Abstract
Ras proteins are distributed in different types of plasma membrane microdomains and endomembranes. However, how microlocalization affects the signals generated by Ras and its subsequent biological outputs is largely unknown. We have approached this question by selectively targeting RasV12 to different cellular sublocalizations. We show here that compartmentalization dictates Ras utilization of effectors and the intensity of its signals. Activated Ras can evoke enhanced proliferation and transformation from most of its platforms, with the exception of the Golgi complex. Furthermore, signals that promote survival emanate primarily from the endoplasmic reticulum pool. In addition, we have investigated the need for the different pools of endogenous Ras in the conveyance of upstream mitogenic and transforming signals. Using targeted RasN17 inhibitory mutants and in physiological contexts such as H-Ras/N-Ras double knockout fibroblasts, we demonstrate that Ras functions at lipid rafts and at the Golgi complex are fully dispensable for proliferation and transformation.
Ras GTPases—H-Ras, N-Ras, and K-Ras 4B/4A—operate as key molecular switches that convey extracellular signals from surface receptors to the interior of the cell, thereby regulating essential processes including proliferation, differentiation, and survival (15, 34). It is well known that Ras must be attached to the inner leaflet of the plasma membrane (PM) to be functional (50). This is accomplished by lipidic additions to the protein C terminus (33), which contains the essential signal for localizing Ras to membranes: the CAAX box (where C is cysteine, A is alyphatic amino acid, and X is serine/methionine). This motif undergoes posttranslational modifications that make it more hydrophobic. The cysteine is farnesylated, the AAX sequence is proteolyzed, and the newly C-terminal cysteine is carboxymethylated (50). However, a second signal is necessary for efficiently positioning Ras in the membrane. This is accomplished by palmitoylation of cysteine 181 in N-Ras, and cysteines 181 and 184 in H-Ras. In the case of K-Ras 4B the second signal is attained by a polybasic motif of six lysines (175 to 180) that interacts electrostatically with the negatively charged membrane (24-26).
Recently, a new twist has been provided by findings indicating that Ras isoforms are distinctively segregated in different PM microdomains with unique biochemical and physicochemical characteristics, H-Ras can be found in bulk membrane and in lipid rafts, both caveolar and noncaveolar. K-Ras is exclusively located in bulk membrane, whereas N-Ras can only be detected in noncaveolar lipid rafts (35, 38-40). Furthermore, recent reports indicate that Ras is also present in endomembranes such as endosomes, endoplasmic reticulum (ER), and the Golgi complex (10, 37, 45). The significance of this distribution seems to go beyond that of a transient event associated to transport and/or recycling. Instead, a pool of Ras appears to reside in these organelles, and therein Ras can productively engage downstream effectors (10, 11, 37, 45). Moreover, at these endomembranes Ras regulation is undertaken by proteins that operate in a location-specific fashion. As such, the guanine nucleotide exchange factor RasGRP specifically regulates Ras activation at the Golgi complex (7, 9), whereas SOS and RasGRF undertake Ras regulation at the ER. Likewise, stimuli such as lysophosphatidic acid preferentially activate the Ras pool at the ER, whereas calcium ionophores are more effective in activating PM Ras (4).
The fact that exogenous stimuli activate Ras distinctively depending on its localization and that Ras regulation at different sites requires the participation of specific intermediaries hints at the necessity for a location-specific control. This, in term, may imply that Ras functions at its different sites may not be totally redundant. Thus, a selective activation of Ras at each of its locations could be intended to generate variability in its biochemical and biological outputs. It is known that Ras regulates numerous cellular functions through the activation of an ever-growing number of effector molecules (15). However, how microlocalization affects the biochemical signals that Ras generates and the biological outputs that it regulates is just beginning to be unfolded (23). Herein, we have addressed that question. We present for the first time a systematic study on the differences in effectors utilization resulting from Ras activity at specific cellular sites. In addition, we also demonstrate how activation or inhibition of Ras restricted to defined membrane systems and microdomains affects biological outcomes such as cellular proliferation, survival, and transformation. Overall, the present study moves one step beyond into understanding how compartmentalization of Ras signals affects its performance and identifies those cellular sites where Ras is essential for cellular proliferation and transformation.
MATERIALS AND METHODS
Constructs.
pEXV H-RasV12 C181,184 S (SS) was provided by J. F. Hancock; pGEX-Ral-RBD was provided by J. L. Bos; and pCEFL-m1 was provided by J. S. Gutkind. Plasmids encoding for v-Sis and v-Src have been described (29). pCEFL hemagglutinin (HA) vectors harboring the targeting signals M1, KDELr, and CD8α have been described (4). KDELr N193D was generated by PCR-directed mutagenesis. Oligonucleotides encoding LCK myristoylation signal were cloned in pCEFL-HA, directly upstream of the HA tag. Tethering constructs expressing FLAG, instead of HA tag, were also synthesized. H-RasV12 SS and H-RasN17 SS were amplified by PCR and cloned into pCEFL-HA or pCEFL-FLAG targeting constructs, respectively. SSS mutants were generated by PCR-directed mutagenesis (QuikChange; Promega), introducing the C186S mutation into the targeted V12 constructs. All constructs were verified by DNA sequencing. Sequences of the oligonucleotides utilized are available upon request. Interleukin-1 and adriamycin were from Preprotech.
Cell culture.
HEK293T, COS-7, MDCK, and NIH 3T3 cells were grown in Dulbecco modified Eagle medium (DMEM)-10% fetal calf serum (calf serum for NIH 3T3). Mouse embryo fibroblasts (MEFs) were grown in DMEM-10% donor calf serum-0.1 mM nonessential amino acids (NEAA) -55 μM β-mercaptoethanol. Stable lines cells were generated by transfection with Lipofectamine (Invitrogen) according to the manufacturer's instructions and selected with 750 μg of G418/ml. 293T and COS-7 cells were transiently transfected with calcium phosphate (46).
Kinase assays.
Extracellular signal-regulated kinase 2 (ERK2) and JNK kinase activities were determined in anti-ERK and anti-JNK immunoprecipitates using as substrates myelin basic protein (Sigma) or glutathione S-transferase (GST)-ATF2, respectively (3). Akt kinase assays were performed as described previously (17) in cells transiently transfected with a plasmid encoding for HA-tagged Akt in anti-HA immunoprecipitates, using histone H2B as substrate.
Antibodies.
Mouse monoclonal anti-HA was from Babco; rabbit polyclonal anti-FLAG was from Invitrogen. Rabbit polyclonal anti-calreticulin was from Calbiochem. Antigiantin mouse monoclonal antibodies were supplied by H. P. Hauri (Basel, Switzerland). Mouse monoclonal anti-transferrin receptor and antitubulin were from Zymed. Mouse monoclonal anti-Ecto-5′-nucleotidase was from BD Biosciences. Rabbit polyclonal anti-Akt and goat polyclonal anti-phospho-Akt were from Cell Signaling, and rabbit polyclonal anti-Raf has been previously described (16). Goat polyclonal antibody anti-Ral A and rabbit polyclonal antibodies anti-HA anti-ERK2, anti-RasGRF1, anticaveolin, anti-PARP, anti-TC21, anti-H-Ras, anti-K-Ras, and anti-N-Ras were from Santa Cruz Laboratories.
Immunoblotting.
Total lysates and immunoprecipitates were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto nitrocellulose filters as described previously (4). Immunocomplexes were visualized by enhanced chemiluminescence detection (Amersham) by using horseradish peroxidase-conjugated secondary antibodies (Cappel).
Focus assays.
NIH 3T3 and MEF cells were cultured in DMEM supplemented with 10% calf serum. NIH 3T3 cells were transfected by using the calcium phosphate technique as described previously (16). MEFs were transfected with Lipofectamine (Invitrogen) according to the manufacturer's instructions. After 10 to 15 days in culture (15 to 20 days for MEFs), cells were fixed and stained, and the foci were scored.
G418-resistant colonies assays.
NIH 3T3 cells were transfected with calcium phosphate and grown in the presence of 750 μg of G418/ml. After 10 to 15 days plates were fixed and stained, and colonies with a diameter greater than 2 mm were scored.
Immunofluorescence.
M1 and KDELr cells lines were washed in phosphate-buffered saline (PBS) and fixed in 3.7% paraformaldehyde. Preparations were sequentially incubated with 0.1 M glycine-PBS for 10 min, permeabilized with 0.5% Triton X-100-PBS for 15 min, and PBS-0.01% Tween 20 for 5 min. CD8 and LCK cell lines were fixed with ice-cold 3.7% paraformaldehyde in PBS and washed with cold PBS without permeabilization. In all cases, preparations were incubated for 1 h with primary antibodies or cholera toxin fluorescein isothiocyanate (FITC; Sigma-Aldrich) that recognizes ganglioside GM1, washed, and incubated for 45 min with secondary antibodies conjugated to FITC or Texas red. Confocal microscopy was performed with a Bio-Rad MRC-1024 at excitation wavelengths of 488 nm (FITC) or 543 nm (Texas red).
Sucrose gradients.
Cells were collected and resuspended in 25 mM Tris (pH 7.4), 150 mM NaCl, 5 mM EDTA, and 0.25% Triton X-100, with rocking at 4°C for 1 h. Lysates were set to a sucrose concentration of 41%. Layers of 8.5 ml of 35% sucrose (in 10 mM Tris [pH 7.4]) and of 2.5 ml of 16% sucrose were sequentially overlaid and centrifuged in a Beckman SW41 rotor for 18 h at 35,000 rpm. We collected 10 to 11 1-ml fractions, precipitated them in 6.5% trichloroacetic acid, resuspended them in loading buffer, and fractionated them by sodium dodecyl sulfate-polyacrylamide gel electrophoresis using 12% gels. Membrane solubilizations in sodium carbonate and subsequent fractionations were performed exactly as described previously (38).
Ras-GTP loading assays.
Performed as described previously (5). Ras-GTP was affinity sequestered by using GST-Raf RBD (amino acids 1 to 149). Immunoblots were performed with anti-HA antibody and quantitated by densitometry using the program NIH Image 1.60. Ras-GTP levels were related to total Ras protein levels as determined by anti-HA immunoblotting in the corresponding total lysates.
Ral GTP assays.
Ral GTP assays were performed as described previously (52). Briefly, cells were lysed in 15% glycerol, 1% NP-40, 50 mM Tris (pH 7.4), 200 mM NaCl, 2.5 mM MgCl2, and protease/phosphatase inhibitors, followed by incubation for 45 min with 10 to 15 μg of GST-Ral RBD. Immunoblots were performed as described above with anti-Ral A antibody and quantitated by densitometry.
Measurement of proliferation and survival rates.
Cells were plated at low density in six-well plates (10,000 cells/per plate) and grown in different serum conditions as indicated. Cells were detached and scored by standard cell counting techniques at the indicated intervals.
RESULTS
Selective tethering of H-Ras V12 to defined sublocalizations.
To investigate the differences in Ras functions at sites where it is present, it was necessary to activate it precisely at, and only at, the desired subcellular localizations. For this purpose we generated constructs that encoded for constitutively active H-RasV12 fused to specific tethering signals. First, we generated a palmitoylation-deficient H-RasV12 by mutating cysteines 181 and 184 to serines (H-RasV12 SS). This mutant is not efficiently retained at the PM and exists in a dynamic equilibrium shuttling between the ER and cytoplasmic pools (10). The palmitoylation signal was then substituted by alternative cues that would specifically direct H-RasV12 SS to the desired locations. To deliver it to the ER, we fused to its N terminus amino acids 1 to 66 of the avian infectious bronchitis virus M protein (M1) (49) (referred to as M1-V12). We sent H-RasV12 SS to the Golgi complex, by means of an N-terminally fused KDEL receptor (KDELr) (9). Since KDELr is an itinerant protein that recycles to the ER, mutation N193D was introduced. This mutation prevents it from redistributing to the ER and renders it a resident Golgi protein (13) (named KDELr-V12). At the PM, we wanted to discriminate between events regulated from bulk membrane and from lipid rafts. As such, H-RasV12 SS was tethered to bulk membrane by placing in its N terminus the transmembrane domain of the CD8α receptor, known to locate exclusively in this type of PM (2) (referred to as CD8-V12 hereafter). Finally, to target H-RasV12 SS to lipid rafts, we utilized an N-terminal myristoylation signal. Unpalmitoylated myristoylation signals, like that of Rasheed sarcoma virus, cannot retain farnesylated Ras at the membrane (8), whereas palmitoylated myristoylation signals, such as the one of LCK (42), act as effective lipid raft anchors (48). An HA tag was included to enable the detection of these proteins.
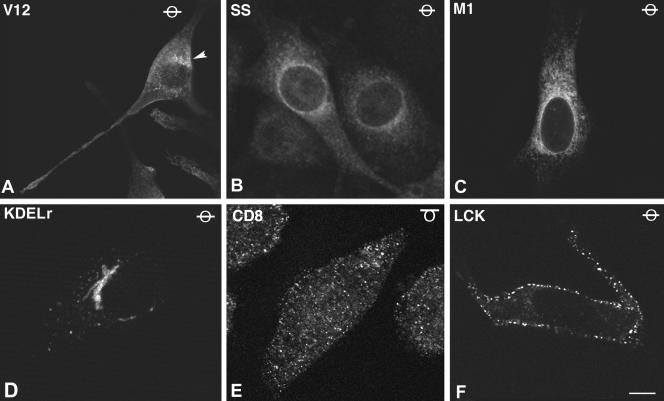
These constructs were stably expressed in NIH 3T3 cells, and polyclonal pools, in which more than 80% of the cells stained positive for HA (data not shown), were utilized for further experimentation. In these, the localization of the targeted Ras proteins was ascertained by immunofluorescence with anti-HA antibodies. Cells expressing H-RasV12 exhibited a typical spindle-shaped morphology. In these, H-RasV12 was evident at the PM outlining the cells, in addition to endomembranes such as the Golgi complex (Fig. 1A, arrowhead) and the ER network, more prominently in the perinuclear area. In contrast, in cells expressing H-RasV12 SS Ras was absent from peripheral PM, and it exhibited a diffuse staining characteristic of cytoplasmic proteins, although staining at the nuclear envelope, typical of ER localization, was also clear (Fig. 1B). On the other hand, ER-tethered M1-V12 displayed a typical reticular distribution with a complete absence of cytoplasmic staining and was undetectable at the PM and at the Golgi complex (Fig. 1C). KDELr-V12 expression was clearly restricted to the Golgi apparatus and was absent from the PM and the ER (Fig. 1D). CD8-V12 was localized in patches throughout the PM, as shown in a tangential confocal plane in Fig. 1E and, within the cell body, in vesicular structures, resembling endosomes. However, it was undetectable at the ER (not shown). Finally, LCK-V12 also exhibited a peripheral membrane staining at defined patches. Small vesicles, probably lipid raft-derived endosomes, were also apparent, but no ER staining was evident (Fig. 1F). Even though Ras proteins are known to be subject to a fast, dynamic shuttling between PM and the Golgi complex (11), we could not detect LCK or CD8-tethered Ras proteins at the Golgi complex (Fig. 1E and F and data not shown). These results pointed to the correct localization of our targeted Ras proteins. Interestingly, these staining patterns, with virtually no mislocalization, were also detected in COS-7 and 293T cells, even though in these cells our plasmids yielded much higher expression levels (data not shown).
FIG.1.
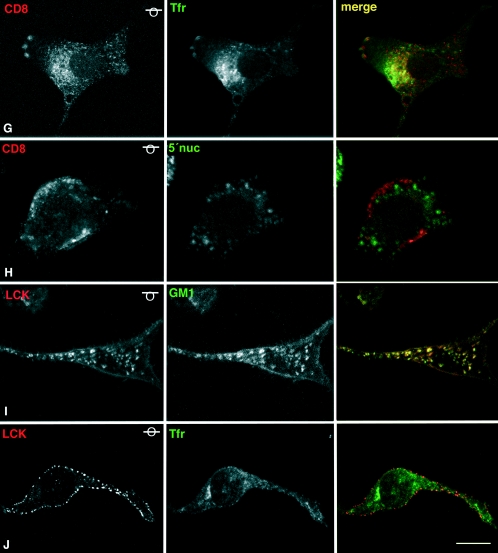
Subcellular localization of targeted Ras proteins. (A to F) Representative confocal micrographs of NIH 3T3 cells stably transfected with the indicated constructs, immunostained with anti-HA. As indicated by the symbol in the upper right corner, all panels show equatorial sections at the cell nucleus level, except panel E, which shows a tangential plane. Scale bars: A, 5 μm; B to F, 10 μm. (G to J) Colocalization of CD8-V12 (G and H) and LCK-V12 (I and J) with lipid rafts and bulk membrane markers. Ras proteins were revealed by anti-HA (red in panels G, H and J; green in panel H), costained with anti-transferrin receptor as a bulk membrane marker (green, panels G and J), and anti-5′-nucleotidase (red, panel H) or FITC-cholera toxin (green, panel I) as raft markers. As indicated by the symbol in the upper right corner, all panels show tangential or semitangential planes, except panel J that shows an equatorial section at the cell nucleus level. Bars: G, I, and J, 20 μm; H, 15 μm.
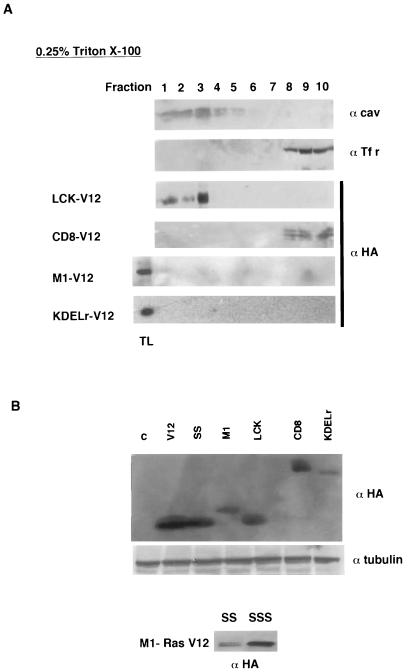
To ascertain beyond doubt that our targeted Ras proteins were correctly localized, we analyzed their colocalization with bona fide markers of the aimed compartments. As we have previously demonstrated, M1- and KDELr-targeted Ras proteins displayed a profuse costaining with the ER marker calreticulin and the Golgi marker giantin, respectively (4; data not shown). In the same fashion, CD8-V12 and LCK-V12 were detected at, and only at, the expected PM microdomains. As such, CD8-V12 markedly colocalized with the transferrin receptor, a marker for disordered membrane (Fig. 1G), but an absolute lack of colocalization with the lipid raft marker 5′ nucleotidase was evident (Fig. 1H). Conversely, LCK-V12 strongly merged with the raft marker ganglioside GM1, as shown in a tangential confocal plane (Fig. 1I), but no costaining with the transferrin receptor was apparent (Fig. 1J). Further proof for the correct localization of these constructs was provided by experiments in which membranes of the targeted-Ras-transfected cells were solubilized in low concentrations of Triton X-100 and fractionated in sucrose gradients. As expected, LCK-V12 cofractionated with the lipid raft marker caveolin in the lighter fractions, whereas CD8-V12 could only be detected in disordered membrane fractions, characterized by the presence of the transferrin receptor. On the other hand, neither M1-V12 nor KDELr-V12 could be detected on either lipid raft or disordered membrane fractions, demonstrating the lack of contamination of the peripheral membrane with the endomembrane-targeted constructs (Fig. 2A). Solubilization in sodium carbonate yielded identical results (data not shown). Overall, these results clearly indicated that our targeted H-RasV12 proteins were specifically localized at the desired microdomains.
FIG. 2.
Localization and expression of targeted Ras proteins. (A) Subcellular fractionation of targeted Ras proteins. NIH 3T3 cells stably expressing the different targeted Ras constructs were solubilized in 0.25% Triton X-100 and partitioned in a sucrose gradient as described in Materials and Methods. The presence of the targeted Ras proteins in the different fractions was analyzed by anti-HA immunoblotting. Anti-caveolin-1 immunoblotting identified the lipid raft fractions; anti-transferrin receptor immunoblotting identified the disordered membrane fractions. In the fractionations corresponding to cells expressing KDELr-V12 and M1-V12, a total lysate (TL) was run alongside as a control. (B) Expression levels of the targeted Ras proteins as determined by anti-HA-immunoblotting in total lysates from the NIH 3T3 cells lines stably expressing the Ras constructs. The middle panel shows tubulin expression levels as a loading control. The bottom panel shows a comparison of the expression levels of M1-V12 unpalmitoylated (SS) versus unpalmitoylated, unfarnesylated (SSS) proteins, as determined by anti-HA-immunoblotting in total lysates from COS-7 cell lines transfected with the indicated constructs.
It was important to acquire some notion of the relative expression levels that each targeted protein attained. By immunoblotting for HA expression it was found that, as expected, all of the targeted-V12 proteins were expressed at lower levels than those of H-Ras V12, widespread throughout all of the sublocalizations, or of H-Ras V12 SS, which accumulated to high levels in the cytoplasm. Noticeably, the PM proteins, LCK-V12 and CD8-V12, were expressed at slightly higher levels than those of M1-V12 and KDELr-V12, located at endomembranes (Fig. 2B). As such, we wanted to know whether these differences were a consequence of variations in the intrinsic expression potentials of each of the constructs. When expressed in vitro in a reticulocyte system, these constructs yielded very similar expression levels (data not shown), something that argued against this concept. Alternatively, the possibility existed that the expression levels of the targeted proteins were a reflection of how much Ras could be inserted on each type of membrane. In our unpalmitoylated constructs, the remaining Ras-specific determinant for membrane localization was their farnesyl moiety. Thus, we introduced the mutation C186S that rendered them farnesylation deficient as well. It was found that, regardless of the targeting signal, these proteins (termed SSS) achieved much higher expression levels than their respective palmitoylation-deficient (SS) constructs, as shown for M1-V12 (Fig. 2B, lower panel). Interestingly, a significant proportion of the SSS proteins had lost their specific localization and was detected at the cytoplasm (data not shown). Overall, these results indicated that a Ras membrane localization determinant was important for defining both the expression levels and the localization of our targeted proteins.
Targeted Ras proteins can effectively engage effectors in vitro and in vivo.
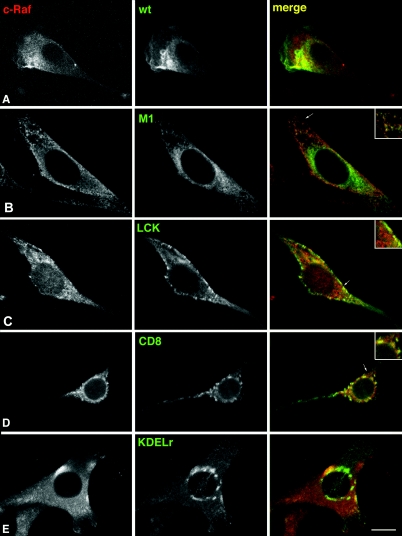
It was essential to verify that the addition of a more or less bulky targeting signal to the N terminus of H-RasV12 did not compromise its GTP-bound state and therefore its ability to engage effectors. Initially, we tested the behavior of the targeted Ras proteins in a standard in vitro effector binding assay; these were expressed in 293T cells, and the GTP-bound fractions were affinity pulled-down by using the c-Raf Ras binding domain (RBD) bound to glutathione beads. It was found that untargeted H-RasV12 and the different location-specific RasV12 proteins bound to GST-RBD with very similar efficiencies (data not shown). Next, we ascertained whether the targeted Ras proteins could also bind to effectors in vivo. In the NIH 3T3 cell lines we examined whether the presence of site-specific RasV12 proteins brought about colocalization with c-Raf at defined sites. In cells expressing H-Ras wild-type, H-Ras could be detected at PM, ER, and Golgi complex, as expected. c-Raf displayed a diffuse staining, typical of a cytoplasmic protein, but no colocalization was apparent at any of these sites under serum starvation conditions (Fig. 3A). In sharp contrast, under the same conditions, in M1-V12-expressing cells a clear Ras/Raf colocalization was present throughout the reticular network (Fig. 3B). A marked costaining was also apparent in LCK-V12- and CD8-V12-expressing cells, but in these cases, merging took place primarily at the peripheral PM and to a lesser extent in endosomes (Fig. 3C and D). Surprisingly, in KDELr-V12-expressing cells the colocalization between Raf and Ras at the Golgi complex was very poor (Fig. 3E), even though, as stated above, KDELr-V12 could effectively bind to c-Raf RBD in vitro. Overall, our results demonstrated that the addition of tethering signals did not compromise RasV12 functionality, both in vitro and at the different subcellular localizations, and validated these constructs for further experimentation.
FIG. 3.
Colocalization of c-Raf with targeted Ras proteins. Serum-starved NIH 3T3 cells expressing the indicated Ras constructs were costained with anti-HA and anti-Raf antibodies. (A) No colocalization with c-Raf is observed in cells expressing H-Ras wild type (wt). (B) M1-V12 displays high colocalization with c-Raf at the ER. (C and D) Colocalization of Raf with LCK- and CD8-V12 at the PM and endosomes. (E) Limited colocalization of c-Raf with KDELr-V12 at the Golgi complex. Confocal sections at the level of the nucleus are shown in all panels. In panels B to D, insets highlight areas of prominent colocalization (arrows). Bars: A, C, and D, 20 μm; B, 10 μm; E, 15 μm.
Distinct pools of RasV12 elicit different biochemical signals.
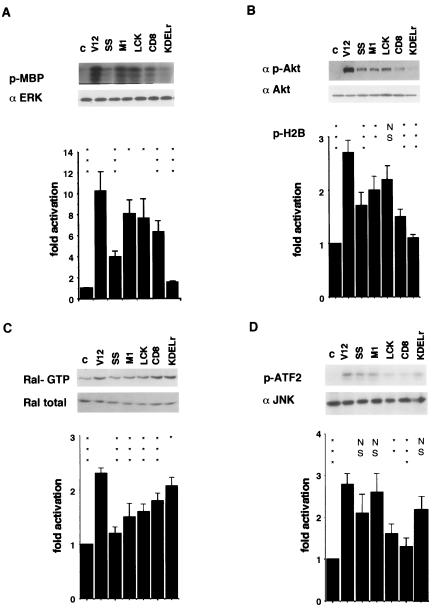
It was of interest to investigate whether signaling pathways triggered by Ras were equally activated irrespective of Ras localization or, alternatively, different cellular pools of Ras evoked distinct signals. For that purpose, in the NIH 3T3 lines stably expressing the targeted RasV12 constructs we analyzed the activation status of several well-known Ras effector pathways. When we studied the activation of the ERK cascade, pronounced differences were detected depending on the cellular site from which the Ras signal originated. H-RasV12 SS, a protein loosely held to the ER, showed a reduced ability to activate ERKs (Fig. 4A), as previously demonstrated (8). However, firmly anchoring Ras to the ER, as was the case for M1-V12, elicited a potent activation of ERKs. LCK-V12, at lipid rafts, was capable of activating ERKs to similar levels, while bulk membrane CD8-RasV12 displayed a lower, though significant, ability to activate this route. Interestingly, very little activation of ERKs was detected when the Ras signal emanated from the Golgi complex, as that one generated by KDELr-V12, in agreement with the low colocalization of Ras and Raf in this organelle. A similar situation was encountered when we studied the PI3K>Akt pathway. In this case, Ras signals originating at the ER and at lipid rafts were the most effective for activating this route (Fig. 4B). Ras capacity to activate Akt from the disordered membrane was noticeably reduced. Once more Ras located at the Golgi complex was the least efficient for activating Akt. Curiously, this pattern was reversed when we looked at the activation of Ral-GDS. In this case, the Golgi complex was the platform from which Ral-GDS was most effectively activated, followed by the bulk membrane (Fig. 4C). Even though the JNK pathway is not considered a bona fide Ras effector route, in NIH 3T3 cells JNK can be activated by Ras to some extent (14). Thus, we also tested how Ras compartmentalization affected the activation of this route. Interestingly, it was found that endomembranes, both ER and Golgi, were the sites preferred by Ras to activate JNK (Fig. 4D), whereas the Ras pools present at PM microdomains were considerably less effective for activating this mitogen-activated protein kinase. Assays similar to those just described were also performed in 293T cells under transient-transfection conditions. Noticeably, the results obtained were identical to those just described (data not shown).
FIG. 4.
Ras effector usage at distinct compartments. (A) Activation of ERK2 by targeted Ras constructs. Serum-starved, NIH 3T3 cells stably expressing the indicated constructs were processed as indicated in Materials and Methods. Kinase assays were performed in anti-ERK2 immunoprecipitates, with myelin basic protein as a substrate. ERK levels were determined by immunoblotting with anti-ERK2 antibodies. Bar chart shows the average ± the standard error of the mean (SEM) of at least five experiments relative to the phosphorylation levels detected in control cells. (B) Activation of Akt. Determined by immunoblotting in serum-starved cells, with antibodies to phosphorylated Akt or by kinase assays with histone H2B as the substrate. The bar chart shows the average ± the SEM of at least five experiments, relative to the H2B phosphorylation levels detected in control cells. (C) Activation of Ral GTPase. Ral GTP pull-down assays were performed in cell lysates as indicated in Materials and Methods and revealed by anti-Ral immunoblotting. Total Ral levels were determined in lysates with the same antibody. The data show the averages ± the SEM of at least five independent experiments relative to the Ral-GTP levels in control cells. (D) Activation of JNK. Kinase assays were performed with GST-ATF2 as substrate. The levels of JNK were determined by immunoblotting with anti-JNK antibody. The data show the results (average ± the SEM) of at least five experiments relative to ATF2 phosphorylation levels found in control cells. analysis of variance was performed by using the Bonferroni multiple comparison test relative to the respective “V12” values. P values: *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, P > 0.05 with a 95% confidence interval.
We needed to ascertain that the signals elicited by the targeted RasV12 proteins were directly generated by Ras itself and were not secondary signals, consequences of autocrine loops induced by Ras activation. To test this, conditioned media from cells expressing the site-specific Ras constructs were collected after 12 and 24 h and added to parental NIH 3T3 cells, and effector activation was assayed. In no case was the conditioned medium able to induce any significant ERK, Akt, or Ral-GDS activation (data not shown), thus demonstrating that the signals detected in the aforementioned experiments were a consequence of RasV12 direct effects.
Distinct pools of RasV12 differentially regulate cellular proliferation, survival, and transformation.
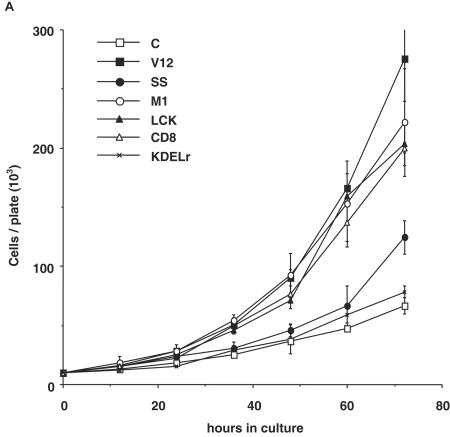
We wanted to determine whether variations in effector usage and in signal magnitude at the distinct Ras signaling platforms translated into different biological responses. First, we evaluated how cellular proliferation was affected by location-restricted Ras signaling by comparing the proliferation rates of the cell lines expressing the targeted Ras constructs when growth occurred under standard culture conditions. We found that most of the pools displayed similar growth curves (Fig. 5A), with a proliferating profile that resembled that one of H-RasV12-transfected cells (doubling time of 13.4 h). The remarkable exception was the line expressing Golgi complex-tethered KDELr-V12, which exhibited much slower growth kinetics (doubling time of 19.6 h), similar to those encountered in control cells. It was important to ascertain to what extent the behavior of the polyclonal cell lines was representative of the clonal population that they were made up of. To this end, the growth kinetics of three isolated clones extracted from each of the polyclonal lines was compared to those of their respective pools. As shown for M1-V12 (Fig. 5B) and for LCK-V12 (Fig. 5C), no significant differences were encountered among the proliferation rates of the pools and of the isolated clones. Likewise, no substantial variations were apparent in the cases of KDELr-V12 and CD8-V12 either (data not shown).
FIG.5.
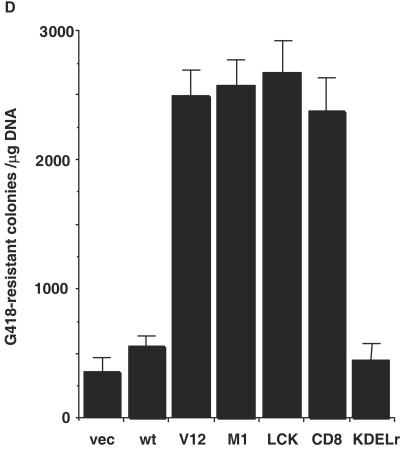
Compartmentalized Ras activation effects on cellular proliferation. (A) Proliferation rate of the NIH 3T3 cells lines expressing the indicated Ras constructs growing in media supplemented with 5% calf serum. (B and C) Proliferation rates of three clones (C1 to C3) isolated from the M1-V12 (B) and the LCK-V12 (C) polyclonal cell lines compared to their respective total pools. (D) Effects of targeted Ras V12 constructs on colony growth. NIH 3T3 fibroblasts were transfected with the indicated constructs (0.25 μg) and selected in the presence of G418 (750 μg/ml). After 2 weeks in culture, colonies were stained, and those with a diameter greater than 0.05 mm were scored. In all cases, results show the average ± the SEM of at least three independent experiments.
The ability of the tethered Ras proteins for supporting proliferation was tested further by colony growth assays. As such, the targeted Ras constructs were transfected into NIH 3T3 cells and after G418 selection the arising colonies were scored. In line with the previous data, all of the targeted Ras construct yielded a number of colonies similar to that of H-Ras V12 (Fig. 5D), with the exception of KDELr-V12, for which the potential to form colonies was similar to that of vector-transfected cells and significantly lower than that of H-Ras wild type.
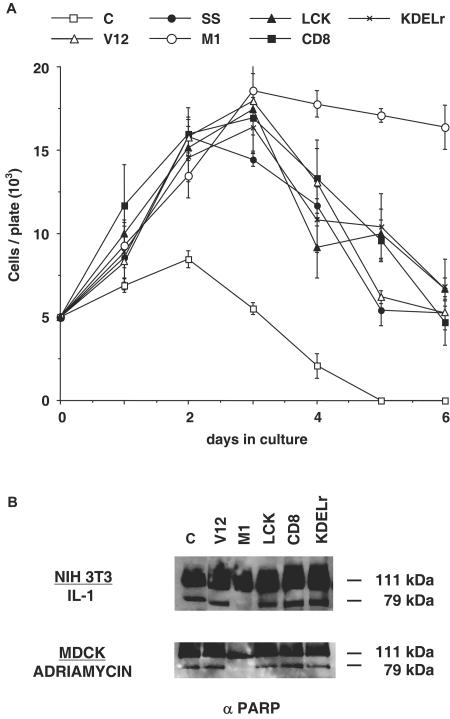
We also wanted to investigate how compartmentalized Ras activity affected cell survival. To this aim, the NIH 3T3 cell lines were deprived of serum, and their ability to survive in these conditions was monitored. As shown in Fig. 6A, in the absence of growth factors RasV12, irrespective of its localization, supported proliferation for almost two complete doubling cycles. Eventually, all lines reached crisis, and cell numbers started dropping with similar kinetics, with the exception of cells harboring ER-tethered M1-V12, in which the rate of cell death was remarkably lower. This behavior was mirrored to some extent by the cell line expressing H-RasV12 SS. Even though this cell line reached crisis before the rest, its death kinetics were noticeably slower.
FIG.6.
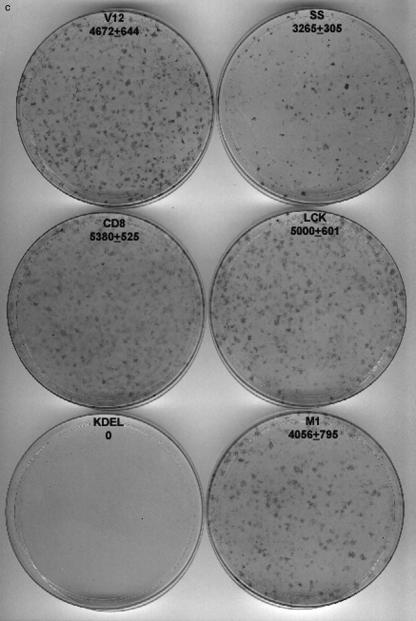
Effects of compartmentalized Ras activation on cellular survival and transformation. (A) Survival rate of the NIH 3T3 cell lines expressing the indicated Ras constructs in the absence of growth factors. Cells were cultured in media supplemented with 0.1% calf serum. Cells were collected at the indicated times and counted. The results represent average ± the SEM of at least three independent experiments. (B) Antiapoptotic effects of site-specific Ras activation. NIH 3T3 and MDCK cells stably expressing the Ras targeted constructs were treated with interleukin-1 (10 μg/ml) for 24 h and with adriamycin (5 μM) for 16 h, respectively. PARP degradation was monitored in total lysates by anti-PARP immunoblotting. (C) Cellular transformation induced by site-specific Ras constructs. NIH 3T3 fibroblasts were transfected with the indicated constructs (100 ng) by the calcium phosphate technique. Foci were stained and scored after 2 weeks in culture. Numbers indicate average number of foci per microgram of DNA ± the SEM of at least three independent experiments.
We extended this observation by analyzing the consequences of compartmentalized Ras activity when cells were challenged with defined apoptogenic stimuli. For this purpose, the NIH 3T3 cell lines were treated with interleukin-1 to induce cell death. It was found that apoptosis, as monitored by the degradation of poly(ADP-ribose) polymerase (PARP), was prominent in the cell lines expressing Ras V12 and the targeted V12 constructs, with the exception of the cells harboring ER-tethered M1-V12, in which apoptosis was remarkably forestalled (Fig. 6B, top panel). To rule out that this phenomenon was limited to a given cell type, we generated lines of MDCK epithelial cells stably expressing the targeted Ras constructs. In this cellular model, Ras activation restricted to the ER also conferred a remarkable resistance to cell death, as induced by treatment with the proapoptotic drug adriamycin (Fig. 6B, bottom panel).
Finally, we looked at whether sublocalization affected H-RasV12 transformation potential. For this purpose, the ability of the targeted RasV12 constructs to generate transformed foci in NIH 3T3 cells was tested. It was found that H-RasV12 was able to robustly induce cellular transformation from most of the cellular localizations where it was targeted (Fig. 6C), slightly more prominently from PM microdomains, both lipid rafts and bulk membrane, than from endomembranes such as the ER. Conversely, Golgi complex-tethered KDELr-V12 was incapable of promoting cellular transformation to any extent. Summing up, these sets of experiments clearly demonstrated that, depending on the subcellular compartment at which it was located, activated H-Ras could trigger proliferation, survival, and transformation to different extents.
Effects of site-specific Ras inhibition on cellular proliferation and transformation.
The aforementioned results clearly supported the notion that sublocalization dictates Ras signal output and its biological consequences. However, these sets of experiments explored how compartmentalization influenced the effects of constitutive Ras signals and did not provide an answer as to how necessary Ras is at its different localizations for the conveyance of upstream signals that drive cellular proliferation or transformation. To address this important point, we designed a strategy to selectively inhibit Ras activity at defined sites. We have previously demonstrated that the inhibitory specificities of Ras N17 dominant inhibitory mutants are related to their microlocalization, in such a way that a N17 mutant acting at a defined microdomain, will inhibit all Ras molecules therein irrespective of the isoform (35). Thus, we reasoned that specifically targeting H-RasN17 to defined localizations, by using the aforementioned tethering cues, could be an effective method to block Ras functions in a site-specific fashion.
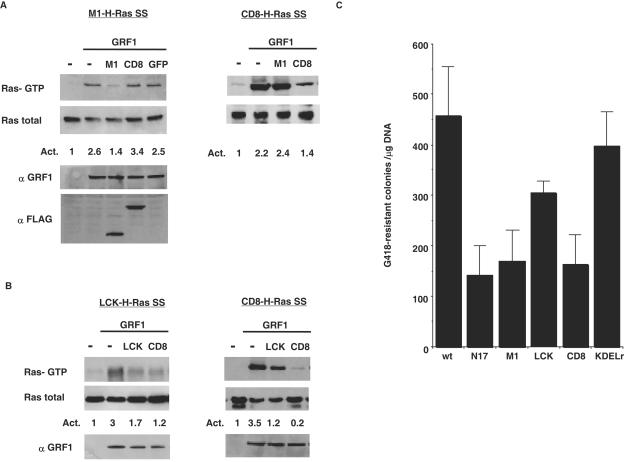
To begin with, we verified the site inhibition specificity of our targeted H-RasN17 constructs. Inhibition at the ER was tested in COS-7 cells by assaying GTP loading in ER-tethered M1-H-Ras, induced by the guanine nucleotide exchange factor RasGRF1, in the presence of the inhibitory constructs M1-N17 and CD8-N17. It was found that GDP/GTP exchange in M1-H-Ras was abrogated by ER-tethered M1-N17, but not by bulk membrane-targeted CD8-N17 (Fig. 7A, left panel). As a negative control M1-green fluorescent protein (GFP) was used to validate that the inhibition was specific for RasN17 and that it was not an unspecific effect due to M1 tethering signal. Conversely, when GTP loading was assayed on bulk membrane-tethered CD8-H-Ras, this was inhibited by CD8-N17 but not by M1-N17 (Fig. 7A, right panel). Similar results were obtained when testing for the cross-inhibitory effects of most N17 constructs (data not shown), thereby demonstrating that the inhibitory effects of the targeted N17 mutants were site specific. Only in one instance cross-inhibition was evident. This was when nucleotide exchange was assayed in lipid raft-tethered LCK-H-Ras that could be inhibited both by LCK-N17 and by CD8-N17 (Fig. 7B, left panel). Likewise, GTP loading in bulk membrane CD8-H-Ras could be inhibited by CD8-N17 and, to a lesser extent, by LCK-N17 as well (Fig, 7B. right panel).
FIG. 7.
Inhibitory specificity of targeted RasN17 mutants. (A) Inhibition of Ras-GRF-induced Ras activation at the ER and bulk membrane by tethered N17 mutants. The left panel shows inhibition of Ras activation at the ER. Cos-7 cells were cotransfected with Ras-GRF (1 μg) and M1-H-Ras (1 μg) in addition to CD8-N17, M1-N17, and M1-GFP (0.5 μg) where indicated. M1-H-Ras GTP levels of a representative experiment are shown, as determined by anti-HA immunoblotting in affinity precipitates using GST-Raf-RBD. Ras-GRF levels were determined in total lysates by using anti-Ras-GRF antibodies, and the levels of CD8- and M1-N17 mutants were monitored with anti-FLAG antibody. The right panel shows inhibition of Ras activation at the bulk membrane. (B) Inhibition of Ras-GRF-induced Ras activation at lipid rafts and bulk membrane by tethered N17 mutants. The left panel shows inhibition of Ras activation at lipid rafts. Cos-7 cells were cotransfected with Ras-GRF (1 μg) and LCK-H-Ras (1 μg) in addition to CD8-N17 and LCK-N17 (0.5 μg) where indicated. LCK-H-Ras GTP levels of a representative experiment are shown. Ras-GRF levels were determined in total lysates using anti-Ras-GRF antibodies. The right panel shows inhibition of Ras activation at the bulk membrane. The figures indicate average levels of activation of at least five independent experiments relative to controls. (C) Effects of targeted Ras N17 mutants on colony growth. NIH 3T3 fibroblasts were transfected with the indicated N17 constructs or with H-Ras (wt) (0.5 μg). Cells were grown in the presence of G418 (750 μg/ml). After 2 weeks in culture, colonies were stained and scored. The results show average ± the SEM of at least five independent experiments.
Next, we studied the effects of the targeted N17 mutants on cellular proliferation by testing their ability to interfere with colony growth. As such, NIH 3T3 cells were transfected with the targeted N17 constructs and, after culture under G418 selection, the resulting colonies were scored. As shown in Fig. 7C, ER-targeted M1-N17 and bulk membrane-targeted CD8-N17 robustly restricted growth, showing more than 66% reduction compared to the number of colonies formed by H-Ras wild-type, a decrease comparable to that one brought about by untargeted H-RasN17. On the other hand, Golgi complex-tethered KDELr-N17 had minimal effects on colony proliferation. Interestingly, LCK-N17, in spite of its cross-interference with Ras activation at bulk membranes, was considerably less efficient than CD8-N17 for restricting colony numbers, reducing it by only 30%. These results hinted that only Ras functions at the ER and at bulk membrane were essential for supporting cellular growth.
We then investigated the effects of site-restricted Ras inhibition on cellular transformation. To do this, we analyzed in NIH 3T3 fibroblasts the transforming potential of different types of oncogenes, when Ras was inhibited at some specific site by cotransfection with the targeted N17 mutants. Since Ras N17 mutants function by unproductively sequestering exchange factors (19), they should not affect Ras V12-evoked events. As such, the concentration of the constructs encoding for the N17 mutants was adjusted so as to minimally interfere with transformation induced by H-RasV12, thereby avoiding nonspecific, inhibitory effects. Under these conditions, transformation by a PM-bound tyrosine kinase such as v-Src proved to be particularly sensitive to Ras inhibition at the PM, in particular at the bulk membrane. Noticeably, Ras inhibition at the ER had little effects on v-Src-induced transformation (Table 1). In the case of v-Sis, encoding for a growth factor, platelet-derived growth factor (PDGF), which acts through a tyrosine kinase receptor, blocking Ras functions at the PM markedly reduced its transforming potential, 73% blockade by bulk membrane CD8-N17 and 53% by LCK-N17 at lipid rafts. On the other hand, inhibiting Ras ER pool had only moderate consequences on v-Sis transformation, preventing it by only 25%. A very similar situation was encountered when we evaluated transformation by the G protein-coupled receptor, m1 muscarinic receptor, an agonist-dependent oncogene, in the presence of its ligand carbachol (20). Interestingly, blocking Ras activation at the Golgi complex by KDELr-N17 had very modest effects on the transforming potential of all of the oncogenes tested (Table 1).
TABLE 1.
Effects of compartmentalized Ras inhibition on cellular transformationa
| Oncogene | Avg no. of foci/μg of oncogene DNA ± SEM (% inhibition)
|
|||||
|---|---|---|---|---|---|---|
| Vector | N17 | M1 N17 | Lck N17 | CD8 N17 | KDELr N17 | |
| H-Ras V12 | 3,270 ± 126 | 3,002 ± 165** (−9) | 2,981 ± 90** (−9) | 3,182 ± 125NS (−3) | 2,943 ± 79** (−10) | 3,197 ± 105NS (−3) |
| v-Src | 402 ± 31 | 304 ± 25** (−25) | 414 ± 46NS (+3) | 246 ± 47*** (−39) | 194 ± 61*** (−52) | 433 ± 38NS (8) |
| v-Sis | 414 ± 83 | 221 ± 29*** (−47) | 322 ± 40* (−23) | 179 ± 34*** (−57) | 112 ± 48*** (−73) | 378 ± 48NS (−9) |
| m1 MAchR | 774 ± 92 | 487 ± 42*** (−38) | 534 ± 75*** (−23) | 480 ± 30*** (−32) | 464 ± 93*** (−41) | 825 ± 33NS (−10) |
NIH 3T3 fibroblasts were cotransfected with H-RasV12 (100 ng), v-Src (1 μg), v-Sis (1.5 μg), or m1 receptor (0.5 μg) in addition to the different N17 targeted constructs (0.5 μg) where indicated. After 2 weeks, cells were stained, and foci were scored. Figures, expressed as foci per micrograms of oncogene DNA, indicate the average ± the SEM of at least four independent transfections. The percent inhibitions relative to vector-transfected plates are shown in parentheses. m1-transfected plates were grown in the presence of 10 μM carbachol. Analysis of variance was determined by Bonferroni multiple comparisons test relative to the respective “vector” values. P values: *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, P > 0.05 with a 95% confidence interval.
Cells devoid of Ras at lipid rafts and at the Golgi complex proliferate normally and can be transformed.
The fact that our N17 constructs tethered to bulk membrane and to lipid rafts exhibited some degree of cross-inhibition precluded us from gaining a clear picture of the necessity for Ras functions at these microdomains. In order to clarify this point, we resorted to an alternative experimental model. This was provided to us by the existence of double-knocked-out mice for H-Ras and N-Ras (18). Cells derived from these mice express only the isoform K-Ras, but since K-Ras is not found in lipid rafts (39) these are completely devoid of all Ras isoforms, thus constituting an unambiguous model for our purposes. In addition, cells derived from H-Ras/N-Ras double-knockout mice also lack Ras at the Golgi complex, since K-Ras is also absent from this organelle (1, 11).
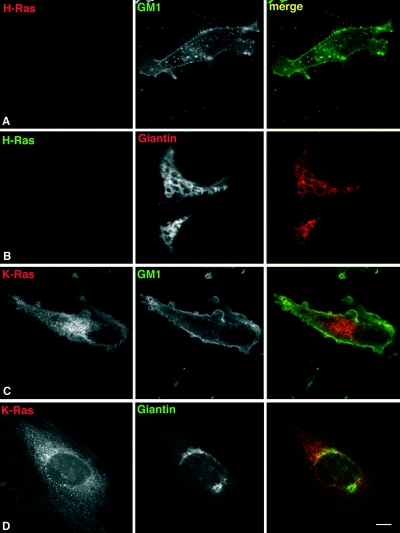
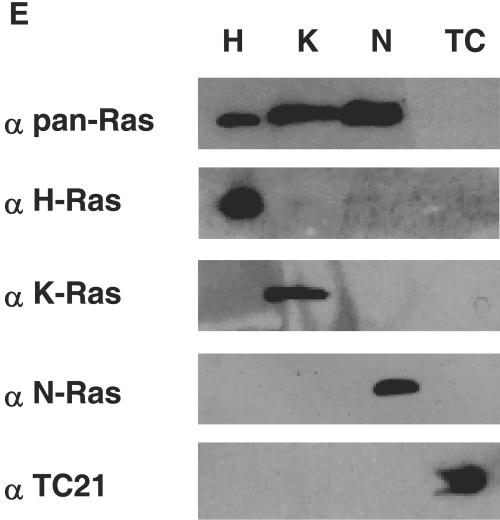
First of all, we ascertained that in MEFs from H-Ras/N-Ras−/− mice, K-Ras had not undergone a redistribution process due to adaptive pressures and lipid rafts and the Golgi complex were indeed devoid of every Ras isoform. Double immunofluorescence analysis of H-Ras with the lipid raft marker GM1 and the Golgi marker giantin verified the complete lack of expression of H-Ras in these cells (Fig. 8A and B). Identical results were obtained when we analyzed the expression of N-Ras (data not shown). When the distribution of K-Ras was analyzed, it was found that this isoform was notoriously present at the ER and at some PM localizations (Fig. 8C). However, upon double staining with GM1 an absolute lack of colocalization was evident, a finding indicative of the absence of K-Ras from lipid rafts. Likewise, coimmunofluorescence with giantin revealed the complete exclusion of K-Ras from the Golgi complex (Fig. 8D). The absence of cross-reactivity among the different Ras isoforms and the GTPase TC-21-specific antibodies utilized in our assays was ascertained to further confirm our data (Fig. 8E). These results clearly corroborated the absence of Ras isoforms from lipid Rafts and the Golgi complex in H-Ras/N-Ras−/− MEFs.
FIG. 8.
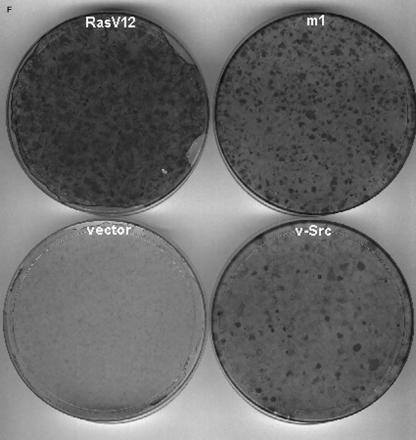
MEFs devoid of Ras at lipid rafts can be transformed. H-Ras/N-Ras−/− MEFs are devoid of Ras isoforms at lipid rafts and the Golgi complex. MEFs were costained with anti-H-Ras antibodies (red, panel A; green, panel B) or anti-K-Ras (red, panels C and D) and cholera toxin to stain lipid rafts (green, panels A and C) or the Golgi marker antigiantin (red, panel B; green, panel D). All panels show equatorial sections at the cell nucleus level. Bars: A and C, 20 μm; B and D, 10 μm. (E) Specificity of the antibodies utilized. Lysates from COS-7 cells transfected with H-Ras (H), K-Ras (K), N-Ras (N), and TC-21 (TC) were immunoblotted with the indicated antibodies. (F) H-Ras/N-Ras−/− MEF transformation induced by oncogenes. MEFs were transfected with vectors encoding for H-Ras V12, v-Src, or m1 muscarinic receptor (1 μg each), in addition to c-Myc (1 μg). After 3 weeks, cells were stained, and the number of foci was scored. m1-transfected plates were grown in the presence of 10 μM carbachol.
The mere fact that H-Ras/N-Ras−/− mice are viable and grow normally (18) was an indisputable proof of Ras functions at lipid rafts and the Golgi complex being completely superfluous for normal cell growth and made further experimentation on this respect unnecessary. So we proceeded to test the need for Ras at these sites in the process of cellular transformation. Thus, we tested the ability of oncogenes to transform H-Ras/N-Ras−/− MEFs. Since MEFs in order to be transformed require the additional participation of a nuclear oncogene, c-Myc was cotransfected in all cases. As expected, H-Ras V12 displayed a potent focus-forming activity. Interestingly, this was also the case for v-Src and for m1 receptor (Fig. 8F). Furthermore, these oncogenes were able to transform H-Ras/N-Ras−/− MEFs in proportions similar to those of the wild-type MEFs (data not shown). Therefore, these results clearly corroborated that the functions undertaken by Ras at lipid rafts and the Golgi complex were fully dispensable for cellular growth and transformation.
DISCUSSION
Today, a large body of evidence supports the notion that Ras proteins are segregated throughout different PM microdomains and internal membrane systems (23). Determining how compartmentalization influences Ras signaling and its subsequent biochemical and biological effects undoubtedly represents a conceptual milestone in the long quest for unraveling the fine points of Ras mechanics. Ideally, one would attempt to selectively activate or inhibit endogenous Ras at a particular compartment. However, our current state of knowledge and technical limitations preclude such an untainted approach. Thus, we are forced to resort to alternative strategies that, though less sophisticated, will enable us to gain a timely insight into this conundrum.
We have approached the problem by selectively targeting H-RasV12 to those cellular compartments where Ras is localized under physiological conditions. This we have achieved by substituting H-Ras palmitoylation signal for N-terminally fused peptides encoding for alternative cues that specifically direct Ras to the desired locations. These proteins have been stably expressed in NIH 3T3 cells, and their correct localization has been extensively corroborated. It is noteworthy the fact that we have encountered very little mislocalization and unwanted intercompartment spillage. This was particularly striking in the case of experiments using transient transfections in COS-7 or 293T cells, in which plasmid episomal replication yields very high protein levels, in spite of which mislocalizations were seldom detected. In this respect, we have observed that there are slight variations in the levels of targeted Ras proteins present at the different localizations. This probably reflects that distinct types of membranes support different amounts of Ras. In support of this notion we have observed that the expression levels of our targeted Ras constructs are dependent on the Ras farnesylation signal. A likely explanation for the absence of mislocalizations could be that once Ras-binding sites at a given location are saturated, excess targeted-Ras proteins, bearing a farnesyl moiety that cannot be inserted into a membrane, are rapidly degraded. In agreement, we have observed that farnesylation-deficient, targeted Ras proteins accumulate in the cytoplasm when overexpressed. Regardless of the targeting signal utilized, there will be a limit to how much protein can be inserted into a given site, after which surplus will accumulate in the cytoplasm and/or will be degraded. In the case of our constructs that limit is set by the availability of Ras-binding sites.
We have thoroughly ascertained the specificity of Ras targeting to bulk membrane and to lipid rafts. This we have monitored by immunofluorescence, detergent solubilization, and lysis in sodium carbonate. The three methods have demonstrated specific targeting and a complete absence of cross-localization. We have observed that the constructs that are targeted to bulk membrane and to lipid rafts can also be detected in small cytoplasmic vesicles, probably bulk membrane and lipid raft-derived endosomes. This phenomenon is highly representative of the internalization and recycling processes that take place at the PM. However, it prevents us from finely discriminating between events strictly taking place at these microdomains and those that occur at their derived endosomes, which would constitute the next level of complexity in our future investigations. At this stage, we have limited ourselves to gain an overall view of the processes regulated by Ras proteins that reside in these membrane subtypes, irrespective of the stage of the membrane turnover cycle. Without interfering with the physiological trafficking and recycling processes that these subdomains are subject to, that could lead to confusing situations.
We have verified that our constructs are functional. In spite of the variable-size peptides that were fused to their N termini, all RasV12 constructs bound with similar efficiencies to an effector molecule in vitro, indicating that the N-terminal structural alterations did not affect their GTP-bound state. This was further corroborated in vivo by their ability to recruit c-Raf to those sites where they had been tethered. A notorious exception was the case of the Golgi complex, in which very little colocalization of c-Raf and KDELr-V12 was observed. This observation is in full agreement with our results showing that activation of the ERK cascade at the Golgi complex was very poor and may reflect either the lack of c-Raf at Golgi microlocalizations where Ras is present or, alternatively, the absence therein of scaffold proteins that intervene in the assembly of the Ras-Raf complex. Even though during mitosis the presence of Raf at the Golgi complex and its participation in Golgi fragmentation has been reported (12), whether Raf is directly activated therein or elsewhere and then shuttles to that organelle has not been demonstrated.
Our results clearly demonstrate that the intensity at which an effector route is activated by Ras is greatly dependent on its localization. It is unlikely that differences in Ras concentrations can account for the observed variations. For example, the potential to engage effectors and the robustness of the biological outcomes of M1-V12 are some of the greatest, even though its expression levels are not as high as those of LCK-V12 or of CD8-V12. Furthermore, at any given compartment some effector routes are very efficiently stimulated, while others are not. Probably, the local intensity of a signal is dictated by the limited availability of some or all of the intermediaries that build up a signaling module or of essential scaffold proteins that optimize signal output through that route. In line with this notion, our results demonstrate that the Ral-GDS route is most efficiently stimulated at the Golgi complex where high concentrations of its cognate GTPase, RalA, are found (31). Likewise, the Raf-ERK pathway is preferentially activated in lipid rafts, reflecting c-Raf high affinity for cholesterol-enriched domains (30). Even though this result may seem contrary to previous reports, indicating that Raf activation is favored in bulk membrane as opposed to lipid rafts (39), Raf activation can be initiated at lipid rafts (44) and lipid raft disruption prevents the activation of the Raf-ERK pathway (43, 47), indicating that the machinery to activate this pathway is readily available at lipid rafts. The fact that we are using fixed Ras constructs, unable to translocate between compartments, could account for the differences in signal intensities reported here and in previous reports (39).
We have found that Ras can effectively support cellular proliferation and transformation from most of its platforms. Since a biological output is the result of the integration of all of the signals generated by a causative stimulus, it is difficult, if not impossible, to discretely associate a biological outcome with the activation of a particular route. However, our results underscore the importance of two essential factors. (i) The first is the signal intensity. How Ras signal strength affects the biological outcome(s) is highlighted by the behavior of the cells harboring M1-V12 and RasV12 SS. Both of them operate at the ER, and both activate the same signaling pathways, M1-V12 more intensively in all cases. As such, M1-V12 is much more effective in promoting proliferation, transformation, and survival. It is noteworthy that signal intensity does not follow a proportional relationship with the resulting biological outcome(s), which hints the existence of signal thresholds over which full biological responses would ensue. (ii) The second factor is the availability of substrates. We show here that only the ER Ras pool can sustain cell survival, even though Ras operating from other platforms, for example, lipid rafts, evokes very similar effectors usage and almost identical signal intensities. One likely explanation for this apparent discrepancy could be that ERK/Akt/Ral substrates mediating in cell survival are confined and available mainly to ER-activated effectors.
The surprising finding of the Ras ER pool being extremely efficient at supporting survival is completely unprecedented. Our results reveal that M1-V12 generates a potent antiapoptotic signal irrespective of the cell type and the apoptogenic stimulus. Interestingly, M1-V12 evokes antiapoptotic events that H-Ras V12 cannot. One possible explanation for this could be that M1-V12 is fixed to the ER, whereas H-Ras V12 is subject to Ras physiological trafficking in and out of the ER. Therefore, it is conceivable that by increasing Ras residence time at this organelle a more efficient pro-survival signaling could be elicited. Even though the exact mechanisms whereby this process takes place are unknown, the ER is a key site directly intervening in the apoptosis-regulating effects of calcium (21). Interestingly, N-Ras, known to provide a potent antiapoptotic signal (51), is particularly abundant at the ER (our unpublished results).
Our results indicate that Ras activation at the Golgi complex cannot promote cellular transformation nor enhance cell proliferation, even though it is competent for sustaining limited cellular growth for short periods under serum starvation conditions. This clearly contrasts with previous reports (7, 10). However, one technical subtlety may likely account for this discrepancy: we utilized here as a Golgi complex-tethering signal KDELr, harboring the mutation N193D that prevents it from translocating to the ER and fixes it to the Golgi complex as a permanent resident protein (13). In contrast, the aforementioned studies used KDELr wild type. As such, proteins tethered with this cue will shuttle between ER and the Golgi complex. Since Ras at the ER can effectively transform, as we and others have shown (10), it is very likely that transformation elicited by KDELr wild-type-tethered Ras is a consequence of its transit through the ER. In full agreement with our results, a former report using yet another Golgi complex-targeting signal, the E1 protein of avian bronchitis virus, detected no transformation induced by Ras at the Golgi complex (28). On the other hand, in light of our data demonstrating the potent activation of RalGDS by Ras at the Golgi complex and of previous studies reporting the importance of this pathway for human oncogenesis (22, 41), it is conceivable that, in contrast to the murine model, Ras at the Golgi complex could play an important role in the transformation of human cells.
The fact that mutationally activated Ras can effectively exert its effects from most of the sites where it is found, provides little information on the relevance and need for endogenous Ras at its different compartments in processes such as physiological proliferation and transformation by upstream stimuli. We have addressed this issue here, specifically blocking endogenous Ras pools by tethering thereto inhibitory N17 mutants. Remarkably, these mutants have yielded highly specific site-restricted inhibitory effects, the sole exception being N17s targeted to lipid rafts and to disordered membrane, which exhibited some degree of cross-inhibition. However, this should not be surprising. The ER and the Golgi complex are tangible, isolated structures, and most of their extension is spatially separated from each other and from the PM. Thus, it is conceivable that mutants targeted thereto can only exert their effect locally, without interfering with Ras GEFs at other confinements sufficiently separated. On the other hand, lipid rafts and bulk membrane are contiguous domains, exhibiting diffuse boundaries. As such, it can be envisioned that lipid raft-targeted N17 can deprive adjacent bulk membrane of its GEFs and vice versa. The fact that CD8-N17 displays a greater cross-inhibitory potential than LCK-N17 could be a reflection of a greater proportion of the PM being constituted by bulk membrane (27).
Using these constructs we have shown that blocking Ras at the ER and at bulk membrane has profound inhibitory effects on cellular proliferation. On the other hand, abrogating Ras activity at lipid rafts or at the Golgi complex had discrete effects on cell growth. These results have been corroborated in an alternative model: embryo fibroblasts derived from H-Ras/N-Ras double-knockout mice (18). As far as our detection limits enabled us to, we ascertained that, in these cells, lipid rafts and Golgi complex were indeed devoid of Ras isoforms. As such, the sheer fact that H-Ras/N-Ras−/− mice grow and reproduce normally proves beyond doubt that Ras functions at these sites are absolutely dispensable for cellular proliferation under physiological conditions. Since these cells only express K-Ras, which is located at disordered membrane (39), these results add further support to the notion that, at the PM, the main Ras signals originate from this microdomain (23). In addition, our results point to an endomembrane, the ER, as another site at which Ras functions are essential for proliferation. In line with our results demonstrating that most effector pathways are effectively engaged from this signaling platform.
Finally, we have also investigated the importance of compartmentalized Ras functions for cellular transformation when induced by upstream stimuli. To do so, we scored the transforming potential of different oncogenes when localized Ras pools were specifically suppressed by N17 mutants. We have observed that some targeted N17 mutants are more effective for inhibiting transformation than untargeted H-RasN17. A possible explanation could be that the expressed H-RasN17 proteins are dispersed between compartments essential and nonessential for transformation, whereas the same amounts of targeted N17s are concentrated at specific sites. This provides an indication of the necessity of Ras at a particular compartment for transformation. As such, our results indicate that Ras at the Golgi complex is not essential for the transmission of upstream transforming signals. Interestingly, even though our previous results pointed to the Ras ER pool as necessary for proliferation, its suppression had only moderate inhibitory effects on transformation. How transforming oncoproteins can circumvent the requirement for Ras ER functions is thus far unknown. With respect to PM microdomains, our results demonstrate the dispensability of Ras lipid raft pool for transformation elicited by upstream oncoproteins. v-Sis cognate receptor PDGFr, Src, and most G protein-coupled receptors are preferentially located in lipid rafts (32, 36, 42). Interestingly, in oligodendrocytes PDGFr is sequestered in a raft domain at a developmental stage in which PDGF does not promote proliferation (6). Our results suggest that these oncoproteins can exert their effects in the absence of Ras in lipid rafts. This could be due to either contiguous bulk membranes being sufficiently close as to be capable of activating Ras therein or to the fact that some active pools of these proteins also reside in bulk membrane. Once again in full agreement with previous results (23), our data highlights the importance of bulk membrane microdomains in Ras signaling.
Overall, our results underscore for the first time the variability of Ras biochemical signals depending on the signaling platform. Noticeably, we demonstrate that mutationally activated Ras can support enhanced cellular proliferation and transformation from most sites. This has special connotations regarding carcinogenesis, indicating that strategies aimed at inhibiting Ras localization at the PM will not prevent Ras transforming effects elicited from alternative sites such as the ER. In addition, our results highlight the importance of Ras signaling at bulk membrane and provide compelling evidence on the dispensability of Ras functions at lipid rafts and the Golgi complex under physiological conditions.
Acknowledgments
We are indebted to J. Bos, J. F. Hancock, and J. S. Gutkind for providing reagents; X. Bustelo and A. Nebreda for fruitful discussions and suggestions; and M. Lafarga for outstanding technical advice.
P.C.'s laboratory is supported by grants BMC2002-01021 and GEN2003-20239-C06-03 from the Spanish Ministry of Education and grant 01-087 from the Association for International Cancer Research. E.S.'s laboratory is supported by grants SAF2003-04177 and GEN2003-20239-C06-02 from Spanish Ministry of Education and FIS PI/021570 from Spanish Ministry of Health. D.M. is an I3P program postdoctoral fellow. V.S. is a Lady Tata Memorial Trust postdoctoral fellow. I.A. is a Fundación Valdecilla postdoctoral fellow. L.A.-I. and F.C. are Spanish Ministry of Education predoctoral fellows.
REFERENCES
- 1.Apolloni, A., I. A. Prior, M. Lindsay, R. G. Parton, and J. F. Hancock. 2000. H-Ras but not K-ras traffics to the plasma membrane through the exocytic pathway. Mol. Cell. Biol. 20:2475-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arcaro, A., C. Gregoire, N. Boucheron, S. Stotz, E. Palmer, B. Malissen, and I. F. Luescher. 2000. Essential role of CD8 palmitoylation in CD8 coreceptor function. J. Immunol. 165:2064-2076. [DOI] [PubMed] [Google Scholar]
- 3.Arozarena, I., D. S. Aaronson, D. Matallanas, V. Sanz, N. Ajenjo, S. Tenbaum, H. Teramoto, T. Ighishi, J. C. Zabala, J. S. Gutkind, and P. Crespo. 2000. The Rho family GTPase Cdc42 regulates the activation of Ras/MAPK pathway by the exchange factor Ras-GRF. J. Biol. Chem. 275:26441-26448. [DOI] [PubMed] [Google Scholar]
- 4.Arozarena, I., D. Matallanas, M. T. Berciano, V. Sanz-Moreno, F. Calvo, M. T. Munoz, G. Egea, M. Lafarga, and P. Crespo. 2004. Activation of H-Ras in the endoplasmic reticulum by the RasGRF family guanine nucleotide exchange factors. Mol. Cell. Biol. 24:1516-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arozarena, I., D. Matallanas, and P. Crespo. 2001. Maintenance of Cdc42 GDP-bound state by Rho-GDI inhibits MAP kinase activation by the exchange factor Ras-GRF. J. Biol. Chem. 276:21878-21884. [DOI] [PubMed] [Google Scholar]
- 6.Baron, W., L. Decker, H. Colognato, and C. ffrench-Constant. 2003. Regulation of integrin growth factor interactions in oligodendrocytes by lipid raft microdomains. Curr. Biol. 13:151-155. [DOI] [PubMed] [Google Scholar]
- 7.Bivona, T. G., I. Perez de Castro, I. M. Ahearn, T. M. Grana, V. K. Chiu, P. J. Lockyer, P. J. Cullen, A. Pellicer, A. D. Cox, and M. R. Philips. 2003. Phospholipase Cγ activates Ras on the Golgi apparatus by means of RasGRP1. Nature 424:694-698. [DOI] [PubMed] [Google Scholar]
- 8.Cadwallader, K. A., H. Paterson, S. G. MacDonald, and J. F. Hancock. 1994. N-terminal myristoylated Ras proteins require palmitoylation or a polybasic domain for plasma membrane localization. Mol. Cell. Biol. 14:4722-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caloca, M. J., J. L. Zugaza, and X. R. Bustelo. 2003. Exchange factors of the RasGRP family mediate Ras activation in the Golgi. J. Biol. Chem. 278:33465-33473. [DOI] [PubMed] [Google Scholar]
- 10.Chiu, V. K., T. Bivona, A. Hach, J. B. Sajous, J. Silletti, H. Wiener, R. L. Johnson, A. D. Cox, and M. R. Philips. 2002. Ras signaling on the endoplasmic reticulum and the Golgi. Nat. Cell. Biol. 4:343-350. [DOI] [PubMed] [Google Scholar]
- 11.Choy, E., V. K. Chiu, J. Silletti, M. Feoktistov, T. Morimoto, D. Michaelson, I. E. Ivanov, and M. R. Philips. 1999. Endomembrane trafficking of Ras: the CAAX motif targets proteins to the ER and Golgi. Cell 98:69-80. [DOI] [PubMed] [Google Scholar]
- 12.Colanzi, A., C. Sutterlin, and V. Malhotra. 2003. RAF1-activated MEK1 is found on the Golgi apparatus in late prophase and is required for Golgi complex fragmentation in mitosis. J. Cell Biol. 161:27-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole, N. B., C. L. Smith, N. Sciaky, M. Terasaki, M. Edidin, and J. Lippincott-Schwartz. 1996. Diffusional mobility of Golgi proteins in membranes of living cells. Science 273:797-801. [DOI] [PubMed] [Google Scholar]
- 14.Coso, O. A., M. Chiariello, J. C. Yu, H. Teramoto, P. Crespo, N. Xu, T. Miki, and J. S. Gutkind. 1995. Rac-1 and cdc42 control the activity of JNK (SAPK) signaling pathway. Cell 81:1137-1146. [DOI] [PubMed] [Google Scholar]
- 15.Crespo, P., and J. Leon. 2000. Ras proteins in the control of the cell cycle and cell differentiation. Cell. Mol. Life Sci. 57:1613-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crespo, P., N. Xu, J. L. Daniotti, J. Troppmair, U. R. Rapp, and J. S. Gutkind. 1994. Signaling through transforming G protein-coupled receptors in NIH 3T3 cells involves c-Raf activation. Evidence for a protein kinase C-independent pathway. J. Biol. Chem. 269:21103-21109. [PubMed] [Google Scholar]
- 17.Datta, K., A. Bellacosa, T. O. Chan, and P. N. Tsichlis. 1996. Akt is a direct target of the phosphatidylinositol 3-kinase: activation by growth factors, v-src and v-Ha-ras, in Sf9 and mammalian cells. J. Biol. Chem. 271:30835-30839. [DOI] [PubMed] [Google Scholar]
- 18.Esteban, L. M., C. Vicario-Arbejon, P. Fernandez-Salguero, A. Fernandez-Melarde, N. Swaminathan, K. Yienger, E. Lopez, R. McKay, J. M. Ward, A. Pellicer, and E. Santos. 2001. Targeted genomic disruption of H-ras and N-ras individually or in combination, reveals the dispensability of both loci for mouse growth and development. Mol. Cell. Biol. 21:1444-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feig, L. A. 1999. Tools of the trade: use of dominant inhibitory mutants of Ras-family GTPases. Nature Cell Biol. 1:25-27. [DOI] [PubMed] [Google Scholar]
- 20.Gutkind, J. S., E. A. Novotny, M. R. Brann, and K. C. Robbins. 1991. Muscarinic acetylcholine receptor subtypes as agonist-dependent oncogenes. Proc. Natl. Acad. Sci. USA 88:4703-4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hajnoczky, G., E. Davies, and M. Madesh. 2003. Calcium signaling and apoptosis. Biochem. Biophys. Res. Commun. 304:445-454. [DOI] [PubMed] [Google Scholar]
- 22.Hamad, N. M., J. H. Elconin, A. E. Karnoub, W. Bai, J. N. Rich, R. T. Abraham, C. J. Der, and C. M. Counter. 2002. Distinct requirements for Ras oncogenesis in human versus mouse cells. Genes Dev. 16:2045-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hancock, J. F. 2003. Ras proteins: different signals from different locations. Nat. Rev. Mol. Cell. Biol. 4:373-384. [DOI] [PubMed] [Google Scholar]
- 24.Hancock, J. F., K. Cadwallader, H. Paterson, and C. J. Marshall. 1991. A CAAX or a CAAL motif and a second signal are sufficient for plasma membrane targeting of Ras proteins. EMBO J. 10:4033-4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hancock, J. F., A. I. Magee, J. E. Childs, and C. J. Marshall. 1989. All Ras proteins are polyisoprenylated but only some are palmytoylated. Cell 57:1167-1177. [DOI] [PubMed] [Google Scholar]
- 26.Hancock, J. F., H. Paterson, and C. J. Marshall. 1990. A polybasic domain or palmytoylation is required in addition to the CAAX motif to localize p21Ras to the membrane. Cell 63:133-139. [DOI] [PubMed] [Google Scholar]
- 27.Harder, T., P. Scheiffele, P. Verkade, and K. Simons. 1998. Lipid domain structure of the plasma membrane revealed by patching of membrane components. J. Cell Biol. 141:929-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hart, K. C., and D. J. Donoghue. 1997. Derivatives of activated H-ras lacking C-terminal lipid modifications retain transforming ability if targeted to the correct subcellular location. Oncogene 14:945-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heidaran, M. A., J. H. Pierce, D. Lombardi, M. Ruggiero, J. S. Gutkind, T. Matsui, and S. A. Aaronson. 1991. Deletion or substitution within the alpha platelet-derived growth factor receptor kinase insert domain: effects on functional coupling with intracellular signaling pathways. Mol. Cell. Biol. 11:134-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hekman, M., H. Hamm, A. V. Villar, B. Bader, J. Kuhlmann, J. Nickel, and U. R. Rapp. 2002. Associations of B- and C-Raf with cholesterol, phosphatidylserine, and lipid second messengers: preferential binding of Raf to artificial lipid rafts. J. Biol. Chem. 277:24090-24102. [DOI] [PubMed] [Google Scholar]
- 31.Hernandez-Munoz, I., M. Benet, M. Calero, M. Jimenez, R. Diaz, and A. Pellicer. 2003. rgr oncogene: activation by elimination of translational controls and mislocalization. Cancer Res. 63:4188-4195. [PubMed] [Google Scholar]
- 32.Liu, P., Y. Ying, Y. Ko, and R. G. W. Anderson. 1996. Localization of platelet-derived growth factor-stimulated phosphorylation cascade to caveolae. J. Biol. Chem. 271:10299-10303. [DOI] [PubMed] [Google Scholar]
- 33.Lowy, D. R., and B. M. Willumsen. 1993. Function and regulation of Ras. Annu. Rev. Biochem. 62:851-891. [DOI] [PubMed] [Google Scholar]
- 34.Malumbres, M., and A. Pellicer. 1998. Ras pathways to cell cycle control and cell transformation. Fronts. Biosci. 3:d887-d912. [DOI] [PubMed] [Google Scholar]
- 35.Matallanas, D., I. Arozarena, M. T. Berciano, D. S. Aaronson, A. Pellicer, M. Lafarga, and P. Crespo. 2003. Differences in the inhibitory specificities of H-Ras, K-Ras and N-Ras (N17) dominant negative mutants are related to their membrane microlocalization. J. Biol. Chem. 278:4572-4581. [DOI] [PubMed] [Google Scholar]
- 36.Ostrom, R. S., and P. A. Insel. 2004. The evolving role of lipid rafts and caveolae in G protein-coupled receptor signaling: implications for molecular pharmacology. Br. J. Pharmacol. 143:235-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perez de Castro, I., T. G. Bivona, M. R. Philips, and A. Pellicer. 2004. Ras activation in Jurkat T cells following low-grade stimulation of the T-cell receptor is specific to N-Ras and occurs only on the Golgi apparatus. Mol. Cell. Biol. 24:3485-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prior, I. A., and J. F. Hancock. 2001. Compartmentalization of Ras proteins. J. Cell Sci. 114:1603-1608. [DOI] [PubMed] [Google Scholar]
- 39.Prior, I. A., A. Harding, J. Yan, J. Sluimer, R. G. Parton, and J. F. Hancock. 2001. GTP-dependent segregation of H-ras from lipid rafts is required for biological activity. Nat. Cell Biol. 3:368-375. [DOI] [PubMed] [Google Scholar]
- 40.Prior, I. A., C. Muncke, R. G. Parton, and J. F. Hancock. 2003. Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J. Cell Biol. 160:165-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rangarajan, A., S. J. Hong, A. Gifford, and R. A. Weinberg. 2004. Species- and cell type-specific requirements for cellular transformation. Cancer Cell 6:171-183. [DOI] [PubMed] [Google Scholar]
- 42.Resh, M. D. 2004. Membrane targeting of lipid modified signal transduction proteins. Subcell. Biochem. 37:217-232. [DOI] [PubMed] [Google Scholar]
- 43.Rizzo, M. A., C. A. Kraft, S. C. Watkins, E. S. Levitan, and G. Romero. 2001. Agonist-dependent traffic of raft-associated Ras and Raf-1 is required for activation of the mitogen-activated protein kinase cascade. J. Biol. Chem. 276:34928-34933. [DOI] [PubMed] [Google Scholar]
- 44.Roy, S., R. Luetterforst, A. Harding, A. Apolloni, M. Etheridge, E. Stang, B. Rolls, J. F. Hancock, and R. G. Parton. 1999. Dominant-negative caveolin inhibits H-ras function by disrupting cholesterol-rich plasma membrane domains. Nat. Cell Biol. 1:98-105. [DOI] [PubMed] [Google Scholar]
- 45.Roy, S., B. Wyse, and J. F. Hancock. 2002. H-Ras signaling and K-Ras signaling are differentially dependent on endocytosis. Mol. Cell. Biol. 22:5128-5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanz-Moreno, V., B. Casar, and P. Crespo. 2003. p38alpha isoform Mxi2 binds to extracellular signal-regulated kinase 1 and 2 mitogen-activated protein kinase and regulates its nuclear activity by sustaining its phosphorylation levels. Mol. Cell. Biol. 23:3079-3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scheel, J., J. Srinivasan, U. Honnert, A. Henske, and T. V. Kurzchalia. 1999. Involvement of caveolin-1 in meiotic cell-cycle progression in Caenorhabditis elegans. Nat. Cell Biol. 1:127-129. [DOI] [PubMed] [Google Scholar]
- 48.Su, M. W., C. L. Yu, S. J. Burakoff, and Y. J. Jin. 2001. Targeting Src homology 2 domain-containing tyrosine phosphatase (SHP-1) into lipid rafts inhibits CD3-induced T-cell activation. J. Immunol. 166:3975-3982. [DOI] [PubMed] [Google Scholar]
- 49.Swift, A. M., and C. E. Machamer. 1991. A golgi retention signal in a membrane-spanning domain of coronavirus E1 protein. J. Cell Biol. 115:19-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Willumsen, B. M., A. Christensen, N. L. Hubbert, A. G. Papageorge, and D. R. Lowy. 1984. The p21 Ras C terminus is required for transformation and membrane association. Nature 310:583-586. [DOI] [PubMed] [Google Scholar]
- 51.Wolfman, J. C., and A. Wolfman. 2000. Endogenous c-N-Ras provides a steady-state antiapoptotic signal. J. Biol. Chem. 275:19315-19323. [DOI] [PubMed] [Google Scholar]
- 52.Wolthuis, R. M., B. Franke, M. van Triest, B. Bauer, R. H. Cool, J. H. Camonis, J. W. Akkerman, and J. L. Bos. 1998. Activation of the small GTPase Ral in platelets. Mol. Cell. Biol. 18:2486-2491. [DOI] [PMC free article] [PubMed] [Google Scholar]