Abstract
secDF mutants of Flavobacterium johnsoniae were deficient in gliding motility and chitin utilization. Cells of the mutants had reduced levels of GldJ protein, which is required for both processes. SecDF is similar to Escherichia coli SecD and SecF, which are involved in protein secretion.
Cells of the bacterium Flavobacterium johnsoniae move rapidly over surfaces in a process called gliding motility. Several models have been proposed to explain this type of gliding, but the mechanism of cell movement remains unknown (16). Genetic techniques have been developed for F. johnsoniae, and genes that are required for motility have been identified (19). gldA, gldF, and gldG encode proteins that are thought to form an ATP-binding cassette transporter that is required for gliding (1, 11). Eight other genes (gldB, -D, -H, -I, -J, -K, -L, and -M) that are required for motility have also been identified (5, 6, 11-13, 17, 18). Cells with mutations in any of these genes are completely nonmotile. They form nonspreading colonies, and individual cells exhibit no movement on agar or glass surfaces. These mutants are also unable to utilize the polysaccharide chitin and are resistant to infection by bacteriophages that infect wild-type cells.
Motile nonspreading (MNS) mutants, which have less severe defects in motility, have also been isolated (7, 9, 10, 26). These mutants form nonspreading colonies like those of the nonmotile gld mutants described above, but individual cells retain some ability to move over glass surfaces. Cells of some MNS mutants move nearly as well as wild-type cells on glass, whereas others are more severely crippled. Nonmotile gld mutants have received considerable attention, but the genetic basis for the motility defects in MNS mutants has not been explored. This paper describes the identification of F. johnsoniae secDF as one gene in which mutations result in the MNS phenotype. Disruption of secDF results in cells that are severely crippled but retain some motility.
Bacterial strains and growth conditions.
F. johnsoniae MM101 (a derivative of F. johnsoniae ATCC 17061) (17) was the wild-type strain used in this study, and all mutants were derived from this strain. The Escherichia coli strains used were DH5αMCR (Invitrogen) and HB101 (4). E. coli strains were grown in Luria-Bertani medium, and F. johnsoniae strains were grown in Casitone-yeast extract (CYE) medium as previously described (19). Antibiotics were used at the following concentrations when needed: ampicillin, 100 μg/ml; chloramphenicol, 35 μg/ml; erythromycin, 100 μg/ml; kanamycin, 30 μg/ml; and tetracycline, 20 μg/ml. To observe colony spreading, F. johnsoniae was grown on PY2 agar medium (1) at 25°C. Wild-type and mutant cells of F. johnsoniae were examined for movement over glass and agar surfaces by phase-contrast microscopy as previously described (18).
Tn4351 mutagenesis and identification of secDF.
F. johnsoniae was mutagenized with Tn4351, and 154 mutants that formed nonspreading colonies were isolated as described previously (13). Thirty-six of the mutants were completely nonmotile and had Tn4351 insertions in gld genes that were previously described (1, 11-13, 18). Another 14 mutants had defects in cell division in addition to loss of motility, similar to those of ftsX mutants (15). These gld mutants and filamentous-nonmotile mutants were not considered further in this study. The remaining 104 mutants formed nonspreading colonies, but individual cells exhibited some motility in wet mounts. Twenty-two of these were selected at random for further study. The sites of the transposon insertions were determined by inverse PCR and DNA sequencing essentially as described previously (11, 20). Two of the mutants, CJ974 and CJ978, that had severe motility defects each had a Tn4351 insertion within a gene that we refer to as secDF (Fig. 1). Most cells of CJ974 and CJ978 displayed no movement, but extended observation revealed rare cells that occasionally exhibited slight movements. Typically one cell out of a field of about 1,000 cells would move.
FIG. 1.
Map of the secDF region of F. johnsoniae. Numbers below the map refer to kilobase pairs of sequence. The sites of Tn4351 insertions are indicated by triangles. mdh encodes a predicted malate dehydrogenase, and fjo28 is a gene of unknown function.
Complementation of secDF mutants.
A 3.6-kbp region of F. johnsoniae DNA which spans secDF was amplified using the Expand high-fidelity PCR system (Roche) and primers 606 (TGCTCAGTTTTCGTAGAAGGCG) and 607 (GTTGTTAAATTCACTTCCGAAGCC). This product was polished using the Perfectly Blunt cloning kit (Novagen) and ligated into the EcoRV site of pBCSK+ (Stratagene) to generate pSN1. pSN1 was digested with KpnI and BamHI and the fragment containing secDF was inserted into the shuttle vector pCP23 (1) which had been cut with the same enzymes to generate pSN2. pSN2 was transferred by conjugation into CJ974 and CJ978 essentially as previously described (13, 19). Introduction of pSN2 restored motility to the mutants. Complemented cells exhibited rapid gliding motility in wet mounts and formed spreading colonies (Fig. 2).
FIG. 2.
Photomicrographs of F. johnsoniae colonies. Colonies were grown for 30 h at 25°C on PY2 agar media. Photomicrographs were taken with a Kodak DC290 digital camera mounted on an Olympus IMT-2 inverted microscope. Bar indicates 0.5 mm. A, Wild-type F. johnsoniae MM101 with pCP23. B, secDF mutant CJ974. C, CJ974 complemented with pSN2, which carries secDF.
The secDF-coding region is 2,976 nucleotides in length. A sequence that matches a putative Bacteroides promoter consensus (TAXXTTTG) (2) starts 137 bp upstream of the predicted start codon, and an inverted repeat (AAAAAGATCCAGTGAAAGCTGGATCTTTTT) that may function as a transcription terminator begins 21 bp downstream of the stop codon. F. johnsoniae secDF encodes a predicted 108.4-kDa membrane protein. HMMTOP analysis (24, 25) predicts that SecDF has 12 transmembrane helices and two large periplasmic domains. The N-terminal region of F. johnsoniae SecDF is similar to E. coli SecD (27% identity over 626 amino acids), and the C-terminal region is similar to E. coli SecF (30% identity over 323 amino acids) (8). A similar arrangement of SecD and SecF modules is found in the SecDF protein of Bacillus subtilis (3). E. coli SecD and SecF and B. subtilis SecDF are involved in protein export across the cytoplasmic membrane (3, 22).
Effect of disruption of secDF on growth rate.
Disruption of secD and secF in E. coli results in severe growth defects that are most pronounced at temperatures below 37°C (22). At 30°C, cells are not viable and fail to give rise to colonies. secDF mutants of F. johnsoniae did not exhibit such severe growth defects but did grow more slowly than wild-type cells at all temperatures tested (16°C, 25°C, and 30°C). Wild-type cells carrying the control plasmid pCP11 (19) had a doubling time of 81 (±3) min in CYE medium containing erythromycin at 30°C, whereas the secDF mutants CJ974 and CJ978 had doubling times of 106 (±1) and 104 (±7) min, respectively. Complementation of the mutants with pSN2 restored the doubling times to wild-type levels. The fact that secDF mutants of F. johnsoniae are not severely impaired in growth indicates that SecDF is not required for translocation of proteins that are essential for viability and growth. Analysis of the nearly complete genome sequence of F. johnsoniae did not identify other genes closely related to secD or secF, so the lack of a dramatic effect on growth does not appear to be the result of redundant SecDF-like proteins.
Bacteriophage resistance of secDF mutants.
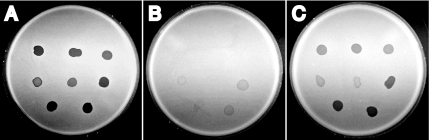
Most nonmotile mutants of F. johnsoniae are resistant to infection by all known F. johnsoniae bacteriophages, including φCj1, φCj13, φCj23, φCj28, φCj29, φCj42, φCj48, and φCj54 (1, 5-7, 11-13, 17, 18, 21, 26). Sensitivity to F. johnsoniae bacteriophages was determined essentially as previously described (13) by spotting 3 μl of phage lysates (109 PFU/ml) onto lawns of cells in CYE overlay agar. The plates were incubated for 24 h at 25°C to observe lysis. Wild-type F. johnsoniae showed complete lysis by all of the bacteriophages described above. The secDF mutants CJ974 and CJ978 were completely resistant to φCj1, φCj13, and φCj23, almost completely resistant to φCj28 and φCj29, and partially resistant to φCj42, φCj48, and φCj54 (Fig. 3). Introduction of pSN2 into CJ974 or CJ978 resulted in restoration of sensitivity to each of the bacteriophages in addition to restoration of wild-type motility.
FIG. 3.
Effect of mutation in secDF on bacteriophage resistance. Bacteriophages (3 μl of lysates containing approximately 109 PFU/ml) were spotted onto lawns of cells in CYE overlay agar. The plates were incubated at 25°C for 24 h to observe lysis. Bacteriophages were spotted in the following order from left to right: top row, φCj1, φCj13, and φCj23; middle row, φCj28, φCj29, and φCj42; and bottom row, φCj48 and φCj54. A, Wild-type F. johnsoniae MM101. B, secDF mutant CJ974. C, CJ974 complemented with pSN2, which carries secDF. Diameter of petri dish is 9 cm.
secDF mutants are defective in chitin utilization.
Wild-type cells of F. johnsoniae digest chitin (23), whereas many nonmotile mutants fail to utilize this insoluble polysaccharide (5-7, 17, 18). The effect of a mutation in secDF on chitin utilization was determined as previously described (18). Cells of the secDF mutant CJ974 were deficient in chitin digestion (Fig. 4). Complementation with pSN2 restored the ability to digest chitin in addition to restoring gliding motility.
FIG. 4.
Effect of mutation in secDF on ability to utilize chitin. Approximately 4 × 107 cells of wild-type F. johnsoniae MM101 (A), of the secDF mutant CJ974 (B), and of CJ974 complemented with pSN2, which carries secDF (C) , were spotted on PY2-chitin medium and incubated for 14 days at 25°C.
Effect of mutations in secDF on Gld protein levels.
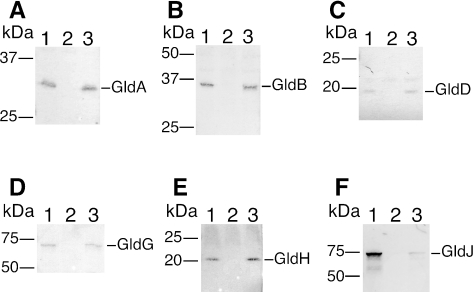
SecDF could play a role in transport of Gld proteins and thus affect motility, chitin utilization, and bacteriophage resistance. We used antibodies to determine the effect of a secDF mutation on the levels of Gld proteins. F. johnsoniae cells were grown to late log phase, and Western blot analysis was performed as previously described (5). CJ974 had normal levels of GldA, GldB, GldD, GldG, and GldH but had dramatically reduced levels of GldJ (Fig. 5). GldJ is required for gliding (6), and the reduction of GldJ levels may explain why secDF mutants are severely crippled. Mutations in gldA, gldB, gldD, gldF, gldG, gldH, and gldI all result in normal levels of gldJ mRNA but decreased levels of GldJ protein (6). We previously speculated that the products of these genes interact and that the absence of individual proteins resulted in the instability of GldJ protein. Mutations in secDF may cause a similar result. SecDF may assist the export of GldJ or of another protein that interacts with and stabilizes GldJ. Introduction of pTB44 (6), which expresses gldJ, did not restore motility or colony spreading to either of the secDF mutants. The partial phage resistance of secDF mutants described above is consistent with a deficiency in GldJ. Most gldJ mutants are completely resistant to bacteriophages, but one frame-shift mutant, UW102-48, displays partial sensitivity to φCj42, φCj48, and φCj54 (6). This pattern is similar to that displayed by the secDF mutants (Fig. 3). UW102-48 might produce a small amount of truncated GldJ, resulting in weak susceptibility to these phages. Further study of phage resistance may help determine the cell surface components required for bacteriophage infection and for gliding motility.
FIG. 5.
Levels of Gld proteins in the secDF mutant CJ974. Western blot analyses of whole-cell extracts were performed using antiserum against GldA (A), GldB (B), GldD (C), GldG (D), GldH (E), and GldJ (F). Lanes 1, wild-type F. johnsoniae MM101. Lanes 2, gldA mutant CJ288 (A), gldB mutant CJ569 (B), gldD mutant CJ282 (C), gldG mutant CJ776 (D), gldH mutant CJ1043 (E), gldJ mutant UW102-48 (F). Lanes 3, secDF mutant CJ978. Total cell protein (20 μg) was loaded in each lane.
Disruption of secDF results in resistance to some bacteriophages and in defects in gliding motility and chitin utilization. The connection between bacteriophage resistance, chitin utilization and gliding motility is not understood. It has been suggested that gliding, bacteriophage sensitivity, and chitin utilization may each rely on one or more transporters that are defective in gld mutants (18). The exact function of SecDF in each of these processes is not known. Given the known roles of E. coli SecD and SecF and of B. subtilis SecDF in protein translocation, it is likely that F. johnsoniae SecDF performs a similar function. SecDF may assist the translocation of components of the motility machinery, such as GldJ, to their sites of assembly and function in the cell envelope and thus be required for efficient gliding motility and chitin utilization. A less likely alternative is that SecDF is directly involved in gliding and that protein export drives cell movement. Such a role for SecDF has been suggested for mycoplasma gliding based on comparative analyses of genomes of motile and nonmotile members of the genus Mycoplasma (14). This role was tentatively suggested since several gliding mycoplasmas lack SecDF, and experimental evidence linking SecDF to mycoplasma motility is not available. Mycoplasma gliding does not appear to be closely related to F. johnsoniae motility since homologs to most of the gld genes that are required for F. johnsoniae gliding are lacking in the sequenced Mycoplasma mobile genome (5, 14). Regardless of the exact role of SecDF in F. johnsoniae motility, it is clearly required for efficient gliding and for the formation of spreading colonies. Further analysis of motility mutants will help determine the mechanism of F. johnsoniae gliding motility.
Nucleotide sequence accession number. The sequence reported in this paper has been deposited in the GenBank database (accession no. AY850226).
Acknowledgments
This research was supported by a grant from the National Science Foundation (MCB-0130967) and by a Milwaukee Foundation Shaw Scientist Award to M.J.M.
Genomic sequence data for F. johnsoniae were obtained from the Joint Genome Institute (http://jgi.doe.gov), Los Alamos National Labs, and the U.S. Department of Energy. We thank D. Saffarini for careful reading of the manuscript.
REFERENCES
- 1.Agarwal, S., D. W. Hunnicutt, and M. J. McBride. 1997. Cloning and characterization of the Flavobacterium johnsoniae (Cytophaga johnsonae) gliding motility gene, gldA. Proc. Natl. Acad. Sci. USA 94:12139-12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bayley, D. P., E. R. Rocha, and C. J. Smith. 2000. Analysis of cepA and other Bacteroides fragilis genes reveals a unique promoter structure. FEMS Microbiol. Lett. 193:149-154. [DOI] [PubMed] [Google Scholar]
- 3.Bolhuis, A., C. Broekhuizen, A. Sorokin, M. van Roosmalen, G. Venema, S. Bron, W. Quax, and J. van Dijl. 1998. SecDF of Bacillus subtilis, a molecular Siamese twin required for the efficient secretion of proteins. J. Biol. Chem. 273:21217-21224. [DOI] [PubMed] [Google Scholar]
- 4.Bolivar, F., and K. Backman. 1979. Plasmids of E. coli as cloning vectors. Methods Enzymol. 68:245-267. [DOI] [PubMed] [Google Scholar]
- 5.Braun, T. F., M. K. Khubbar, D. A. Saffarini, and M. J. McBride. 2005. Flavobacterium johnsoniae gliding motility genes identified by mariner mutagenesis. J. Bacteriol. 187:6943-6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun, T. F., and M. J. McBride. 2005. Flavobacterium johnsoniae GldJ is a lipoprotein that is required for gliding motility. J. Bacteriol. 187:2628-2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, L. Y. E., J. L. Pate, and R. J. Betzig. 1984. Isolation and characterization of nonspreading mutants of the gliding bacterium Cytophaga johnsonae. J. Bacteriol. 159:26-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gardel, C., K. Johnson, A. Jacq, and J. Beckwith. 1990. The secD locus of E. coli codes for two membrane proteins required for protein export. EMBO J. 10:3209-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Godchaux, W., III, M. A. Lynes, and E. R. Leadbetter. 1991. Defects in gliding motility in mutants of Cytophaga johnsonae lacking a high-molecular-weight cell surface polysaccharide. J. Bacteriol. 173:7607-7614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorski, L., W. Godchaux III, E. R. Leadbetter, and R. R. Wagner. 1992. Diversity in surface features of Cytophaga johnsonae motility mutants. J. Gen. Microbiol. 138:1767-1772. [Google Scholar]
- 11.Hunnicutt, D. W., M. J. Kempf, and M. J. McBride. 2002. Mutations in Flavobacterium johnsoniae gldF and gldG disrupt gliding motility and interfere with membrane localization of GldA. J. Bacteriol. 184:2370-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunnicutt, D. W., and M. J. McBride. 2001. Cloning and characterization of the Flavobacterium johnsoniae gliding motility genes gldD and gldE. J. Bacteriol. 183:4167-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunnicutt, D. W., and M. J. McBride. 2000. Cloning and characterization of the Flavobacterium johnsoniae gliding-motility genes gldB and gldC. J. Bacteriol. 182:911-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaffe, J. D., N. Stange-Thomann, C. Smith, D. DeCaprio, S. Fisher, J. Butler, S. Calvo, T. Elkins, M. G. FitzGerald, N. Hafez, C. D. Kodira, J. Major, S. Wang, J. Wilkinson, R. Nicol, C. Nusbaum, B. Birren, H. C. Berg, and G. M. Church. 2004. The complete genome and proteome of Mycoplasma mobile. Genome Res. 14:1447-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kempf, M. J., and M. J. McBride. 2000. Transposon insertions in the Flavobacterium johnsoniae ftsX gene disrupt gliding motility and cell division. J. Bacteriol. 182:1671-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McBride, M. J. 2001. Bacterial gliding motility: multiple mechanisms for cell movement over surfaces. Annu. Rev. Microbiol. 55:49-75. [DOI] [PubMed] [Google Scholar]
- 17.McBride, M. J., and T. F. Braun. 2004. GldI is a lipoprotein that is required for Flavobacterium johnsoniae gliding motility and chitin utilization. J. Bacteriol. 186:2295-2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McBride, M. J., T. F. Braun, and J. L. Brust. 2003. Flavobacterium johnsoniae GldH is a lipoprotein that is required for gliding motility and chitin utilization. J. Bacteriol. 185:6648-6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McBride, M. J., and M. J. Kempf. 1996. Development of techniques for the genetic manipulation of the gliding bacterium Cytophaga johnsonae. J. Bacteriol. 178:583-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ochman, H., A. S. Gerber, and D. L. Hartl. 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120:621-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pate, J. L., S. J. Petzold, and L.-Y. E. Chang. 1979. Phages for the gliding bacterium Cytophaga johnsonae that infect only motile cells. Curr. Microbiol. 2:257-262. [Google Scholar]
- 22.Pogliano, J. A., and J. Beckwith. 1994. SecD and SecF facilitate protein export in Escherichia coli. EMBO J. 13:554-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stanier, R. Y. 1947. Studies on nonfruiting myxobacteria. I. Cytophaga johnsonae, n. sp., a chitin-decomposing myxobacterium. J. Bacteriol. 53:297-315. [PMC free article] [PubMed] [Google Scholar]
- 24.Tusnady, G. E., and I. Simon. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849-850. [DOI] [PubMed] [Google Scholar]
- 25.Tusnady, G. E., and I. Simon. 1998. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J. Mol. Biol. 283:489-506. [DOI] [PubMed] [Google Scholar]
- 26.Wolkin, R. H., and J. L. Pate. 1985. Selection for nonadherent or nonhydrophobic mutants co-selects for nonspreading mutants of Cytophaga johnsonae and other gliding bacteria. J. Gen. Microbiol. 131:737-750. [Google Scholar]