Abstract
Human SIRT1 is an enzyme that deacetylates the p53 tumor suppressor protein and has been suggested to modulate p53-dependent functions including DNA damage-induced cell death. In this report, we used EX-527, a novel, potent, and specific small-molecule inhibitor of SIRT1 catalytic activity to examine the role of SIRT1 in p53 acetylation and cell survival after DNA damage. Treatment with EX-527 dramatically increased acetylation at lysine 382 of p53 after different types of DNA damage in primary human mammary epithelial cells and several cell lines. Significantly, inhibition of SIRT1 catalytic activity by EX-527 had no effect on cell growth, viability, or p53-controlled gene expression in cells treated with etoposide. Acetyl-p53 was also increased by the histone deacetylase (HDAC) class I/II inhibitor trichostatin A (TSA). EX-527 and TSA acted synergistically to increase acetyl-p53 levels, confirming that p53 acetylation is regulated by both SIRT1 and HDACs. While TSA alone reduced cell survival after DNA damage, the combination of EX-527 and TSA had no further effect on cell viability and growth. These results show that, although SIRT1 deacetylates p53, this does not play a role in cell survival following DNA damage in certain cell lines and primary human mammary epithelial cells.
SIRT1 is an enzyme that catalyzes the deacetylation of acetyl-lysine residues by a mechanism in which NAD+ is cleaved and a unique product, O-acetyl ADP-ribose, is generated (4, 19, 21). In addition, the reaction results in the release of nicotinamide, which acts as an end product inhibitor (3). SIRT1 is distinct from the class I and II histone deacetylase (HDAC) enzymes that remove acetyl groups without hydrolysis of NAD+ (13). Indeed, SIRT1 catalytic activity is not affected by the class I and II HDAC inhibitor trichostatin A (TSA) (19).
SIRT1 plays a role in a wide variety of processes including stress resistance, metabolism, differentiation, and aging (4). Overexpression of SIRT1 orthologs in Saccharomyces cerevisiae, Caenorhabditis elegans, and Drosophila melanogaster leads to an increased life span of these organisms (23, 43, 49), and this has excited speculation that SIRT1 might also regulate life span in mammals (5). SIRT1 binds to and regulates the activity of several transcription factors, including FOXO1, FOXO3a, and FOXO4 (8, 16, 37, 38, 51), HES-1 and HEY-2 (48), MyoD (15), CTIP2 (46), PPARγ (40), NF-κB (55), and PGC1α (42). SIRT1 has also been shown to interact with and deacetylate the p53 tumor suppressor protein (25, 30, 53). p53 is a key transcriptional regulator of genes involved in cell cycle progression, apoptosis, and DNA repair. Indeed, many human tumors have inactivated p53 protein (54). p53 becomes acetylated after DNA damage, and the acetylated form has been reported to have increased transcriptional activity, promote coactivator recruitment, and enhance site-specific DNA binding (2). Acetylation of p53 is also believed to increase p53 stability by preventing ubiquitination of key lysine residues and subsequent proteasomal degradation (27).
Compounds that activate p53 function have been successfully used to treat tumors in vivo (9, 20, 52). Therefore, small-molecule inhibitors of SIRT1 might function as anticancer agents if SIRT1 acts as an inhibitor of p53. This hypothesis is supported by studies in cell lines in which overexpression of a catalytically impaired variant of SIRT1 decreased survival and increased p53 target gene expression after DNA damage (53). Overexpression of wild-type SIRT1 had the opposite effect of increasing cell survival after DNA damage (30). Moreover, thymocytes derived from mice lacking Sir2α (mouse SIRT1) exhibit increased sensitivity to γ-radiation although, interestingly, embryonic stem cells and fibroblasts from these mice showed no altered resistance to DNA damage-induced stress (10, 33).
In this study, the role of SIRT1 on p53 acetylation and function is examined using a novel, potent, and specific small-molecule inhibitor of SIRT1 catalytic activity, designated EX-527. In cells subjected to DNA damage, inhibition of SIRT1 with EX-527 results in increased p53 acetylation but surprisingly does not result in detectable effects on p53-controled gene expression, cell survival, or cell proliferation. In contrast, inhibition of class I/II HDAC-mediated p53 deacetylation with TSA increases p53 acetylation after DNA damage, with reduced cell survival and growth. Although p53 acetylation is synergistically increased by inhibition of both HDAC and SIRT1, concomitant treatment with EX-527 and TSA does not further affect cell proliferation or survival after DNA damage.
MATERIALS AND METHODS
Materials and cell culture.
Etoposide was obtained from Calbiochem. TSA, nicotinamide, adriamycin, hydroxyurea, hydrogen peroxide, and other reagent quality chemicals were from Sigma. Tissue culture plastics were obtained from Corning except where noted. NCI-H460, MCF-7, IMR-90, 293T, and U-2 OS cells were obtained from the American Type Tissue Collection. NCI-H460 cells were cultured in RPMI 1640 medium with 10 mM HEPES, 1 mM sodium pyruvate, and 10% fetal bovine serum (FBS). MCF-7, IMR-90, and 293T cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% FBS. U-2 OS cells were cultured in McCoy's 5a medium with 1.5 mM l-glutamine and 10% FBS. Primary human mammary epithelial cells (HMEC, derived from normal adult mammary glands) and complete serum-free medium were obtained from Cell Applications, Inc. HMEC were passaged no more than three times before use in experiments.
Deacetylation assays and inhibitory compounds.
Deacetylation was measured using the Fluor de Lys kit (AK-555; Biomol) using a fluorogenic peptide encompassing residues 379 to 382 of p53, acetylated on lysine 382 (KI-177; Biomol). The acetylated lysine residue was coupled to an aminomethylcoumarin moiety. The peptide was deacetylated by SIRT1, followed by the addition of a proteolytic developer that released the fluorescent aminomethylcoumarin. Briefly, enzyme preparations were incubated with 170 μM NAD+ and 100 μM p53 fluorogenic peptide for 45 min at 37°C followed by incubation in developer for 15 min at 37°C. Fluorescence was measured by excitation at 360 nm and emission at 460 nm and enzymatic activity was expressed in relative fluorescence units.
Human SIRT5 (N-terminal glutathione S-transferase [GST] tag) was expressed in Escherichia coli and purified by affinity chromatography with glutathione-Sepharose (G.E. Healthcare). Deacetylation of p53 lysine 382 was measured using the Fluor de Lys kit (as described above). SIRT5 activity was also measured by the deacetylation of cytochrome c, prepared by chemical acetylation with [3H]acetate. First, 1 mg cytochrome c (Sigma) was acetylated for 2 h at room temperature in 140 μl of 100 mM morpholineethanesulfonic acid (MES), pH 5.0, with 250 μCi [3H]acetate (ICN) and 2.2 mg/ml 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride (Pierce). Unincorporated [3H]acetate was removed on a G-50 microspin column (G.E. Healthcare). The labeled protein was precipitated with 20% trichloroacetic acid for 20 min on ice, washed three times with ice-cold acetone, air dried, and resuspended at 1 mg/ml in distilled water. The deacetylation reaction was performed as described previously (45) using 10 μg SIRT5.
EX-527 (see Fig. 1A) and additional SIRT1-inhibitory compounds (EX-519, EX-586, EX-589, EX-622, and EX-635) were identified in a high-throughput screen using the Fluor de Lys assay (J. Hixon, T. McDonagh, R. Curtis, P. S. DiStefano, A. Napper, T. Hesterkamp, R. Thomas, K. Keavey, and J. Pons, Soc. Biomol. Screen. 10th Annu. Meet., poster P10045, 2004). EX-527 was found to be 200- to 500-fold more selective for SIRT1 than SIRT2 and SIRT3. EX-527 is a racemic mixture, and its optical isomers were separated by chiral high-performance liquid chromatography and designated EX-242 and EX-243. When tested in the deacetylation assay, EX-243 was active in SIRT1 inhibition, whereas EX-242 was inactive. The details of the synthesis and separation of EX-527 are described elsewhere (37a). Compounds were stored at −20°C in dimethyl sulfoxide and were retested in the deacetylation assay prior to use in cell-based assays.
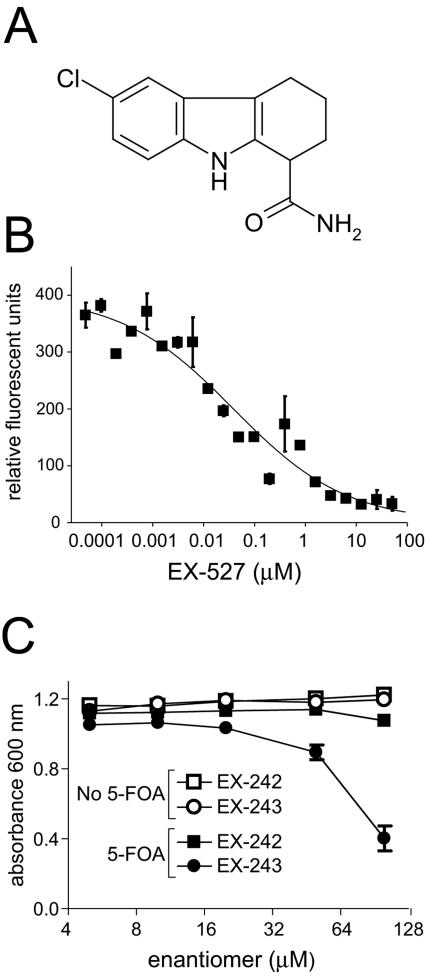
FIG. 1.
EX-527 is a potent inhibitor of SIRT1 that also inhibits Sir2-dependent silencing in yeast. (A) Structure of EX-527. (B) EX-527 inhibits deacetylase activity of purified human SIRT1. Deacetylase activity of GST-SIRT1 in the presence of increasing concentrations of EX-527 was measured by the Fluor de Lys assay. Samples were assayed in triplicate and analyzed using Prism (Graphpad Software, Inc.). (C) The active optical isomer of EX-527, EX-243, and not the inactive optical isomer, EX-242, inhibits Sir2-dependent silencing in yeast. A yeast strain constructed with a URA3 gene inserted near a telomere was grown in the presence of compounds with or without 5-FOA. Growth was measured by light scattering at 600 nm.
Inhibition of GST-SIRT1 deacetylase activity.
293T cells were transiently transfected with GST-tagged human SIRT1 in the pDEST27 Gateway vector (Invitrogen) using FuGENE-6 (Roche). After 48 h, the cells were lysed with 50 mM Tris, pH 8.0, 120 mM NaCl, 1 mM EDTA, and 0.5% Nonidet P-40, supplemented with Complete Mini protease inhibitor cocktail tablets (Roche). GST-SIRT1 was purified from lysates using glutathione-Sepharose beads (G.E. Healthcare) and washed extensively in the above buffer. The deacetylation assay was performed with approximately 30 ng of GST-SIRT1 in the presence of EX-527 (48 pM to 100 μM).
Yeast Sir2 silencing assay.
The effect of compounds on Sir2-dependent silencing was assessed by measuring the growth of a yeast strain harboring a telomere-proximal URA3 gene in the presence or absence of 5-fluoroorotic acid (5-FOA) (17). Strain SL8c was constructed by mating PSY316AUTa to W303ARUTα (35). Cells were grown overnight in SD medium (2% glucose, 0.67% yeast nitrogen base, 0.39% Casamino Acids, 0.005% adenine, 0.005% tryptophan). The culture was split 1:20 into fresh yeast extract-peptone-dextrose medium containing 2% glucose and grown for 5 h. Cells were plated in 96-well plates (Costar) in 150 μl of SD containing 0.005% uracil either with or without 0.1% 5-FOA, containing 5 to 100 μM EX-242 or EX-243. Plates were cultured at 30°C for 18 to 24 h, and growth of resuspended cells was measured by light scattering at 600 nm in a spectrophotometer.
Determination of acetylated p53 levels.
NCI-H460 cells, MCF-7 cells, U-2 OS cells, or HMEC (1 to 2 million) were plated in six-well tissue culture plates or 10-cm dishes. After 1 day (NCI-H460, MCF-7, and U-2 OS cells) or 2 days (HMEC), p53 acetylation was induced for 6 h with one of the following DNA damaging agents: etoposide (20 μM), hydrogen peroxide (400 μM), hydroxyurea (1 mM), or adriamycin (0.2 μg/ml for NCI-H460, MCF-7, and U-2 OS cells; 0.8 μg/ml for HMEC). To determine the effects of deacetylase inhibitors on p53 acetylation levels, cells were treated with 6.25 to 400 nM TSA and/or 1 μM EX-527. In some experiments, other SIRT1 inhibitors, EX-635, EX-622, EX-519, EX-586, and EX-589, or the optical isomers, EX-242 and EX-243, were used at 1 μM instead of EX-527. After washing with phosphate-buffered saline, cells were lysed in 50 mM Tris, pH 7.8, 137 mM NaCl, 10 mM NaF, 1 mM EDTA, 1% Triton X-100, 0.2% Sarkosyl, 1 mM dithiothreitol, and 10% glycerol containing Complete Mini protease inhibitor cocktail tablets and supplemented with 10 μM TSA and 5 mM nicotinamide to prevent deacetylation after cell lysis.
Immunoprecipitation of total p53 was performed with agarose-conjugated p53 monoclonal antibody (Ab-6; Oncogene Research Products) for 2 h at 4°C, followed by incubation with protein A-Sepharose (Pharmacia) for 1 h at 4°C. Beads were washed extensively with lysis buffer. Cell lysates or immunoprecipitates were resolved on 10% Criterion Tris-HCl gels (Bio-Rad). p53 acetylation was detected with anti-acetylated-Lys382 p53 antibody (Cell Signaling Technology), and total p53 levels were determined with antibodies Ab-6 or Ab-7 (Oncogene Research Products). Bound antibodies were visualized with horseradish peroxidase-conjugated secondary antibody and SuperSignal West Femto maximum sensitivity substrate (Pierce). Luminescence was quantified by a Bioimaging system (UVP).
SIRT1 immunoblotting.
SIRT1 in cell lysates was visualized by immunoblotting using antibody 07-131 (Upstate Cell Signaling Solutions). Gels, secondary antibodies, and horseradish peroxidase substrate were used as described above.
Cell viability and proliferation assays.
NCI-H460 cells, MCF-7 cells, U-2 OS cells, or HMEC were plated at 2,000 cells per well in opaque-walled 96-well plates (Corning) for the viability assay and 800 cells per well in 96-well Cytostar-T scintillating microplates (Amersham) for the proliferation assay. Cells were incubated for 1 day (NCI-H460) or 2 days (MCF-7, U-2 OS, and HMEC) prior to exposure to DNA-damaging agents and deacetylase inhibitors. All experiments were performed in triplicate.
For viability assays, cells were treated with the indicated compounds for 48 h. Cell viability was then determined using the Cell Titer-Glo luminescent assay (Promega), which measures total ATP levels as an index of cell number. Luminescence was measured on a Luminoskan Ascent (ThermoLabSystems).
For the proliferation assay, 0.5 μCi/ml of [14C]thymidine was added to the medium immediately after the genotoxins and deacetylase inhibitors. Plates were counted at 48 h (HMEC) or 72 h (NCI-H460, MCF-7, and U-2 OS cells) in a Microbeta liquid scintillation counter (Perkin-Elmer). Thymidine incorporated by the cells was detected by proximity to the scintillant in the base of the Cytostar-T tissue culture plate. Values were graphed as means ± standard deviations.
Effects on p53-controlled gene expression.
NCI-H460 cells, MCF-7 cells, U-2 OS cells, or HMEC (0.5 to 2 million) were plated in 10-cm tissue culture plates. After 1 to 2 days, cells were treated with various concentrations of etoposide (0 to 100 μM) in the presence or absence of 1 μM EX-527 for 6 h. RNA was isolated from cells using the RNAqueous 4-PCR kit (Ambion). cDNA was generated using the First-Strand cDNA synthesis kit (Roche). Quantitative PCR was performed to determine mRNA levels using Lightcycler (Roche) and the following primers: p21 forward, GCGACTGTGATGCGCTAATGG; p21 reverse, GCGTTTGGAGTGGTAGAAATCTG; BAX forward, GCGAGTGTCTCAAGCGC; BAX reverse, GCACCAGTTTGCTGGCA; hypoxanthine phosphoribosyltransferase forward, TAGCCCTCTGTGTGCT; hypoxanthine phosphoribosyltransferase reverse, CTTCGTGGGGTCCTTT.
To detect p21 protein, NCI-H460 cells were treated with adriamycin (0.2 μg/ml) or etoposide (20 μM) in the presence or absence of 1 μM EX-527 for 6 h and then lysed in 50 mM Tris buffer, pH 8.0, 120 mM NaCl, 1 mM EDTA, and 0.5% Nonidet P-40, supplemented with Complete Mini protease inhibitor cocktail tablets. The samples were resolved on Criterion 10% Tris-HCl gels, and blots were probed with anti-p21 antibody F-5 (Santa Cruz Biotechnology) at a 1:1,000 dilution. Tubulin was visualized with antibody B-5-1-2 (Sigma) at a 1:5,000 dilution as a protein loading control. Secondary antibodies and horseradish peroxidase substrate were used as described above.
SIRT1 overexpression.
Constructs that express either SIRT1 or green fluorescent protein (GFP) were made using the pLP-LNCX vector (Clontech). The constructs were packaged into virus by cotransfecting them using FuGENE 6 into 293T cells (500,000 cells per 60-mm dish) with the pVPack-VSV-G and pVPack-GP vectors (Stratagene). After 24 h, virus was collected from the supernatant and filtered through a 0.45-μm filter, and Polybrene (Sigma) was added to a final concentration of 8 μg/ml. IMR-90 and MCF-7 cells cultured in six-well plates were infected with virus-containing medium for 24 h, after which the medium was replaced with fresh medium containing G418. Following selection, cells were plated in 96-well plates at 5,000 cells per well and treated with 200 to 1,600 μM hydrogen peroxide for 24 h, and the cell viability was determined using the Cell Titer-Glo kit (Promega) as described above.
RESULTS
Characterization of a potent inhibitor of SIRT1.
EX-527 (Fig. 1A) is an inhibitor of SIRT1 enzymatic activity (50% inhibitory concentration [IC50], 98 nM), identified in a high-throughput screen using bacterially expressed human SIRT1 (37a). To confirm the activity of EX-527 against SIRT1 produced in mammalian cells, the potency of EX-527 was determined using the in vitro Fluor de Lys deacetylation assay. The activity of GST-tagged human SIRT1, purified from 293T cells, was measured in the presence of EX-527 (48 pM to 100 μM). As shown in Fig. 1B, EX-527 inhibited SIRT1 in a concentration-dependent manner with an IC50 of 38 nM, in agreement with the activity on bacterially expressed SIRT1.
EX-527 has much lower potency against SIRT2 (IC50, 19.6 μM) or SIRT3 (IC50, 48.7 μM) but does not inhibit class I/II HDAC activity at concentrations up to 100 μM (37a). We and others (39) have found that SIRT5 possesses low, albeit significant, deacetylase activity, and we sought to determine if this can be inhibited by EX-527. SIRT5 had no effect on deacetylation of p53 lysine 382 peptide in the Fluor de Lys assay (data not shown). Therefore, the release of [3H]acetate from chemically acetylated cytochrome c was measured. SIRT5-mediated deacetylation of cytochrome c was not inhibited by EX-527 at concentrations up to 50 μM (data not shown). No deacetylase activity was detected for SIRT4, SIRT6, and SIRT7. Thus, EX-527 is selective for SIRT1 compared to other SIRT family members and class I/II HDAC enzymes.
To confirm that EX-527 acts on NAD-dependent deacetylase enzymes, the compound was tested in the well-characterized Sir2-dependent silencing assay in yeast. SIRT1 is the closest human ortholog of the yeast silent information regulator-2 protein, Sir2 (12). Sir2 regulates transcriptional silencing at the DNA near telomeres, the silent mating type locus, and in the region encoding rRNA by deacetylating lysines on histone tails (36). Inhibition of yeast Sir2 activity by the pure optical isomers of EX-527 was determined (Fig. 1C). The optical isomers are EX-243, which inhibits SIRT1 catalytic activity, and EX-242, which does not. The inactive isomer serves as a control for cellular effects of the compound that are unrelated to the inhibition of catalytic activity. Sir2 activity was assayed by analyzing the silencing of a URA3 gene inserted near a telomere (17). 5-FOA in the medium blocks the growth of yeast only when the URA3 gene is expressed. Sir2 activity silences the URA3 gene near the telomere, and the cells grow normally in the presence of 5-FOA. However, if Sir2-dependent silencing is inhibited, the URA3 gene is expressed and growth in the presence of 5-FOA is reduced. As expected, in the absence of 5-FOA, neither enantiomer had any growth inhibitory effect, indicating that EX-243 and EX-242 are not toxic at the concentrations tested (Fig. 1C). In the presence of 5-FOA, the active enantiomer EX-243 inhibited growth, whereas EX-242 had no effect, indicating that Sir2 silencing of URA3 is reduced (Fig. 1C). EX-527 gave results similar to those of EX-243 (data not shown). These results demonstrate that EX-527 can inhibit NAD-dependent deacetylation in a cellular environment.
Inhibition of SIRT1 enhances p53 acetylation in response to DNA damaging agents.
Previous studies have shown that the acetylation of lysine 382 of p53 is modulated by SIRT1 (25, 30, 53). To determine if inhibition of SIRT1 resulted in an increase in p53 acetylation, NCI-H460 cells were treated with EX-527. NCI-H460 cells were chosen because they have previously been shown to express abundant SIRT1 protein, as well as wild-type p53, and they have an intact p53 response to DNA damage (24, 30). EX-527 (1 μM) had no detectable effect on the acetylation of lysine 382 in the absence of genotoxic insult (data not shown). In contrast, in cells treated with the DNA damaging agent etoposide, EX-527 produced a concentration-dependent increase in the amount of acetylated p53, with a maximal effect at 1 μM (Fig. 2A). Nicotinamide (5 mM, a concentration commonly used to inhibit SIRT1) was less effective at increasing p53 acetylation than 1 μM EX-527 (Fig. 2A).
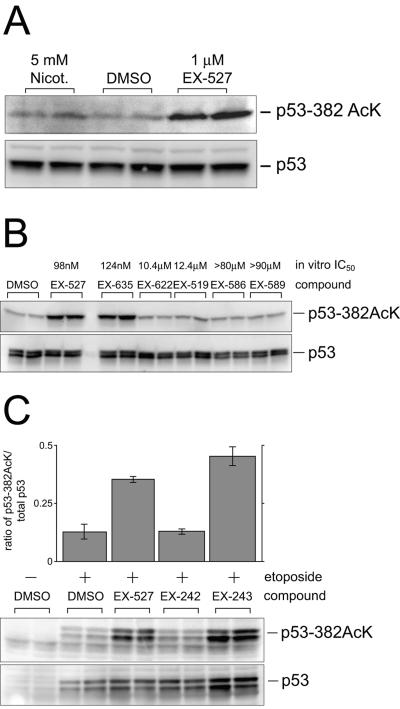
FIG. 2.
Inhibition of SIRT1 catalytic activity increases p53 acetylation after DNA damage. (A) Effect of EX-527 on p53 acetylation after DNA damage. NCI-H460 cells were treated with etoposide in combination with either nicotinamide (Nicot.), EX-527, or dimethyl sulfoxide vehicle (DMSO) for 6 h. Blots were probed with an anti-acetylated p53 Lys 382 antibody (upper panel) or p53 antibody (Ab-7) that recognizes all forms of human p53 (lower panel). (B) Effect of EX-527 and a series of closely related compounds on p53 acetylation after DNA damage. Cells were treated with 1 μM concentrations of the indicated compounds for 6 h, and samples were prepared as described for panel A, except that cells were treated additionally with 25 nM TSA to inhibit class I/II HDAC activity. (C) Effect of enantiomers of EX-527 on p53 acetylation after DNA damage. Cells were treated with EX-242 or EX-243 for 6 h. p53 was immunoprecipitated and immunoblotted as described for panel A. +, present; −, absent.
To eliminate the possibility that EX-527 might influence p53 acetylation via nonspecific effects on p53, several compounds from the same chemical series as EX-527 were tested. These compounds varied in their ability to inhibit SIRT1 in the in vitro deacetylation assay (IC50, 124 nM to >80 μM). Significantly, inhibition of SIRT1 catalytic activity correlated well with the ability to increase p53 acetylation in etoposide-treated cells when the compounds were tested at 1 μM in the presence of 25 nM TSA (Fig. 2B). For additional validation, the optical isomers of EX-527 were tested, with the inactive isomer EX-242 serving as a control for SIRT1-independent cellular effects. One micromolar EX-243, the active isomer, enhanced the acetylation of p53 in the presence of etoposide, whereas 1 μM EX-242 had no effect (Fig. 2C). These results confirm that EX-527 regulates p53 acetylation directly by inhibition of SIRT1 catalytic activity.
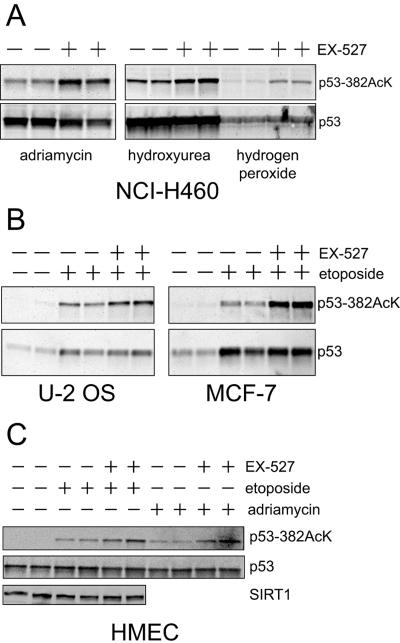
Acetylation of p53 is known to occur in response to a wide variety of DNA damaging agents (1). To determine whether the catalytic activity of SIRT1 plays a general role in controlling DNA damage-induced acetylation, NCI-H460 cells were treated with 1 μM EX-527 in combination with a variety of genotoxic agents. EX-527 increased acetylation of lysine 382 in response to adriamycin, hydroxyurea, and hydrogen peroxide (Fig. 3A) as well as etoposide (Fig. 2). We also investigated whether inhibition of SIRT1 with EX-527 can regulate acetyl-p53 in other cell lines as well as primary human cells. In addition to NCI-H460 cells, EX-527 inhibited p53 deacetylation in U-2 OS and MCF-7 cells treated with etoposide (Fig. 3B). Both of these cell lines express SIRT1 as assessed by immunoblotting (data not shown).
FIG. 3.
Inhibition of SIRT1 catalytic activity enhances p53 lysine 382 acetylation after a variety of DNA-damaging agents and is not cell type specific. (A) NCI-H460 cells were treated with adriamycin, hydroxyurea, or hydrogen peroxide for 6 h. p53 was immunoprecipitated and immunoblotted with an anti-acetylated p53 Lys 382 antibody or p53 antibody (Ab-7). (B) U-2 OS and MCF-7 cells were treated with etoposide in the presence or absence of 1 μM EX-527 for 6 h. Immunoprecipitation and immunoblotting was performed as described for panel A. (C) HMEC were treated with etoposide or adriamycin in the presence or absence of 1 μM EX-527 for 6 h. Immunoprecipitation and immunoblotting of p53 was performed as described for panel A. Total SIRT1 levels were measured by immunoblotting using antibody 07-131 (Upstate Cell Signaling Solutions). +, present; −, absent.
HMEC were treated with etoposide or adriamycin in the presence or absence of EX-527 (Fig. 3C). These cells express SIRT1, as shown by immunoblotting, and inhibition with EX-527 increased acetylation of p53 lysine 382 in response to both etoposide and adriamycin. These results show that SIRT1 regulates p53 acetylation in several cell types, including primary cells, under a broad range of conditions.
Effects of inhibition of SIRT1 on p53-controlled gene expression.
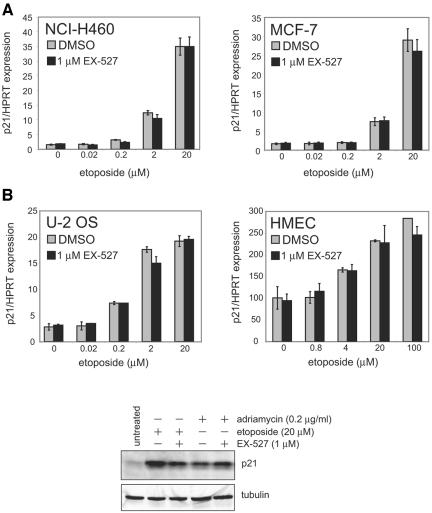
As p53 carries out many of its effects by functioning as a transcription factor, for instance, increasing the expression of p21 and BAX (26), we assessed the role of SIRT1-mediated deacetylation in p53-controlled gene expression. NCI-H460, MCF-7, and U-2 OS cells were treated with various concentrations of etoposide (0 to 20 μM) with or without 1 μM EX-527, and then p21 mRNA was measured by quantitative reverse transcription-PCR. As expected, p21 transcripts showed a progressive increase with 0.2 μM, 2 μM, and 20 μM etoposide. Primary HMEC were also treated with etoposide (0.8 to 100 μM), as higher concentrations were needed to induce p21 in these cells. SIRT1 inhibition by 1 μM EX-527 had no effect on p21 mRNA levels in the absence of etoposide (no DNA damage) or at any concentration of etoposide in any of the cells tested (Fig. 4A). We also found no effect of SIRT1 inhibition on the regulation of BAX mRNA in the same samples (data not shown). p21 protein was not affected by 1 μM EX-527 in NCI-H460 cells treated with either etoposide (20 μM) or adriamycin (0.2 μg/ml) (Fig. 4B). These data suggest that the levels of two specific p53 target genes are not regulated by SIRT1-mediated deacetylation of p53 at lysine 382, following etoposide-induced DNA damage.
FIG. 4.
p53-controlled gene expression is not altered by inhibition of SIRT1 in several p53-positive cell lines. (A) NCI-H460, MCF-7, U-2 OS, and HMEC were treated with various concentrations of etoposide for 6 h in the presence of 1 μM EX-527 or dimethyl sulfoxide vehicle (DMSO). Levels of p21 mRNA were measured using quantitative PCR and normalized to the expression levels of hypoxanthine phosphoribosyltransferase (HPRT). (B) NCI-H460 cells were treated with etoposide or adriamycin in the presence or absence of EX-527 for 6 h. Cell lysates were prepared for immunoblotting and were probed with antibodies directed against p21 and tubulin. +, present; −, absent.
Effects of SIRT1 inhibition on cell viability and growth.
Previous studies have suggested that acetylation regulates p53 activity, although this is controversial (41). If increased p53 acetylation leads to increased p53 activity, then the ability of cells to survive and proliferate after DNA damage might be altered by treatment with SIRT1 inhibitors. To test this idea, NCI-H460 and U-2 OS cells were treated with 0 to 20 μM etoposide in the presence or absence of 1 μM EX-527 and cell survival and growth were measured. NCI-H460 cells were also treated with 0 to 20 μM etoposide and 5 mM nicotinamide. In addition, MCF-7 cells showed limited responses to etoposide at these concentrations and were therefore treated with adriamycin (0 to 1,000 ng/ml) to induce DNA damage in the same assays.
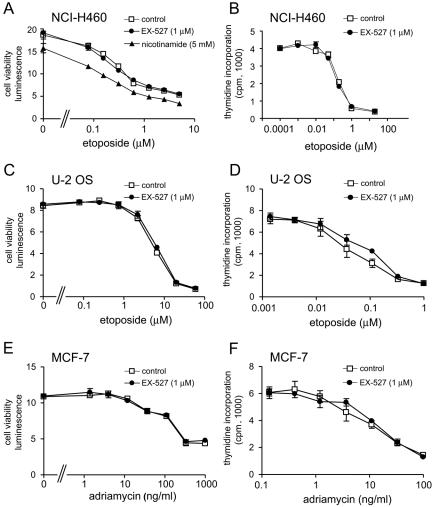
First, cell viability was reduced in a concentration-dependent manner by 48 h of exposure to etoposide or adriamycin (Fig. 5A, C, and E). There was no effect of 1 μM EX-527 on cell viability in the absence of etoposide or adriamycin, indicating that 1 μM EX-527 is not toxic to these cells (Fig. 5A, C, and E). In contrast, exposure to 5 mM nicotinamide for 48 h reduced NCI-H460 cell viability in the absence of etoposide, suggesting that nicotinamide has toxic effects not caused by SIRT1 inhibition (Fig. 5A). At all concentrations of etoposide tested, EX-527 had no effect on cell viability in NCI-H460 or U-2 OS cells (Fig. 5A and C). Likewise, EX-527 had no effect on cell viability in MCF-7 cells at any concentration of adriamycin (Fig. 5E). Next, the effect of EX-527 on cell proliferation was determined by measuring incorporation of [14C]thymidine. Cell proliferation was reduced by treatment with etoposide (NCI-H460 and U-2 OS cells) or adriamycin (MCF-7 cells) in a concentration-dependent manner, but EX-527 had no effect on proliferation (Fig. 5B, D, and F). These results suggest that survival and cell proliferation following DNA damage are unaffected by inhibition of SIRT1 catalytic activity.
FIG. 5.
Effect of inhibition of SIRT1 catalytic activity on cell viability and proliferation following DNA damage in cell lines. NCI-H460, U-2 OS, and MCF-7 cells were treated with increasing concentrations of etoposide or adriamycin in the presence or absence of 1 μM EX-527. NCI-H460 cells were additionally treated with 5 mM nicotinamide. (A, C, and E) Cell viability was measured at 48 h; (B, D, and F) cell proliferation at 72 h was measured by monitoring [14C]thymidine incorporation.
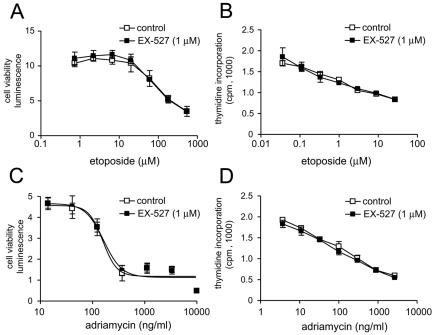
We also tested the effects of 1 μM EX-527 on the survival and growth of primary HMEC (Fig. 6). In preliminary experiments, we found that higher concentrations of DNA-damaging agents were needed to affect primary cells compared to tumor cell lines. HMEC exposed to 0 to 600 μM etoposide or 0 to 10 μg/ml adriamycin showed a concentration-dependent decrease in cell viability and proliferation. However, 1 μM EX-527 had no effect on survival or proliferation in response to etoposide (Fig. 6A and B) or adriamycin (Fig. 6C and D), suggesting that the response to DNA damage in these cells is not regulated by SIRT1-mediated deacetylation.
FIG. 6.
Effect of inhibition of SIRT1 catalytic activity on cell viability and proliferation following DNA damage in primary human cells. HMEC were treated with increasing concentrations of etoposide or adriamycin in the presence or absence of 1 μM EX-527. (A and C) Cell viability was measured at 48 h; (B and D) cell proliferation at 48 h was measured by monitoring [14C]thymidine incorporation.
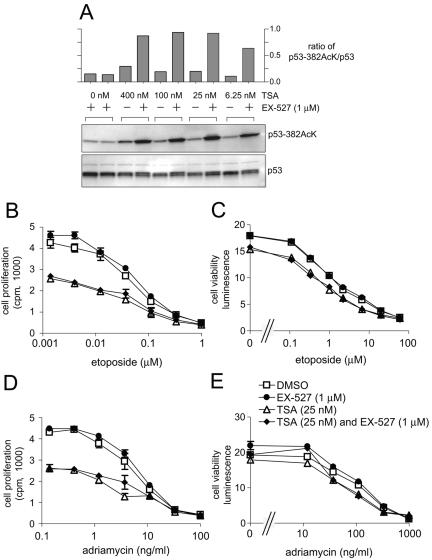
SIRT1 is not the only p53 deacetylase, as it has been established that HDAC1 also binds to and deacetylates p53 (7, 22). It is possible that the level of p53 acetylation caused by inhibition of SIRT1 alone is insufficient to produce phenotypic effects. It has been shown previously that addition of TSA, a potent HDAC inhibitor, enhances the effects of dominant-negative SIRT1 or nicotinamide on p53 acetylation (29, 53). Therefore, the effects of SIRT1 inhibition on p53 might only be apparent when both SIRT1 and HDAC1 activities are inhibited. To test this hypothesis, etoposide-induced p53 acetylation was measured in NCI-H460 cells treated with 6.25 to 400 nM TSA in the presence or absence of 1 μM EX-527 (Fig. 7A). TSA at concentrations of 6.25 to 25 nM had comparable effects to 1 μM EX-527 on p53 acetylation, whereas combining EX-527 and TSA led to a synergistic increase in acetyl-p53. This confirms the model that HDAC1 and SIRT1 coordinately regulate p53 acetylation and that TSA can enhance the effects of SIRT1 inhibition. Furthermore, these data establish that a combination of 1 μM EX-527 and 25 nM TSA consistently increases acetyl-p53 to higher levels than either compound alone.
FIG. 7.
Effect of inhibiting SIRT1 and HDAC1 on p53 lysine 382 acetylation and cell growth. (A) NCI-H460 cells were treated for 6 h with various concentrations of TSA and 1 μM EX-527 in the presence of 20 μM etoposide. Cell lysates were analyzed by immunoblotting with an anti-acetylated p53 Lys 382 antibody or p53 antibody (Ab-7). The graph shows the quantification of bands on the gel shown, but this is representative of the results from two such experiments. +, present; −, absent. (B to E) NCI-H460 cells were treated with increasing concentrations of etoposide or adriamycin and deacetylase inhibitors as indicated. Dimethyl sulfoxide (DMSO) was used as the vehicle control. (B and D) Cell proliferation at 72 h was measured by monitoring [14C]thymidine incorporation; (C and E) cell viability was measured at 48 h.
To determine the effect of high acetyl-p53 levels achieved with the combination of EX-527 and TSA, cell viability and proliferation were measured after DNA damage (Fig. 7B to E). NCI-H460 cells were treated with etoposide (0 to 60 μM) or adriamycin (0 to 1,000 ng/ml) in the presence of 1 μM EX-527 and/or 25 nM TSA (Fig. 7B to E). Twenty-five nanomolar TSA decreased cell viability and proliferation in conjunction with both DNA-damaging agents. However, combining EX-527 and TSA had no further effects on viability or proliferation compared to TSA alone. Furthermore, EX-527 alone did not affect cell viability and proliferation in response to etoposide (Fig. 7B and C) or adriamycin (Fig. 7D and E). These results suggest that SIRT1 inhibition in NCI-H460 cells does not significantly alter p53 function with regard to cell survival and proliferation relative to the effects of TSA alone.
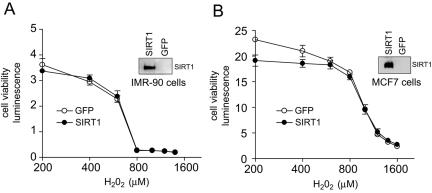
Modulation of SIRT1 protein levels by overexpression or gene knockout technology provides conflicting results, suggesting that SIRT1 can either affect apoptosis (8, 30) or has no effect on this process (53). As inhibition of SIRT1 catalytic activity with EX-527 had no effects on cell viability, we tested the ability of SIRT1 overexpression to regulate cell survival (Fig. 8). Stable cell lines were generated which overexpressed either human SIRT1 or GFP. SIRT1 overexpression was confirmed by immunoblot analysis (Fig. 8), and the ectopically expressed SIRT1 protein was functional, as the levels of acetylated p53 lysine 382 in response to etoposide were strikingly reduced in the overexpressing cells. In MCF7 cells overexpressing SIRT1, the ratio of acetylated p53 to total p53 was 0.038, a sevenfold reduction from the ratio observed in GFP-expressing control cells (0.275). However, no effect of SIRT1 overexpression on cell viability was detected after exposure to 200 to 1,600 μM hydrogen peroxide in either IMR-90 or MCF-7 cells.
FIG. 8.
Effect of overexpression of SIRT1 on hydrogen peroxide-induced cell death. Cell viability was measured in IMR-90 (A) or MCF-7 (B) cells overexpressing SIRT1 or GFP treated with increasing concentrations of hydrogen peroxide for 24 h. Overexpression of SIRT1 in these cells was confirmed using immunoblot analysis (insets). Endogenous SIRT1 is not visible because very small amounts of protein (2 to 4 μg) were needed to obtain a clean signal from cells overexpressing SIRT1 protein.
DISCUSSION
We have used a novel small-molecule inhibitor of SIRT1 to determine the role of SIRT1 catalytic activity in p53 function. EX-527 inhibits the deacetylase activity of SIRT1 purified from human cells (IC50, 38 nM). Thus, EX-527 is approximately 1,000-fold more potent than nicotinamide (IC50, 40 to 50 μM) (3, 31) and other SIRT1 inhibitor compounds, including splitomicin (IC50, 60 μM) (3), M15, and sirtinol (IC50, 40 to 70 μM) (17). Moreover, EX-527 is selective for SIRT1 and is more than 200- to 500-fold less active against the closely related enzymes SIRT2 and SIRT3 (37a). We found that 1 μM EX-527 is nontoxic and remains active in cell culture for several days, enabling its use in longer term studies (data not shown). The potency, specificity, stability, and low toxicity suggest that EX-527 is a more useful reagent than other inhibitors of SIRT1 catalytic activity identified to date.
EX-527 was used to examine the effects of inhibiting SIRT1 catalytic activity on p53 acetylation and cell survival following treatment with a diverse set of DNA-damaging agents. In the presence of EX-527, acetylation at lysine 382 is increased, indicating that SIRT1 is a physiological regulator of p53 acetylation. The effects of EX-527 were due to inhibition of SIRT1 rather than nonspecific activities, as the active enantiomer EX-243 increased acetyl-p53 levels, whereas the inactive isomer did not. In other words, acetyl-p53 levels are raised by the inhibition of SIRT1 catalytic activity and not by any nonspecific activity. The most likely mechanism by which EX-527 increases acetylation is by inhibition of SIRT1 acting directly on p53 as a substrate. It is also possible that inhibition of SIRT1 increases p53 acetylation indirectly by effects on p300 acetyltransferase activity (6). Although EX-527 inhibits p53 deacetylation, this did not affect expression of some p53 target genes in three cell lines and primary HMEC. Furthermore, EX-527 did not affect cell viability or cell proliferation following DNA damage in the same cells. Addition of TSA and EX-527 caused a synergistic increase in p53 acetylation in NCI-H460 cells; however, no additional effects on cell proliferation or viability were detected compared to TSA alone. Taken together, these data are consistent with the notion that acetylation at lysine 382 does not alter the activity of p53. Indeed, the role of p53 acetylation at lysine 382 remains unclear. Fibroblasts and embryonic stem cells obtained from SIRT1 knockout mice are not sensitized to genotoxic stress in spite of increased p53 acetylation (10, 34). In a recent report, mouse embryonic fibroblasts were engineered so that endogenous p53 could not be acetylated because six lysine residues were mutated, including lysine 379 (the mouse equivalent of human lysine 382). Strikingly, p53 activity was not altered in response to DNA damage in these cells (14).
These results contrast with previous studies in which overexpression of a catalytically impaired variant, SIRT1H363Y, decreased cell survival following DNA damage (53). Importantly, the effects of SIRT1H363Y could be reversed by overexpression of dominant-negative p53, demonstrating that the effect of SIRT1H363Y on cell viability requires active p53. EX-527, like SIRT1H363Y, caused a significant increase in p53 acetylation; however, no consequences on cell viability were observed. This suggests that the effects of SIRT1H363Y are not due solely to effects on its catalytic activity. Perhaps, in addition to reducing catalytic activity, the SIRT1H363Y mutation causes a change in the conformation of SIRT1 that affects its interaction with other proteins. This raises the possibility that, in some cases, the interactions of SIRT1 with other proteins may be more important than its catalytic activity.
Overexpression of mouse SIRT1 has been reported to increase cell survival after exposure to hydrogen peroxide and reduce apoptosis after exposure to etoposide (30). However, we did not observe any effect of overexpression of human SIRT1 on cell viability after exposure to hydrogen peroxide. These data are consistent with the work of Vaziri et al., who showed that SIRT1 overexpression in BJ cells had no effect on cell survival after exposure to gamma irradiation (53).
Nicotinamide, as well as EX-527, was able to increase p53 acetylation in the presence of DNA damage. However, in contrast to EX-527, nicotinamide reduced cell viability in the absence of DNA damage. This suggests that nicotinamide has effects that are independent of inhibition of SIRT1 deacetylase activity. Nicotinamide is a known inhibitor of the enzyme poly(ADP-ribose) polymerase, whose inhibition can result in cell cycle arrest or apoptosis (44). Nicotinamide also inhibits SIRT2 (39) and SIRT3 (45) (Hixon et al., Soc. Biomol. Screen. 10th Annu. Meet.). The effects of nicotinamide on poly(ADP-ribose) polymerase, SIRT2, or SIRT3 may be responsible for the observed reduction in cell viability. These data suggest that cell-based experiments using nicotinamide as a means to selectively inhibit SIRT1 catalytic activity must be interpreted with caution.
The HDAC1 inhibitor TSA increased p53 acetylation after DNA damage and, interestingly, reduced cell survival and proliferation. Although HDAC1 may be a more important regulator of p53 than SIRT1, it is more likely that inhibition of HDAC1 affects cellular function via hyperacetylation of proteins other than p53. The effects of 25 nM TSA on survival and proliferation in the absence of etoposide are consistent with stimulation of non-p53-dependent pathways, as 1 μM EX-527 induced equivalent levels of acetylation on p53 lysine 382 without reducing cell survival or proliferation. Indeed, TSA inhibits all class I and II HDAC enzymes, and it has been shown that reduced levels of HDAC2 lead to p53-independent increases in p21 expression and decreased cell viability (18).
Our data do not preclude a role for SIRT1 in the regulation of p53. First, physical interactions between SIRT1 and p53 may play a role in p53 function; therefore, inhibition of SIRT1 catalytic activity may not reveal the importance of SIRT1. Second, SIRT1 may be an important regulator of p53 function in cell types that we have not yet examined. Intriguingly, thymocytes from SIRT1 knockout mice have been shown to be sensitized to the effects of gamma irradiation (10). It will be interesting to determine whether this effect is dependent on p53, as a recent report demonstrates that SIRT1 is involved in p53-independent responses to DNA damage (32). SIRT1 has been shown to have a role in the regulation of p53 during senescence (11, 25). This suggests that SIRT1, which is expressed in a wide range of adult tissues, may regulate other functions of p53, including differentiation, DNA repair, and aging (25, 28, 47, 50).
Acknowledgments
We are indebted to members of the Elixir laboratory for critical reading of the manuscript and helpful discussions. We thank Su-Ju Lin for providing yeast strain SL8c.
All of the authors are employees of Elixir Pharmaceuticals, Inc.
REFERENCES
- 1.Appella, E., and C. W. Anderson. 2001. Post-translational modifications and activation of p53 by genotoxic stresses. Eur. J. Biochem. 268:2764-2772. [DOI] [PubMed] [Google Scholar]
- 2.Barlev, N. A., L. Liu, N. H. Chehab, K. Mansfield, K. G. Harris, T. D. Halazonetis, and S. L. Berger. 2001. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell 8:1243-1254. [DOI] [PubMed] [Google Scholar]
- 3.Bitterman, K. J., R. M. Anderson, H. Y. Cohen, M. Latorre-Esteves, and D. A. Sinclair. 2002. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J. Biol. Chem. 277:45099-45107. [DOI] [PubMed] [Google Scholar]
- 4.Blander, G., and L. Guarente. 2004. The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 73:417-435. [DOI] [PubMed] [Google Scholar]
- 5.Bordone, L., and L. Guarente. 2005. Calorie restriction, SIRT1 and metabolism: understanding longevity. Nat. Rev. Mol. Cell Biol. 6:298-305. [DOI] [PubMed] [Google Scholar]
- 6.Bouras, T., M. Fu, A. A. Sauve, F. Wang, A. A. Quong, N. D. Perkins, R. T. Hay, W. Gu, and R. G. Pestell. 2005. SIRT1 deacetylation and repression of p300 involves lysine residues 1020/1024 within the cell cycle regulatory domain 1. J. Biol. Chem. 280:10264-10276. [DOI] [PubMed] [Google Scholar]
- 7.Brooks, C. L., and W. Gu. 2003. Ubiquitination, phosphorylation and acetylation: the molecular basis for p53 regulation. Curr. Opin. Cell Biol. 15:164-171. [DOI] [PubMed] [Google Scholar]
- 8.Brunet, A., L. B. Sweeney, J. F. Sturgill, K. F. Chua, P. L. Greer, Y. Lin, H. Tran, S. E. Ross, R. Mostoslavsky, H. Y. Cohen, L. S. Hu, H. L. Cheng, M. P. Jedrychowski, S. P. Gygi, D. A. Sinclair, F. W. Alt, and M. E. Greenberg. 2004. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303:2011-2015. [DOI] [PubMed] [Google Scholar]
- 9.Bykov, V. J., N. Issaeva, A. Shilov, M. Hultcrantz, E. Pugacheva, P. Chumakov, J. Bergman, K. G. Wiman, and G. Selivanova. 2002. Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat. Med. 8:282-288. [DOI] [PubMed] [Google Scholar]
- 10.Cheng, H. L., R. Mostoslavsky, S. Saito, J. P. Manis, Y. Gu, P. Patel, R. Bronson, E. Appella, F. W. Alt, and K. F. Chua. 2003. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc. Natl. Acad. Sci. USA 100:10794-10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chua, K. F., R. Mostoslavsky, D. B. Lombard, W. W. Pang, S. Saito, S. Franco, D. Kaushal, H. L. Cheng, M. R. Fischer, N. Stokes, M. M. Murphy, E. Appella, and F. W. Alt. 2005. Mammalian SIRT1 limits replicative life span in response to chronic genotoxic stress. Cell Metab. 2:67-76. [DOI] [PubMed] [Google Scholar]
- 12.Denu, J. M. 2003. Linking chromatin function with metabolic networks: Sir2 family of NAD(+)-dependent deacetylases. Trends Biochem. Sci. 28:41-48. [DOI] [PubMed] [Google Scholar]
- 13.de Ruijter, A. J., A. H. van Gennip, H. N. Caron, S. Kemp, and A. B. van Kuilenburg. 2003. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 370:737-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng, L., T. Lin, H. Uranishi, W. Gu, and Y. Xu. 2005. Functional analysis of the roles of posttranslational modifications at the p53 C terminus in regulating p53 stability and activity. Mol. Cell. Biol. 25:5389-5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fulco, M., R. L. Schiltz, S. Iezzi, M. T. King, P. Zhao, Y. Kashiwaya, E. Hoffman, R. L. Veech, and V. Sartorelli. 2003. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol. Cell 12:51-62. [DOI] [PubMed] [Google Scholar]
- 16.Giannakou, M. E., and L. Partridge. 2004. The interaction between FOXO and SIRT1: tipping the balance towards survival. Trends Cell Biol. 14:408-412. [DOI] [PubMed] [Google Scholar]
- 17.Grozinger, C. M., E. D. Chao, H. E. Blackwell, D. Moazed, and S. L. Schreiber. 2001. Identification of a class of small molecule inhibitors of the sirtuin family of NAD-dependent deacetylases by phenotypic screening. J. Biol. Chem. 276:38837-38843. [DOI] [PubMed] [Google Scholar]
- 18.Huang, B. H., M. Laban, C. H. Leung, L. Lee, C. K. Lee, M. Salto-Tellez, G. C. Raju, and S. C. Hooi. 2005. Inhibition of histone deacetylase 2 increases apoptosis and p21(Cip1/WAF1) expression, independent of histone deacetylase 1. Cell Death Differ. 12:395-404. [DOI] [PubMed] [Google Scholar]
- 19.Imai, S., C. M. Armstrong, M. Kaeberlein, and L. Guarente. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403:795-800. [DOI] [PubMed] [Google Scholar]
- 20.Issaeva, N., P. Bozko, M. Enge, M. Protopopova, L. G. Verhoef, M. Masucci, A. Pramanik, and G. Selivanova. 2004. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat. Med. 10:1321-1328. [DOI] [PubMed] [Google Scholar]
- 21.Jackson, M. D., and J. M. Denu. 2002. Structural identification of 2′- and 3′-O-acetyl-ADP-ribose as novel metabolites derived from the Sir2 family of beta-NAD+-dependent histone/protein deacetylases. J. Biol. Chem. 277:18535-18544. [DOI] [PubMed] [Google Scholar]
- 22.Juan, L. J., W. J. Shia, M. H. Chen, W. M. Yang, E. Seto, Y. S. Lin, and C. W. Wu. 2000. Histone deacetylases specifically down-regulate p53-dependent gene activation. J. Biol. Chem. 275:20436-20443. [DOI] [PubMed] [Google Scholar]
- 23.Kaeberlein, M., M. McVey, and L. Guarente. 1999. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 13:2570-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai, S. L., R. P. Perng, and J. Hwang. 2000. p53 gene status modulates the chemosensitivity of non-small cell lung cancer cells. J. Biomed. Sci. 7:64-70. [DOI] [PubMed] [Google Scholar]
- 25.Langley, E., M. Pearson, M. Faretta, U. M. Bauer, R. A. Frye, S. Minucci, P. G. Pelicci, and T. Kouzarides. 2002. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 21:2383-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levine, A. J. 1997. p53, the cellular gatekeeper for growth and division. Cell 88:323-331. [DOI] [PubMed] [Google Scholar]
- 27.Li, M., J. Luo, C. L. Brooks, and W. Gu. 2002. Acetylation of p53 inhibits its ubiquitination by Mdm2. J. Biol. Chem. 277:50607-50611. [DOI] [PubMed] [Google Scholar]
- 28.Lin, T., C. Chao, S. Saito, S. J. Mazur, M. E. Murphy, E. Appella, and Y. Xu. 2005. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat. Cell Biol. 7:165-171. [DOI] [PubMed] [Google Scholar]
- 29.Luo, J., M. Li, Y. Tang, M. Laszkowska, R. G. Roeder, and W. Gu. 2004. Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc. Natl. Acad. Sci. USA 101:2259-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo, J., A. Y. Nikolaev, S. Imai, D. Chen, F. Su, A. Shiloh, L. Guarente, and W. Gu. 2001. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell 107:137-148. [DOI] [PubMed] [Google Scholar]
- 31.Marcotte, P. A., P. R. Richardson, J. Guo, L. W. Barrett, N. Xu, A. Gunasekera, and K. B. Glaser. 2004. Fluorescence assay of SIRT protein deacetylases using an acetylated peptide substrate and a secondary trypsin reaction. Anal. Biochem. 332:90-99. [DOI] [PubMed] [Google Scholar]
- 32.Matsushita, N., Y. Takami, M. Kimura, S. Tachiiri, M. Ishiai, T. Nakayama, and M. Takata. 2005. Role of NAD-dependent deacetylases SIRT1 and SIRT2 in radiation and cisplatin-induced cell death in vertebrate cells. Genes Cells 10:321-332. [DOI] [PubMed] [Google Scholar]
- 33.McBurney, M. W., X. Yang, K. Jardine, M. Bieman, J. Th'ng, and M. Lemieux. 2003. The absence of SIR2alpha protein has no effect on global gene silencing in mouse embryonic stem cells. Mol. Cancer Res. 1:402-409. [PubMed] [Google Scholar]
- 34.McBurney, M. W., X. Yang, K. Jardine, M. Hixon, K. Boekelheide, J. R. Webb, P. M. Lansdorp, and M. Lemieux. 2003. The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol. Cell. Biol. 23:38-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mills, K. D., D. A. Sinclair, and L. Guarente. 1999. MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell 97:609-620. [DOI] [PubMed] [Google Scholar]
- 36.Moazed, D. 2001. Common themes in mechanisms of gene silencing. Mol. Cell 8:489-498. [DOI] [PubMed] [Google Scholar]
- 37.Motta, M. C., N. Divecha, M. Lemieux, C. Kamel, D. Chen, W. Gu, Y. Bultsma, M. McBurney, and L. Guarente. 2004. Mammalian SIRT1 represses forkhead transcription factors. Cell 116:551-563. [DOI] [PubMed] [Google Scholar]
- 37a.Napper, A. D., J. Hixon, T. McDonagh, K. Keavey, J.-F. Pons, J. Barker, W. T. Yau, P. Amouzegh, A. Flagg, E. Hamelin, R. J. Thomas, M. Kates, S. Jones, M. A. Navia, J. O. Saunders, P. S. DiStefano, and R. Curtis. 9 November 2005, posting date. Discovery of indoles as potent and selective inhibitors of the deacetylase SIRT1. J. Med. Chem. doi: 10.121/jm050522v. [DOI] [PubMed]
- 38.Nemoto, S., M. M. Fergusson, and T. Finkel. 2004. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science 306:2105-2108. [DOI] [PubMed] [Google Scholar]
- 39.North, B. J., B. L. Marshall, M. T. Borra, J. M. Denu, and E. Verdin. 2003. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell 11:437-444. [DOI] [PubMed] [Google Scholar]
- 40.Picard, F., M. Kurtev, N. Chung, A. Topark-Ngarm, T. Senawong, R. Machado De Oliveira, M. Leid, M. W. McBurney, and L. Guarente. 2004. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature 429:771-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prives, C., and J. L. Manley. 2001. Why is p53 acetylated? Cell 107:815-818. [DOI] [PubMed] [Google Scholar]
- 42.Rodgers, J. T., C. Lerin, W. Haas, S. P. Gygi, B. M. Spiegelman, and P. Puigserver. 2005. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature 434:113-118. [DOI] [PubMed] [Google Scholar]
- 43.Rogina, B., and S. L. Helfand. 2004. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc. Natl. Acad. Sci. USA 101:15998-16003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saldeen, J., L. Tillmar, E. Karlsson, and N. Welsh. 2003. Nicotinamide- and caspase-mediated inhibition of poly(ADP-ribose) polymerase are associated with p53-independent cell cycle (G2) arrest and apoptosis. Mol. Cell. Biochem. 243:113-122. [DOI] [PubMed] [Google Scholar]
- 45.Schwer, B., B. J. North, R. A. Frye, M. Ott, and E. Verdin. 2002. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J. Cell Biol. 158:647-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Senawong, T., V. J. Peterson, D. Avram, D. M. Shepherd, R. A. Frye, S. Minucci, and M. Leid. 2003. Involvement of the histone deacetylase SIRT1 in chicken ovalbumin upstream promoter transcription factor (COUP-TF)-interacting protein 2-mediated transcriptional repression. J. Biol. Chem. 278:43041-43050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sengupta, S., and C. C. Harris. 2005. p53: traffic cop at the crossroads of DNA repair and recombination. Nat. Rev. Mol. Cell Biol. 6:44-55. [DOI] [PubMed] [Google Scholar]
- 48.Takata, T., and F. Ishikawa. 2003. Human Sir2-related protein SIRT1 associates with the bHLH repressors HES1 and HEY2 and is involved in HES1- and HEY2-mediated transcriptional repression. Biochem. Biophys. Res. Commun. 301:250-257. [DOI] [PubMed] [Google Scholar]
- 49.Tissenbaum, H. A., and L. Guarente. 2001. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 410:227-230. [DOI] [PubMed] [Google Scholar]
- 50.Tyner, S. D., S. Venkatachalam, J. Choi, S. Jones, N. Ghebranious, H. Igelmann, X. Lu, G. Soron, B. Cooper, C. Brayton, S. Hee Park, T. Thompson, G. Karsenty, A. Bradley, and L. A. Donehower. 2002. p53 mutant mice that display early ageing-associated phenotypes. Nature 415:45-53. [DOI] [PubMed] [Google Scholar]
- 51.van der Horst, A., L. G. Tertoolen, L. M. de Vries-Smits, R. A. Frye, R. H. Medema, and B. M. Burgering. 2004. FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2(SIRT1). J. Biol. Chem. 279:28873-28879. [DOI] [PubMed] [Google Scholar]
- 52.Vassilev, L. T., B. T. Vu, B. Graves, D. Carvajal, F. Podlaski, Z. Filipovic, N. Kong, U. Kammlott, C. Lukacs, C. Klein, N. Fotouhi, and E. A. Liu. 2004. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303:844-848. [DOI] [PubMed] [Google Scholar]
- 53.Vaziri, H., S. K. Dessain, E. Ng Eaton, S. I. Imai, R. A. Frye, T. K. Pandita, L. Guarente, and R. A. Weinberg. 2001. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107:149-159. [DOI] [PubMed] [Google Scholar]
- 54.Vogelstein, B., and K. W. Kinzler. 2004. Cancer genes and the pathways they control. Nat. Med. 10:789-799. [DOI] [PubMed] [Google Scholar]
- 55.Yeung, F., J. E. Hoberg, C. S. Ramsey, M. D. Keller, D. R. Jones, R. A. Frye, and M. W. Mayo. 2004. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 23:2369-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]