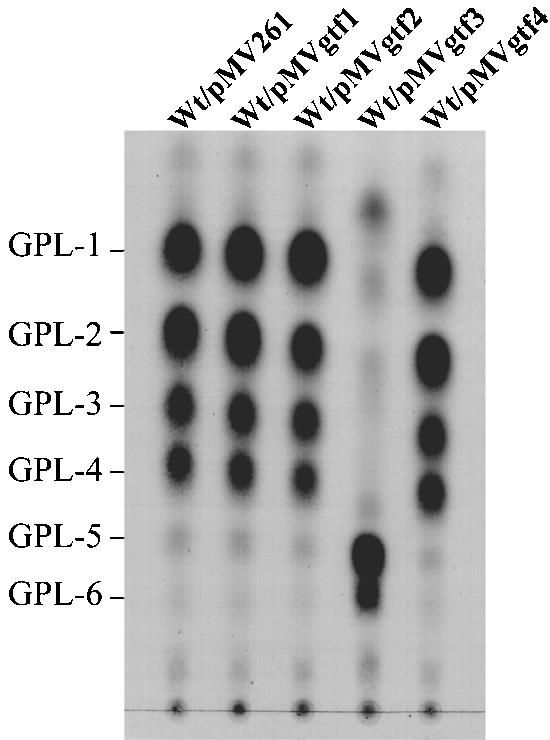
TLC analyses of crude GPL extracts from the M. smegmatis mc2155 strain (Wt) transformed with gtf expression vectors. Total lipid fraction after mild alkaline hydrolysis was spotted on plates and developed in CHCl3-CH3OH (9:1 [vol/vol]). GPLs were visualized by spraying with 10% H2SO4 and charring. Each total lipid fraction was extracted from an equal weight of harvested cells.
Characterization of M. avium gtfA and gtfB.
We showed that both M. smegmatis gtf1 and gtf2 were responsible for glycosylation of the fatty acyl-tetrapeptide core. Comparison of the genome sequences encompassing the GPL biosynthetic gene cluster among several species of M. avium have shown that gtfA and gtfB (GenBank accession no. AF125999.1) are very similar to M. smegmatis gtf1 and gtf2, respectively, in the corresponding putative amino acid sequences and might contribute to the glycosylation of the fatty acyl-tetrapeptide core (13). However, the function of each gene has not been thoroughly analyzed (13). Therefore, to confirm the role of gtfA and gtfB, we complemented Δgtf1 and Δgtf2 with the gtf expression vectors carrying gtfA (pMVgtfA) and gtfB (pMVgtfB). As shown in Fig. 8, TLC analyses revealed that gtfA and gtfB restored the production of wild-type GPLs in Δgtf1 and Δgtf2, respectively, whereas transformants with reverse vectors (Δgtf1/pMVgtfB and Δgtf2/pMVgtfA) did not produce wild-type GPLs. These results suggested that the function of M. avium gtfA and gtfB is the same as that of M. smegmatis gtf1 and gtf2, respectively.
FIG. 8.
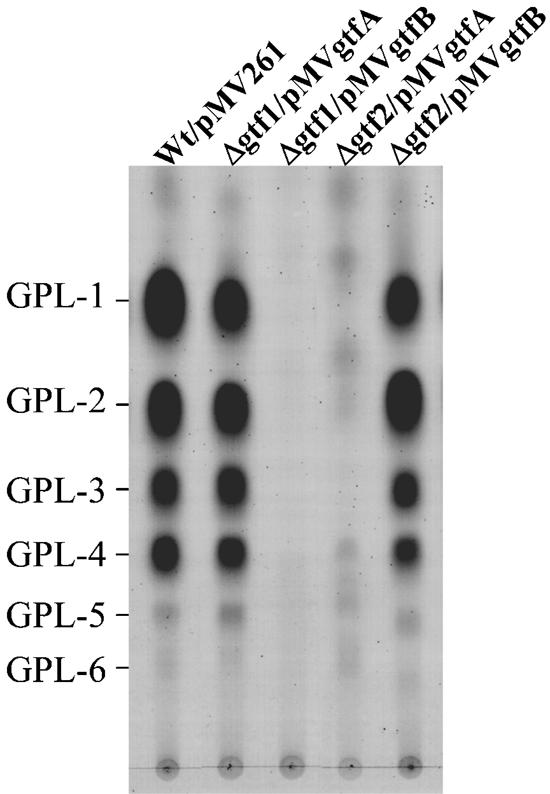
TLC analyses of crude GPL extracts from the M. smegmatis mc2155 strain (Wt) and its gene disruptants transformed with M. avium gtfA and gtfB. Total lipid fraction after mild alkaline hydrolysis was spotted on plates and developed in CHCl3-CH3OH (9:1 [vol/vol]). GPLs were visualized by spraying with 10% H2SO4 and charring. Each total lipid fraction was extracted from an equal weight of harvested cells.
DISCUSSION
It has been shown that the rtfA gene of M. avium encodes a rhamnosyltransferase which synthesizes ssGPLs, while other genes involved in the glycosylation of the fatty acyl-tetrapeptide core remain unknown (12). In this study, we focused on the four genes of M. smegmatis, which show high similarity to rtfA, and generated their disruptants to characterize the role in the GPL biosynthesis.
In the early glycosylation steps of the fatty acyl-tetrapeptide core, we observed that the disruption of gtf1 abolished the whole GPLs and led to the accumulation of O-Me-Rha derivatives without 6-d-Tal in Δgtf1 (Fig. 3B). Thus, we propose that the gtf1 gene product catalyzes the transfer of 6-d-Tal to fatty acyl-tetrapeptide core. It is reported that the M. avium 104Rg strain, which has a spontaneous deletion in the genome region including gtfA, also accumulated O-methylated and nonmethylated Rha without 6-d-Tal (13, 30). This property is directly supported by our result that the gtfA could complement Δgtf1 (Fig. 8). However, M. avium 104Rg mainly contained nonmethylated Rha, whereas Δgtf1 derived from M. smegmatis mc2155 contained only O-Me-Rha. These different observations may be due to differences in the substrate specificity of methyltransferase, because 2,3,4-tri-O-Me-Rha was present in M. smegmatis mc2155 but was not identified in M. avium species (8, 25).
When the gtf2 gene was disrupted, we detected 6-d-Tal without Rha derivatives in GC/MS analysis, which demonstrates that the gtf2 gene contributes to the transfer of Rha to the fatty acyl-tetrapeptide core (Fig. 3C). In addition, complementation revealed that the gtfB gene of M. avium had the same function as gtf2 (Fig. 8). In the previous studies of GPL biosyntheses, the mutant accumulating 6-d-Tal-containing derivatives without the Rha residue have not been isolated from GPL-producing species so far. Our results directly indicated for the first time that 6-d-Tal-containing derivatives could be an intermediate for the biosynthetic pathways of GPLs.
As for the order of glycosylation steps regulated by gtf1 and gtf2, we cannot determine which step takes place earlier, since both disruptants accumulated the intermediates having different component (Fig. 3B and C). For M. avium serovar 2, Eckstein et al. proposed a pathway in which the transfer of the Rha residue to the fatty acyl-tetrapeptide core occurred prior to that of 6-d-Tal, because a mutant strain, 104Rg, having the gtfA region deleted, accumulated the fatty acyl-tetrapeptide core with only the Rha residue (13). However, our results lead to the interesting possibility that there are two alternative glycosylation pathways for the formation of nsGPLs (Fig. 9). If the glycosylation should occur in a single pathway, we would expect the accumulation of a nonglycosylated intermediate in either of the disruptants, because one of the genes, gtf1 or gtf2, would be responsible for the first step of glycosylation converting the fatty acyl-tetrapeptide core to a glycosylated intermediate. Thus, the detection of glycosylated intermediates from both Δgtf1 and Δgtf2 suggests that (i) the fatty acyl-tetrapeptide core could be the substrate for both Gtf1 and Gtf2 and (ii) the glycosylated intermediates could also be the substrates for both Gtf1 and Gtf2. We prove here that Gtf1 and Gtf2 have broad substrate specificity and propose that the fatty acyl-tetrapeptide core is glycosylated by Gtf1 and Gtf2 at the same time and then converted to the nsGPLs having both 6-d-Tal and O-Me-Rha via cross-glycosylations (Fig. 9).
FIG. 9.
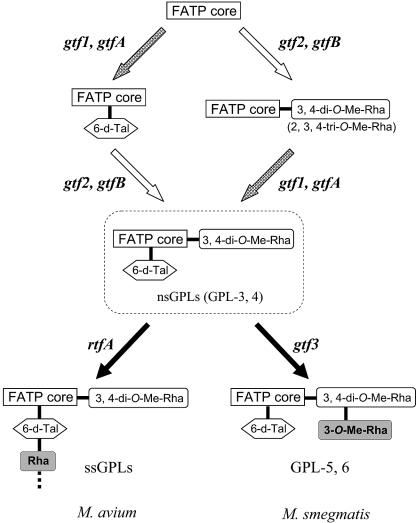
Proposed biosynthetic pathways for GPLs of M. smegmatis and M. avium. FATP core, fatty acyl-tetrapeptide core.
Structural determination of GPL-5 and GPL-6 revealed that l-alaninol of the fatty acyl-tetrapeptide core was glycosylated with disaccharide (3-O-Me- and 3,4-di-O-Me-Rha), which was structurally different from GPLs including GPL-1 to -4 and ssGPLs (Fig. 6). However, it is reported that M. fortuitum complex produced GPLs which are glycosylated as in GPL-5 and GPL-6 as major components (19, 20). Therefore, these observations suggest that this type of glycosylation is not specific for M. smegmatis. GC/MS analyses of GPL-5 and GPL-6 indicated the presence of 3-O-Me-Rha in addition to 3,4-di-O-Me-Rha, and analyses of perdeuteriomethylated GPL-5 and GPL-6 showed that position C-1 of 3-O-Me-Rha is linked to position C-2 of 3,4-di-O-Me-Rha. Recent studies have shown that M. smegmatis mc2155 newly produces two polar GPLs which contained two units of 3,4-di-O-Me-Rha at l-alaninol of the fatty acyl-tetrapeptide core with no 3-O-Me-Rha at any other position when cultured in carbon-limited medium (23, 24). However, the reason for not being able to detect 3-O-Me-Rha remains unknown.
In the gtf3-overexpressed strain Wt/pMVgtf3, the productivities of GPL-5 and GPL-6 were much higher than those of other GPLs (Fig. 7). So, we can speculate that the expression level of gtf3 is usually repressed and could be regulated by some environmental factors, such as the nutrient condition or the gene encoding sigma factor (23, 24). GC/MS analyses showed that GPL-5 and GPL-6 have the structures in which 3-O-Me-Rha is linked to GPL-3 and GPL-4. These results suggest that GPL-3 and GPL-4 could be the precursors of GPL-5 and GPL-6, respectively, and in Wt/pMVgtf3, overexpression of gtf3 resulted in 2-O-rhamnosylation of 3,4-di-O-Me-Rha in GPL-3 and GPL-4 instead of 2-O-methylation for converting to GPL-1 and GPL-2, so that GPL-5 and GPL-6 were synthesized.
Figure 9 represents proposed glycosylation steps related to M. smegmatis and M. avium. We showed that the functions of gtf1 and gtf2 corresponded to those of gtfA and gtfB, respectively. This finding demonstrates that the biosynthetic pathway for nsGPLs, which is the glycosylation of the fatty acyl-tetrapeptide core with the 6-d-Tal and Rha residues, is common between M. smegmatis and M. avium. Moreover, the biochemical characterization of Δgtf2 and Δgtf1 suggested that the glycosylation pathways for nsGPLs might not be stringent. On the other hand, it has been shown that the rtfA gene of M. avium triggers the biosynthesis of ssGPLs by transfer of Rha to 6-d-Tal of nsGPLs (12). In M. smegmatis, our results indicated that the gtf3 gene plays a role in synthesis of 3-O-Me-rhamnosyl-(1→2)-3,4-di-O-Me-Rha linked to l-alaninol of the fatty acyl-tetrapeptide core by transfer of an extra Rha residue to nsGPLs. Thus, the rtfA and gtf3 genes have the ability to confer the biosynthetic differences between M. avium and M. smegmatis, suggesting that these genes may be responsible for the phylogenetic distinctions in the two species of mycobacteria.
Acknowledgments
We thank W. R. Jacobs, Jr. (Albert Einstein College of Medicine, N.Y.), for providing us with the specialized transducing phage system.
This work was supported in part by grants from Health Science Research Grants—Research on Emerging and Re-emerging Infectious Diseases, Grant-in-Aid for Research on HIV/AIDS, the Ministry of Health, Labor and Welfare, Japan.
REFERENCES
- 1.Aspinall, G. O., D. Chatterjee, and P. J. Brennan. 1995. The variable surface glycolipids of mycobacteria: structures, synthesis of epitopes, and biological properties. Adv. Carbohydr. Chem. Biochem. 51:169-242. [DOI] [PubMed] [Google Scholar]
- 2.Bardarov, S., S. Bardarov, Jr., M. S. Pavelka, Jr., V. Sambandamurthy, M. Larsen, J. Tufariello, J. Chan, G. Hatfull, and W. R. Jacobs, Jr. 2002. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG, and M. smegmatis. Microbiology 148:3007-3017. [DOI] [PubMed] [Google Scholar]
- 3.Barrow, W. W., T. L. Davis, E. L. Wright, V. Labrousse, M. Bachelet, and N. Rastogi. 1995. Immunomodulatory spectrum of lipids associated with Mycobacterium avium serovar 8. Infect. Immun. 63:126-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belisle, J. T., K. Klaczkiewicz, P. J. Brennan, W. R. Jacobs, Jr., and J. M. Inamine. 1993. Rough morphological variants of Mycobacterium avium. Characterization of genomic deletions resulting in the loss of glycopeptidolipid expression. J. Biol. Chem. 268:10517-10523. [PubMed] [Google Scholar]
- 5.Billman-Jacobe, H., M. J. McConville, R. E. Haites, S. Kovacevic, and R. L. Coppel. 1999. Identification of a peptide synthetase involved in the biosynthesis of glycopeptidolipids of Mycobacterium smegmatis. Mol. Microbiol. 33:1244-1253. [DOI] [PubMed] [Google Scholar]
- 6.Bjorndal, H., C. G. Hellerqvist, B. Lindberg, and S. Svensson. 1970. Gas-liquid chromatography and mass spectrometry in methylation analysis of polysaccharides. Angew. Chem. Int. Ed. 9:610-619. [Google Scholar]
- 7.Brennan, P. J., and H. Nikaido. 1995. The envelope of mycobacteria. Annu. Rev. Biochem. 64:29-63. [DOI] [PubMed] [Google Scholar]
- 8.Chatterjee, D., and K. H. Khoo. 2001. The surface glycopeptidolipids of mycobacteria: structures and biological properties. Cell. Mol. Life Sci. 58:2018-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciucanu, I., and F. Kerek. 1984. A simple and rapid method for the permethylation of carbohydrates. Carbohydr. Res. 131:209-217. [Google Scholar]
- 10.Daffe, M., M. A. Laneelle, and G. Puzo. 1983. Structural elucidation by field desorption and electron-impact mass spectrometry of the C-mycosides isolated from Mycobacterium smegmatis. Biochim. Biophys. Acta 751:439-443. [DOI] [PubMed] [Google Scholar]
- 11.Daffe, M., and P. Draper. 1998. The envelope layers of mycobacteria with reference to their pathogenicity. Adv. Microb. Physiol. 39:131-203. [DOI] [PubMed] [Google Scholar]
- 12.Eckstein, T. M., F. S. Silbaq, D. Chatterjee, N. J. Kelly, P. J. Brennan, and J. T. Belisle. 1998. Identification and recombinant expression of a Mycobacterium avium rhamnosyltransferase gene (rtfA) involved in glycopeptidolipid biosynthesis. J. Bacteriol. 180:5567-5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckstein, T. M., J. T. Belisle, and J. M. Inamine. 2003. Proposed pathway for the biosynthesis of serovar-specific glycopeptidolipids in Mycobacterium avium serovar 2. Microbiology 149:2797-2807. [DOI] [PubMed] [Google Scholar]
- 14.Etienne, G., C. Villeneuve, H. Billman-Jacobe, C. Astarie-Dequeker, M. A. Dupont, and M. Daffe. 2002. The impact of the absence of glycopeptidolipids on the ultrastructure, cell surface and cell wall properties, and phagocytosis of Mycobacterium smegmatis. Microbiology 148:3089-3100. [DOI] [PubMed] [Google Scholar]
- 15.Horgen, L., E. L. Barrow, W. W. Barrow, and N. Rastogi. 2000. Exposure of human peripheral blood mononuclear cells to total lipids and serovar-specific glycopeptidolipids from Mycobacterium avium serovars 4 and 8 results in inhibition of TH1-type responses. Microb. Pathog. 29:9-16. [DOI] [PubMed] [Google Scholar]
- 16.Jeevarajah, D., J. H. Patterson, M. J. McConville, and H. Billman-Jacobe. 2002. Modification of glycopeptidolipids by an O-methyltransferase of Mycobacterium smegmatis. Microbiology 148:3079-3087. [DOI] [PubMed] [Google Scholar]
- 17.Kano, H., T. Doi, Y. Fujita, H. Takimoto, I. Yano, and Y. Kumazawa. 2005. Serotype-specific modulation of human monocyte functions by glycopeptidolipid (GPL) isolated from Mycobacterium avium complex. Biol. Pharm. Bull. 28:335-339. [DOI] [PubMed] [Google Scholar]
- 18.Krzywinska, E., S. Bhatnagar, L. Sweet, D. Chatterjee, and J. S. Schorey. 2005. Mycobacterium avium 104 deleted of the methyltransferase D gene by allelic replacement lacks serotype-specific glycopeptidolipids and shows attenuated virulence in mice. Mol. Microbiol. 56:1262-1273. [DOI] [PubMed] [Google Scholar]
- 19.Lopez Marin, L. M., M. A. Laneelle, D. Prome, M. Daffe, G. Laneelle, and J. C. Prome. 1991. Glycopeptidolipids from Mycobacterium fortuitum: a variant in the structure of C-mycoside. Biochemistry 30:10536-10542. [DOI] [PubMed] [Google Scholar]
- 20.Lopez-Marin, L. M., N. Gautier, M. A. Laneelle, G. Silve, and M. Daffe. 1994. Structures of the glycopeptidolipid antigens of Mycobacterium abscessus and Mycobacterium chelonae and possible chemical basis of the serological cross-reactions in the Mycobacterium fortuitum complex. Microbiology 140:1109-1118. [DOI] [PubMed] [Google Scholar]
- 21.Minami, H. 1998. Promotion of phagocytosis and prevention of phagosome-lysosome (P-L) fusion in human peripheral blood monocytes by serotype specific glycopeptidolipid (GPL) antigen of Mycobacterium avium complex (MAC). Kekkaku 73:545-556. [PubMed] [Google Scholar]
- 22.Miyamoto, Y., T. Mukai, F. Takeshita, N. Nakata, Y. Maeda, M. Kai, and M. Makino. 2004. Aggregation of mycobacteria caused by disruption of fibronectin-attachment protein-encoding gene. FEMS Microbiol. Lett. 236:227-234. [DOI] [PubMed] [Google Scholar]
- 23.Mukherjee, R., M. Gomez, N. Jayaraman, I. Smith, and D. Chatterji. 2005. Hyperglycosylation of glycopeptidolipid of Mycobacterium smegmatis under nutrient starvation: structural studies. Microbiology 151:2385-2392. [DOI] [PubMed] [Google Scholar]
- 24.Ojha, A. K., S. Varma, and D. Chatterji. 2002. Synthesis of an unusual polar glycopeptidolipid in glucose-limited culture of Mycobacterium smegmatis. Microbiology 148:3039-3048. [DOI] [PubMed] [Google Scholar]
- 25.Patterson, J. H., M. J. McConville, R. E. Haites, R. L. Coppel, and H. Billman-Jacobe. 2000. Identification of a methyltransferase from Mycobacterium smegmatis involved in glycopeptidolipid synthesis. J. Biol. Chem. 275:24900-24906. [DOI] [PubMed] [Google Scholar]
- 26.Recht, J., and R. Kolter. 2001. Glycopeptidolipid acetylation affects sliding motility and biofilm formation in Mycobacterium smegmatis. J. Bacteriol. 183:5718-5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911-1919. [DOI] [PubMed] [Google Scholar]
- 28.Stover, C. K., V. F. de la Cruz, T. R. Fuerst, J. E. Burlein, L. A. Benson, L. T. Bennett, G. P. Bansal, J. F. Young, M. H. Lee, G. F. Hatfull, S. B. Snapper, R. G. Barletta, W. R. Jacobs, Jr., and B. R. Bloom. 1991. New use of BCG for recombinant vaccines. Nature 351:456-460. [DOI] [PubMed] [Google Scholar]
- 29.Tassell, S. K., M. Pourshafie, E. L. Wright, M. G. Richmond, and W. W. Barrow. 1992. Modified lymphocyte response to mitogens induced by the lipopeptide fragment derived from Mycobacterium avium serovar-specific glycopeptidolipids. Infect. Immun. 60:706-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torrelles, J. B., D. Ellis, T. Osborne, A. Hoefer, I. M. Orme, D. Chatterjee, P. J. Brennan, and A. M. Cooper. 2002. Characterization of virulence, colony morphotype and the glycopeptidolipid of Mycobacterium avium strain 104. Tuberculosis (Edinburgh) 82:293-300. [DOI] [PubMed] [Google Scholar]
- 31.Vergne, I., and M. Daffe. 1998. Interaction of mycobacterial glycolipids with host cells. Front. Biosci. 3:d865-d876. [DOI] [PubMed] [Google Scholar]
- 32.Villeneuve, C., G. Etienne, V. Abadie, H. Montrozier, C. Bordier, F. Laval, M. Daffe, I. Maridonneau-Parini, and C. Astarie-Dequeker. 2003. Surface-exposed glycopeptidolipids of Mycobacterium smegmatis specifically inhibit the phagocytosis of mycobacteria by human macrophages. Identification of a novel family of glycopeptidolipids. J. Biol. Chem. 278:51291-51300. [DOI] [PubMed] [Google Scholar]