Abstract
YB-1 is a broad-specificity RNA-binding protein that is involved in regulation of mRNA transcription, splicing, translation, and stability. In both germinal and somatic cells, YB-1 and related proteins are major components of translationally inactive messenger ribonucleoprotein particles (mRNPs) and are mainly responsible for storage of mRNAs in a silent state. However, mechanisms regulating the repressor activity of YB-1 are not well understood. Here we demonstrate that association of YB-1 with the capped 5′ terminus of the mRNA is regulated via phosphorylation by the serine/threonine protein kinase Akt. In contrast to its nonphosphorylated form, phosphorylated YB-1 fails to inhibit cap-dependent but not internal ribosome entry site-dependent translation of a reporter mRNA in vitro. We also show that similar to YB-1, Akt is associated with inactive mRNPs and that activated Akt may relieve translational repression of the YB-1-bound mRNAs. Using Affymetrix microarrays, we found that many of the YB-1-associated messages encode stress- and growth-related proteins, raising the intriguing possibility that Akt-mediated YB-1 phosphorylation could, in part, increase production of proteins regulating cell proliferation, oncogenic transformation, and stress response.
More than two decades ago, several abundant proteins within the size range of ∼50 to 60 kDa were identified in complexes with maternal mRNA in Xenopus laevis oocytes and reported to be involved in translational “masking” of mRNA during early metazoan development (9, 34). Subsequently, these proteins initially cloned as FRGY1 and FRGY2 (frog Y-box proteins 1 and 2) (42) turned out to be common for male and female germ cells in all organisms studied, including mammals (27, 37). In somatic mammalian cells, the closely related 50-kDa protein (>96% amino acid identity), first designated p50 and most recently YB-1 (Y-box-binding protein 1), was shown to be a predominant component of translationally inactive messenger ribonucleoprotein particles (mRNPs) (14, 28). Interestingly, YB-1 was independently cloned as a transcription factor that specifically binds to the Y-box promoter element of major histocompatibility complex class II genes (11). It is now well established that YB-1 and related proteins are involved in regulation of both transcription and translation by virtue of sequence-specific and nonspecific binding to nucleic acids (45). The DNA and RNA sequence specificity of YB-1 is mediated through an evolutionarily conserved cold shock domain (CSD), which contains the RNA-binding motifs RNP1 and RNP2. The C terminus of YB-1 possesses alternating basic and acidic clusters and is implicated in both nonspecific DNA or RNA binding and protein-protein interactions (12, 24). YB-1 functions as a structural protein involved in spatial organization of mRNPs (36). It is also known to bind in close proximity to the mRNA cap structure and to displace the initiation factors eukaryotic translation initiation factor 4E (eIF4E) and eIF4G, thereby causing mRNA translational silencing and stabilization (13, 30). Consistent with its inhibitory role in translation, YB-1 is mainly associated with nonpolysomal inactive mRNPs, whereas active mRNPs derived from polysomes contain significantly lower YB-1 levels (29). Accordingly, activation of stored mRNPs in germinal and somatic cells is accompanied by dissociation of YB-1 and related proteins (29, 33, 34). However, the mechanism regulating binding of these proteins to mRNA remains elusive. Initially, phosphorylation of FRGY1 and FRGY2 by casein kinase II was shown to increase their binding to mRNAs and thus considered a potential mechanism for mRNA silencing during Xenopus oogenesis (27, 37). We also found that YB-1 is efficiently phosphorylated by casein kinase II; however, no effect of this phosphorylation event on the ability of YB-1 to bind to RNA was observed (35). Recently, another mechanism involving a YB-1-interacting protein called YBAP1 has been proposed (26), although the functional relevance of this finding in vivo remains to be established.
In our efforts to determine how YB-1 activities in transcription and translation might be regulated, we identified the serine/threonine kinase Akt as a direct interactor with YB-1. We found that Akt-mediated phosphorylation of YB-1 in vitro occurs at Ser-102. Treatment of quiescent NIH 3T3 cells with insulin-like growth factor I (IGF-I) induced phosphorylation of the wild-type YB-1 protein, but not a Ser-102-to-Ala mutant YB-1 protein, suggesting an importance of this site for YB-1 phosphorylation in vivo. Elevation of Akt activity in the cell did not affect expression levels of YB-1, its subcellular localization, or general RNA-binding ability. However, phosphorylated YB-1 was less capable of cross-linking to the mRNA cap structure and of inhibiting cap-dependent translation of a reporter mRNA. These data suggest that YB-1 phosphorylation by Akt weakens its cap-binding capability, thereby facilitating translational activation of silenced mRNA species.
MATERIALS AND METHODS
Antibodies and expression constructs.
The following antibodies were purchased from Cell Signaling: phosphorylated Akt, cyclin E and D1, phosphorylated glycogen synthase kinase 3β, phosphorylated FKHR, phosphorylated mammalian target of rapamycin (mTOR), total mTOR, phosphorylated MEK1/2, total MEK1/2, phosphorylated extracellular signal-regulated kinase 1/2 (Erk1/2), and eIF4E-binding protein 1 (4E-BP1). Total Akt and actin antibodies were from Sigma, c-jun antibodies were from Upstate, antihemagglutinin (anti-HA) was from Babco, and anti-YB-1 antibodies were described earlier (10). pcDNA3-HA-YB-1, pET-15b-YB-1, and pGEX-based YB-1 expression constructs were described previously (13, 14). Constructs coding for the T80S and S102A YB-1 point mutants were created by site-directed mutagenesis with the QuikChange kit (Stratagene) using pBSK-YB-1 plasmid (14) as a template. The primers used for the T80S and S102A YB-1 point mutants were as follows: for the T80A mutant, forward primer 5′-C AAC AGG AAT GAC GCC AAG GAA GAT GTA and reverse 5′-TAC ATC TTC CTT GGC GTC ATT CCT GTT G; for the S102A mutant, forward primer 5′-G AAG TAC CTT CGC GCT GTA GGA GAT GGA G and reverse primer 5′-C TCC ATC TCC TAC AGC GCG AAG GTA CTT C (point mutations underlined). The NdeI-EcoRI fragments containing the corresponding mutations were then subcloned into pGEX-AP/CSD, pET15b-YB-1, or pcDNA3-HA-YB-1 plasmid to replace the wild-type sequence.
Cell cultures, transient transfections, and metabolic labeling.
NIH 3T3 and MCF-7 cells were obtained from ATCC and cultured according to the suppliers' recommendations. Nontransformed or K-Ras-transformed NIH 3T3 (K-Ras-NIH 3T3) cells expressing HA-tagged YB-1 (HA-YB-1) or vector alone were generated by retroviral transduction as previously described (39). For transient-transfection experiments, 1 × 106 NIH 3T3 cells were plated onto a 10-cm dish 24 h prior to transfection of 5 μg of pcDNA3-HA plasmids coding for the wild-type or S102A mutant YB-1 proteins. Cells were transfected using Lipofectamine 2000 (Invitrogen). For orthophosphate labeling, cells were treated for 2 h with different drugs or pharmacological inhibitors in the presence of 0.1 mCi/ml of [32P]orthophosphate (3,000 mCi/mmol; NEN). For [35S]methionine labeling, K-Ras-NIH 3T3 cells expressing HA-YB-1 or vector alone were treated with IGF-I or wortmannin for 2 h. For the last 60 min, the Dulbecco modified Eagle medium was free of l-methionine but contained 0.1 mCi/ml of l-[35S]methionine (>1,000 Ci/mmol; Amersham). The cells were washed with phosphate-buffered saline, lysed, and utilized for immunoprecipitation (IP) as described below.
mRNP isolation, IP, and IB.
Cells were scraped and lysed with buffer containing 20 mM HEPES-KOH (pH 7.8), 100 KCl, 5 mM MgCl2, 2 mM dithiothreitol (DTT), 0.25% Nonidet P-40, 2 μg/ml leupeptin, 2 μg/ml pepstatin, 0.25 mM Na3VO4, 10 nM okadaic acid, and 10 μg/ml cycloheximide. Nuclei and mitochondria were removed by centrifugation at 10,000 rpm for 15 min, and cytosolic extracts were then either directly utilized for IP or spun at 100,000 rpm in a TLA-100 centrifuge (Beckman) for 15 min to separate postpolysomal supernatant and polysomes. Polysomal fractions were then additionally purified from resuspended pellets by spinning through a 30% sucrose cushion for 15 min at 100,000 rpm. For IP, cell extracts were pretreated with RNase A (20 μg/ml) and then incubated with the corresponding antibodies (5 μg of each) immobilized on protein A/G-Sepharose beads (Invitrogen) for 2 h at 4°C. Postpolysomal and polysomal mRNPs were isolated from the corresponding fractions by oligo(dT)-cellulose chromatography as previously described (10, 14). Briefly, polysomes were dissociated using 33 mM EDTA, pH 8.0, and poly(A)+ mRNPs were adsorbed to the column using lysis buffer containing 300 mM of KCl. After extensive washing with the same buffer, mRNPs were eluted with 2× Laemmli buffer, boiled, and analyzed by sodium dodecyl sulfate (SDS)-10% polyacrylamide gel electrophoresis (PAGE) and autoradiography or immunoblotting (IB).
In vitro Akt kinase assay and GST pull-down assays.
Recombinant YB-1 or its derivatives (0.2 μg of each) were incubated with the activated or inactive Akt forms (0.5 μg; Upstate) in a 20-μl reaction mixture containing 25 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 2 mM DTT, 5 mM β-glycerophosphate, 0.1 mM Na3VO4, 2 μM ATP, and 5 μCi [γ-32P]ATP (3,000 Ci/mmol; NEN). Where indicated, phosphorylation was performed in the presence of 0.5 mM of cold ATP. After incubation for 20 min at 37°C, reactions were stopped by adding 2× Laemmli buffer, boiled, and analyzed by SDS-10% PAGE and autoradiography. For the experiments in Fig. 5, YB-1 proteins were additionally purified after phosphorylation using heparin-Sepharose equilibrated with 10 mM Tris-HCl, pH 7.5, and 200 mM KCl. After extensive washes, YB-1 proteins were eluted with 2 M KCl and then dialyzed against 10 mM Tris-HCl, pH 7.5, and 200 mM KCl. For pull-down assays, the recombinant glutathione S-transferase (GST)-tagged YB-1 proteins (1 μg of each) were immobilized on glutathione-Sepharose beads and then incubated for 60 min at 4°C with 1 μg of the activated or inactive form of Akt in 100 μl of binding buffer (150 mM NaCl, 5 mM EDTA, 1 mM DTT, 10 mM Tris-HCl [pH 7.6], 1% Nonidet P-40, 0.05% sodium deoxycholate). After extensive washes, samples were eluted with 2× Laemmli buffer, boiled, and analyzed by SDS-10% PAGE and silver staining.
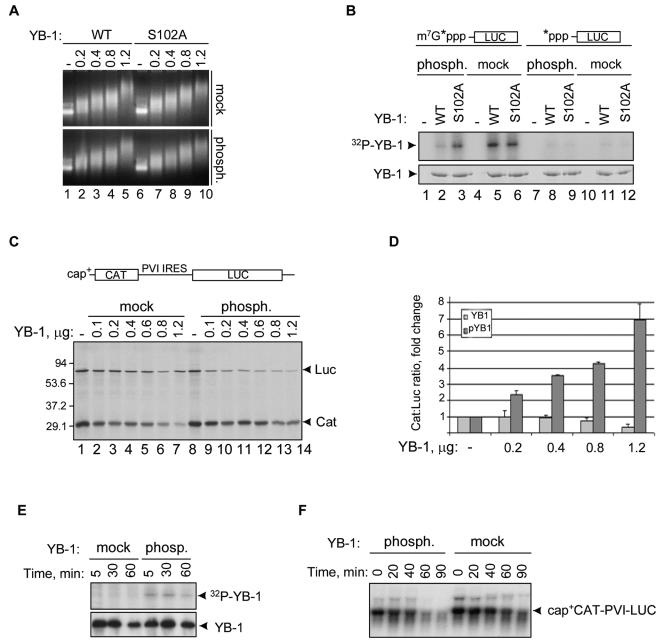
FIG. 5.
Phosphorylation by Akt reduces the abilities of YB-1 to bind to the capped 5′ end of mRNA and to inhibit cap-dependent translation. (A) Gel retardation assay using uncapped CAT mRNA (1 μg) and increasing amounts (in micrograms) of the wild-type (WT) or recombinant YB-1 proteins phosphorylated (phosph.) by the inactive or activated Akt forms in the presence of cold ATP. Ethidium bromide staining of 1% agarose gel is shown. mock, mock phosphorylated. (B) UV cross-linking of YB-1 proteins to the cap-labeled or 5′-labeled LUC mRNAs (top). Wild-type YB-1 (WT) or S102A mutant were phosphorylated (phosph.) by the activated or inactive Akt forms in the presence of cold ATP prior to incubation with the corresponding 32P-labeled LUC mRNAs. Coomassie blue staining of the same gel is shown below. mock, mock phosphorylated. (C) Translation of capped (cap+) CAT-PVI IRES-LUC mRNA (0.25 μg) in rabbit reticulocyte lysate in the presence of [35S]methionine and indicated amounts of YB-1 phosphorylated (phosph.) by the inactive or activated Akt forms as in panel A. After phosphorylation, YB-1 proteins were additionally purified using heparin-Sepharose to eliminate any traces of Akt. Following 60-min incubation of the translation reaction mixture, proteins were resolved by SDS-10% PAGE and visualized by autoradiography. The positions of LUC and CAT are shown to the right of the gel, and the positions of molecular mass markers (in kilodaltons) are shown to the left of the gel. (D) The results of panel C as measured from three independent experiments by phosphorimaging software with the means ± standard deviations from the means (error bars) shown. pYB1, phosphorylated YB-1. (E) YB-1 was phosphorylated by the inactive or activated forms of Akt in the presence of [γ-32P]ATP, purified using heparin-Sepharose, and incubated in rabbit reticulocyte cell-free translation system as described above for panel C. IB of the same membrane is shown below. (F) Northern blotting showing degradation kinetics of cap+ CAT-PVI-LUC in the presence of mock-phosphorylated or phosphorylated YB-1 proteins. This experiment was done as described for panel C except for [35S]methionine. Total RNAs were recovered from translation reactions at the times indicated and analyzed by Northern blot hybridization using the 32P-labeled CAT cDNA probe as described earlier (13).
UV cross-linking.
Cap-labeled LUC mRNA was generated from uncapped LUC mRNA in the presence of 0.4 mM S-adenosyl-l-methionine (Sigma) and 100 μCi [α-32P]GTP using vaccinia virus guanylyltransferase (Gibco-BRL). 5′-labeled LUC mRNA was generated in the presence of 50 μCi [γ-32P]ATP using T4 polynucleotide kinase (New England Biolabs). Cross-linking assay using YB-1 proteins (0.5 μg) and the corresponding 32P-labeled LUC mRNAs (∼0.1 μg; 40,000 cpm of each) was done in parallel as described earlier (13), except that YB-1 proteins were phosphorylated by the activated or inactive Akt form in the presence of cold ATP prior to incubation with LUC mRNAs.
Affymetrix microarrays and data analysis.
Isolation of RNAs bound to YB-1 was done essentially as described previously (43). Briefly, IPs of HA-YB-1-RNA complexes from K-Ras-NIH 3T3 cells ectopically expressing HA-YB-1 were performed in lysis buffer containing 250 mM KCl and 20 units of RNasin (Promega) using the anti-HA antibody immobilized on protein G-Sepharose beads. To control for nonspecific binding, RNAs from counterpart K-Ras-NIH 3T3 cells transduced with vector alone were isolated in parallel using anti-HA antibody beads. RNAs bound to the beads were phenol/chloroform deproteinized and precipitated with ethanol. Duplicated samples of total RNA or RNAs bound to anti-HA antibody beads (5 μg of each) were then subjected to microarray analysis on MOE-430A MurineGenome chips (containing probe sets for 22,626 murine genes) using the Affymetrix platform (Santa Clara, CA). Synthesis of cDNA, biotin-labeled cRNA, target hybridization, washing, staining, and scanning were performed using standard Affymetrix protocols. The relative levels of mRNA were calculated from the probe-specific hybridization intensities, using the Probe Profiler probe-modeling algorithm (19). This algorithm uses a principal component analysis to generate the weight for each probe in a probe set on the basis of the performance of the probe across all samples in the data set. Expression levels were normalized to ensure an approximately normal distribution of values. Profiles were analyzed for significant differences between samples using Genetrix software (Epicenter Software). The initial analyses were based on assessment of changes in pair-wise comparisons using a change plot for each gene. Genes exceeding a predefined threshold (a minimum expression level of 100 Affymetrix units) were selected as a subset list for further analysis. The specificity of signals generated from the HA-YB-1-bound mRNAs was enhanced by excluding those genes that were nonspecifically bound to anti-HA antibody beads in the absence of HA-YB-1 (see above). Functional annotations for genes in the filtered list were obtained using the Gene Ontology database (http://www.geneontology.org).
Semiquantitative reverse transcription-PCR (RT-PCR) and RNase protection assay.
Reverse transcription was carried out using Superscript II reverse transcriptase (Invitrogen) and total RNA or RNA recovered from YB-1 immunocomplexes (2 μg of each) as templates. All primer sets for PCR shown in Table 1 were designed to span exon boundaries to eliminate the risk of template contamination by genomic DNA. PCR was performed using Taq polymerase (Invitrogen) under the following conditions: 5 μl cDNA; 200 nM primer mix; 100 μM of all four deoxynucleoside triphosphates; 2 mM MgCl2; and 30 cycles, with 1 cycle consisting of 30 s at 94°C, 1 min at 54°C, and 1 min at 72°C. RNAs extracted from the immunocomplexes with YB-1 or from polysomes were also analyzed by RNase protection by using the RiboQuant RNase protection assay kit (BD Biosciences) according to the manufacturer's protocols.
TABLE 1.
Primer sets utilized for semiquantitative RT-PCR
| Primer set | Gene | Primer sequencea | Product size (bp) |
|---|---|---|---|
| 1 | GADD45β | 5′-TCTGGTCGCACGGGAAGGT (F) | 365 |
| 5′-ATGTCATTGTCGCAGCAGAACG (R) | |||
| 2 | cyclin D1 | 5′-GCCGCCATGACTCCCCACGATTTC (F) | 699 |
| 5′-GCACCCCCTGGCTCCCTACTCTCA (R) | |||
| 3 | L32 | 5′-GGCGGAAACCCAGAGGCATTGA (F) | 281 |
| 5′-CCTGGCGTTGGGATTGGTGACTCT (R) | |||
| 4 | c-jun | 5′-ACGCCGCCCCTGTCCCCTATCG (F) | 466 |
| 5′-CGCGCCACGTCCTTCCCACTCC (R) | |||
| 5 | mertk | 5′-TCCCGAAGCCCATGGTGATTTT (F) | 463 |
| 5′-CTGTGGCCGTGGAGAAGGTAGTCG (R) | |||
| 6 | PDGFR-β | 5′-GGCGGCCAGGAATGTGCT (F) | 525 |
| 5′-AGAGGGGATCGGAGGCTGTG (R) | |||
| 7 | TGFβi4 | 5′-CAGTGGCGATGGATCTAGGAGTTTACC (F) | 337 |
| 5′-GCCGGAGGGGAGCCAGTCT (R) | |||
| 8 | VEGF-C | 5′-CTCTCACAAGGCCCCAAACCAGTCACAA (F) | 607 |
| 5′-GCTTCAGTCGATTCGCACACGGTCTTCT (R) |
The forward (F) and reverse (R) primers are shown.
RESULTS
YB-1 phosphorylation is increased in response to certain growth stimuli.
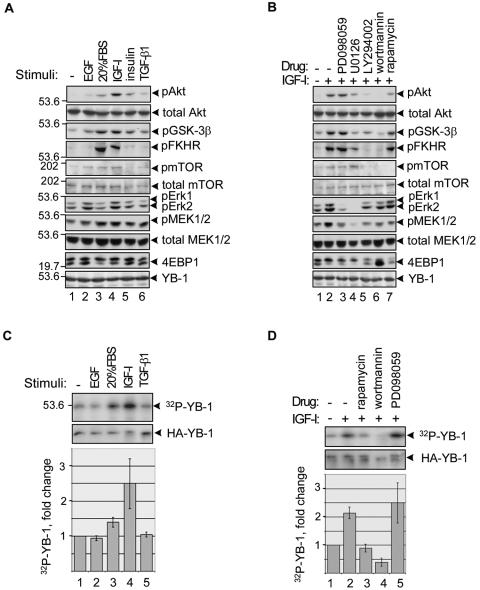
Protein synthesis is up-regulated in response to various signals, including growth factors, hormones, and cytokines, and is accompanied by recruitment of translationally inactive mRNAs into polysomes (18, 31). Since these signals exert their effects in part via phosphorylation of a large number of proteins involved in translational regulation (16), we tested whether YB-1 phosphorylation status might be altered in response to proliferative stimuli. For a model for these studies, we utilized nontransformed NIH 3T3 cells, as they display low basal activity of the major proliferative pathways, such as Ras-mitogen-activated protein kinase (MAPK) and phosphatidylinositol 3-kinase (PI3K)-Akt. To monitor YB-1, these cells were engineered to ectopically express hemagglutinin-tagged YB-1 protein. This allowed us to use rabbit anti-YB-1 antibodies for immunoprecipitation and mouse anti-HA antibodies for immunoblotting or vice versa, thus avoiding overlapping signals from heavy-chain immunoglobulins. To establish the optimal treatment conditions, we monitored activation (phosphorylation) of the key molecules of the Ras-MAPK and PI3K-Akt cascades. As seen in Fig. 1A, in our experimental system the strongest induction of both pathways was achieved by using IGF-I, which stimulated phosphorylation of Akt and its downstream targets glycogen synthase kinase 3β, FKHR, and mTOR as well as phosphorylated MEK1/2 and phosphorylated Erk1/2 (lane 4). To distinguish the input of PI3K-Akt or MAPK pathways, we utilized pharmacological inhibitors of MEK1/2 (PD098059 and UO126) and PI3K (LY294002 and wortmannin). We also tested rapamycin, a specific inhibitor of the mTOR serine/threonine kinase whose activity is regulated via Akt phosphorylation (17). As expected, PD098059 specifically inhibited phosphorylation of MEK1/2 and Erk1/2 to the basal levels, while having no effect on phospho-Akt and its targets (Fig. 1B, lane 3). For unknown reasons, UO126 partially inhibited phosphorylation of certain proteins in the PI3K-Akt pathway (Fig. 1B, lane 4) and thus was excluded from further experiments. We also found that LY294002 slightly reduced phospho-MEK1/2 in this cell line (Fig. 1B, lane 5), so only wortmannin was utilized hereafter. To rigorously test the effects of growth stimulation or pharmacological blockade on YB-1 phosphorylation, NIH 3T3 cells were treated with various growth factors or inhibitors as described above and simultaneously labeled with [32P]orthophosphate. Phosphorylated YB-1 was then immunoprecipitated and detected by autoradiography. As shown in Fig. 1C, phosphorylation of YB-1 was significantly induced by 20% fetal bovine serum and IGF-I but not by epidermal growth factor (EGF) or transforming growth factor β1 (TGF-β1). We also found that pretreatment with wortmannin but not with PD098059 completely abolished YB-1 phosphorylation (Fig. 1D, compare lanes 4 and 5), indicating involvement of the PI3K-Akt pathway. Although the levels of phosphorylated YB-1 were also reduced by rapamycin (Fig. 1D, lane 3), Akt phosphorylation was also slightly inhibited by rapamycin to a degree that parallels the decrease in YB-1 phosphorylation (Fig. 1B, lane 7); thus, the specific contribution of mTOR is unclear. These results suggest that YB-1 activities may depend on growth conditions and are mainly regulated via the PI3K-Akt pathway.
FIG. 1.
YB-1 phosphorylation is increased following activation of the PI3K-Akt pathway. (A and B) NIH 3T3 cells stably expressing HA-YB-1 were grown to ∼80% confluence, serum starved for 24 h, and then either not stimulated (−) or stimulated with 100 ng/ml EGF, 20% dialyzed fetal bovine serum (FBS), 50 ng/ml IGF-I, or 10 ng/ml TGF-β1 for 2 h (A) or treated with 20 ng/ml rapamycin, 0.1 μM wortmannin, 100 μM LY294002, 50 μM U0126, or 100 μM PD098059 for 1 h prior to stimulation with IGF-I (+) (B). The whole-cell extracts were analyzed by IB. The positions of phosphorylated Akt (pAkt), phosphorylated glycogen synthase kinase 3β (pGSK-3β), phosphorylated FKHR (pFKHR), phosphorylated mTOR (pmTOR), phosphorylated Erk1 (pErk1), pErk2, phosphorylated MEK1/2 (pMEK1/2), 4E-BP1, and YB-1 are shown to the right of the gels. The positions of molecular mass markers (in kilodaltons) are shown to the left of the gels. (C and D) Another set of plates was treated as described above for panel A in the presence of [32P]orthophosphate. YB-1 was immunoprecipitated from cytosolic cell extracts with anti-YB-1 antibodies and detected by autoradiography or IB using anti-HA antibodies. The bars in the graphs below the gels indicate the relative amounts of recovered 32P-labeled YB-1 (32P-YB-1) as measured from two independent experiments by phosphorimaging software and normalized to the total amounts of YB-1 in the immunoprecipitates.
YB-1 is phosphorylated by Akt in vitro and in vivo.
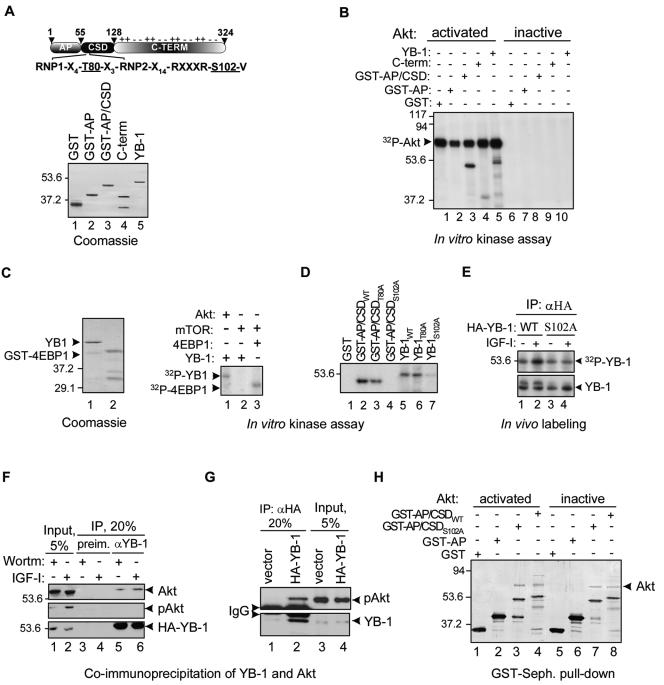
A search for potential phosphorylation motifs in YB-1 using a motif scanner prediction tool (http://scansite.mit.edu) highlighted Thr-80 and Ser-102 within the CSD as potential phosphorylation sites of the Akt kinase (Fig. 2A). To test whether YB-1 could be phosphorylated by Akt, we first performed in vitro kinase assays using recombinant full-length YB-1 or deletion mutants (Fig. 2A) and commercially available inactive and activated forms of Akt. As seen in Fig. 2B, full-length YB-1 and an N-terminal fragment containing both the Ala-Pro-rich domain and the CSD were efficiently phosphorylated by activated Akt (lanes 3 and 5). Phosphorylation was extremely weak or absent when the C-terminal portion or the Ala-Pro domain of YB-1 alone was tested (Fig. 2B, lanes 2 and 4), consistent with the notion that the CSD contains a major Akt phosphorylation site(s). Activated Akt by itself appears to be phosphorylated (Fig. 2B, lanes 1 to 5), probably due to autophosphorylation (44). To test the possibility that mTOR might also be involved in YB-1 phosphorylation, we utilized an mTOR immunoprecipitate as a source of kinase activity to phosphorylate YB-1 or a classical mTOR substrate, 4E-BP1 (5, 15). As seen in Fig. 2C, 4E-BP1 was readily phosphorylated in the presence of mTOR immunoprecipitates (lane 3). In contrast, YB-1 was phosphorylated by activated Akt but not by mTOR, implying that YB-1 is unlikely to be a direct target of mTOR.
FIG. 2.
YB-1 interacts with and is phosphorylated by Akt. (A) The domain organization of YB-1 with the indicated RNP1 and RNP2 consensus motifs and potential phosphorylation sites is shown at the top. The positions of the Ala-Pro-rich domain (AP), CSD, and C-terminal (C-TERM) portion are indicated. The electrical charges of portions of the C-terminal region are shown above the schematic. YB-1 and its derivatives (1 μg of each) used in the kinase assay are shown below. GST-AP/CSD, N-terminal fragment containing both the Ala-Pro-rich domain (AP) and CSD. The positions of molecular mass markers (in kilodaltons) are shown to the left of the gel. (B) In vitro kinase assay using YB-1 derivatives and activated or inactive Akt forms, as indicated (+, present; −, absent). (C) In vitro kinase assay using activated Akt or mTOR immunoprecipitate as a source of kinase activity to phosphorylate YB-1 or GST-tagged 4E-BP1 (right). Coomassie blue staining of recombinant YB-1 and GST-tagged 4E-BP1 (1 μg of each) utilized in the assay is shown to the left. (D) In vitro kinase assay using activated Akt and the recombinant wild-type (WT) or mutant YB-1 proteins. (E) NIH 3T3 cells were transiently transfected with constructs expressing the HA-tagged wild-type YB-1 (WT) or S102A mutant YB-1 protein. Following 24 h posttransfection, cells were treated with IGF-I (+) and simultaneously labeled with [32P]orthophosphate for 2 h. Cytosolic cell extracts were then subjected to IP using anti-HA antibodies (αHA). YB-1 proteins were detected by autoradiography (top) or IB (bottom). (F to H) YB-1 interacts with Akt in vivo and in vitro. MCF-7 (F) or K-Ras-NIH 3T3 (G) cytosolic cell extracts were utilized for IP using anti-HA (αHA) or anti-YB-1 (αYB-1) antibodies, as indicated. preim., preimmune rabbit antibodies; Wortm, wortmannin; pAkt, phosphorylated Akt; IgG, immunoglobulin G. (H) The GST-tagged YB-1 proteins were immobilized on glutathione-Sepharose and incubated with the activated or inactive Akt forms (+). Silver staining of proteins bound to the beads is shown. GST-Seph., GST-Sepharose.
To further address the potential involvement of Akt in YB-1 phosphorylation, predicted phosphorylation sites on YB-1 were mutated to alanine, and the resulting mutant proteins (T80A and S102A) were tested using an in vitro kinase assay. While no effect was seen with the YB-1 T80A mutant, the Ser-102-to-Ala mutation abolished YB-1 phosphorylation (Fig. 2D), thereby confirming Ser-102 as an Akt phosphorylation site in vitro. The wild-type YB-1 protein or YB-1 S102A mutant protein was then transiently expressed in NIH 3T3 cells and then metabolically labeled with [32P]orthophosphate. Stimulation of these cells with IGF-I increased phosphorylation of wild-type YB-1 but had no effect on the YB-1 S102A mutant (Fig. 2E), demonstrating the responsiveness of the Ser-102 site to IGF-I and implicating Akt as a primary candidate kinase responsible for YB-1 phosphorylation at Ser-102. It is noticeable, however, that in contrast to wortmannin (Fig. 1D), the S102 mutation did not block completely YB-1 phosphorylation. This could be due to additional phosphorylation of YB-1 by some other kinases of the PI3K-Akt cascade, such as p70S6.
Next, we tested potential interactions between Akt and YB-1 by screening for Akt—YB-1 complexes within cells. Using anti-YB-1 antibodies, we were able to coimmunoprecipitate Akt from cytosolic extracts of MCF-7 cells regardless of whether they were treated with IGF-I or wortmannin (Fig. 2F). Although phosphorylated Akt was undetectable in immunocomplexes with YB-1 obtained from these cells (Fig. 2F, lanes 5 and 6), this might be due to the lower sensitivity of phospho-Akt versus total Akt antibodies. Indeed, using K-Ras-transformed NIH 3T3 cells, which possess significantly higher levels of phosphorylated Akt, we could detect phospho-Akt in complexes with YB-1 (Fig. 2G). The IP experiments were performed in the presence of RNase A to avoid a possibility of indirect interaction between YB-1 and Akt through the RNA molecule. Direct interaction between Akt and YB-1 was confirmed by pull-down experiments of recombinant Akt with the GST-tagged YB-1 proteins immobilized on glutathione-Sepharose. Notably, Akt was pulled down with both wild-type and S102A mutant YB-1 proteins (Fig. 2H), suggesting that the Akt binding site on YB-1 may differ from the site of phosphorylation. This possibility is currently under investigation. The YB-1-Akt association was also independent of whether activated or inactive Akt forms were used for pull-down experiments (Fig. 2H, compare lanes 3 and 4 to lanes 7 and 8), confirming the results of co-IP experiments. Nevertheless, due to limitations of the in vitro pull-down assays and IP experiments discussed above, none of these experimental approaches may precisely determine the affinity of phosphorylated versus nonphosphorylated Akt forms to YB-1, and therefore, the possibility of differential association of YB-1 with these Akt forms in vivo cannot be excluded. We conclude that Akt binds to and phosphorylates YB-1 in vitro and in vivo in a Ser-102-dependent manner.
Akt is associated with postpolysomal mRNPs.
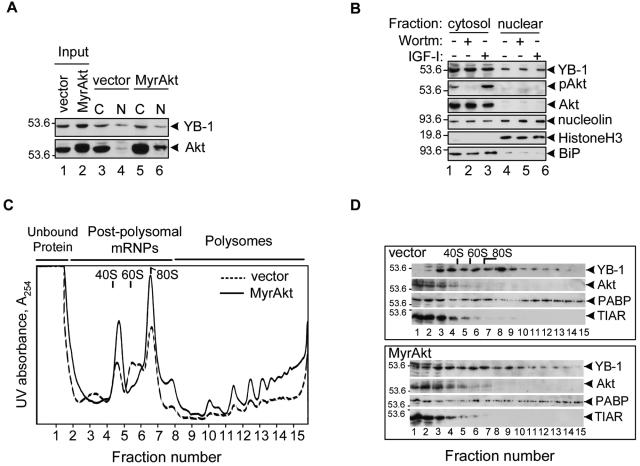
Recently, Sutherland and colleagues reported phosphorylation of YB-1 by Akt and identified Ser-102 as an in vitro Akt phosphorylation site on YB-1 (41). Their results suggest that Akt-mediated YB-1 phosphorylation may be a potential mechanism regulating nucleocytosolic shuttling of YB-1. In other studies, Bader and colleagues reported that YB-1 functions downstream of Akt to selectively suppress PI3K- and Akt-mediated oncogenic transformation; however, they found that YB-1 is transcriptionally down-regulated in chicken embryo fibroblasts expressing a constitutively active myristylated form of Akt (MyrAkt) (1). To clarify the potential biological significance of YB-1 phosphorylation by Akt, we utilized Rat1a embryo fibroblasts ectopically expressing MyrAkt or vector alone. Consistent with previous reports (3, 20, 40), YB-1 exhibited predominantly cytosolic localization (Fig. 3A). Despite high expression levels and elevated nuclear localization of MyrAkt, neither the levels of endogenous YB-1 nor its subcellular localization was changed. Also, we did not observe any difference in YB-1 levels or localization using MCF-7 cells (data not shown) or K-Ras-transformed NIH 3T3 fibroblasts treated with wortmannin or IGF-I (Fig. 3B).
FIG. 3.
YB-1 association with mRNAs but not its cellular localization or protein levels may be affected by activated Akt. (A) Whole-cell extracts from Rat1a cells expressing vector alone or MyrAkt (input) or cytosolic (C) and nuclear (N) fractions were analyzed by IB using anti-Akt or anti-YB-1 antibodies. The positions of YB-1 and Akt are shown to the right of the gel. The positions of molecular mass markers (in kilodaltons) are shown to the left of the gel. (B) Cytosolic and nuclear fractions from K-Ras-NIH 3T3 cells treated with wortmannin (Wortm) (0.1 μM) or IGF-I (50 ng/ml) for 2 h (+) were analyzed by IB. Histone H3 and BiP were chosen as nuclear and cytosolic markers, respectively. Nucleolin is an example of a nucleocytoplasmic shuttling RNA-binding protein with predominantly nuclear localization. pAkt, phosphorylated Akt. (C) Polysomal profiles derived from Rat1a cells expressing vector alone or MyrAkt. Cytosolic cell extracts were layered on 15 to 50% (wt/vol) sucrose gradients in detergent-free lysis buffer and centrifuged at 4°C in a Beckman SW41 rotor for 3 h at 50,000 rpm. Gradients were fractionated from the bottom of the gradient and scanned for absorbance at 254 nm (A254). (D) Following fractionation, 15 fractions were collected, concentrated by precipitation with ice-cold 5% trichloroacetic acid, and analyzed by IB. Note that in contrast to vector alone, in MyrAkt cells YB-1 is detected in the top fractions containing unbound protein (lanes 1 and 2). PABP, poly(A)-binding protein.
Because cytosolic YB-1 is almost entirely bound to mRNAs (10), we next tested whether its association with postpolysomal or polysomal mRNAs might be affected by Akt. To this end, we performed sucrose gradient fractionation to compare the distribution of YB-1 within polysomal profiles obtained from Rat 1a cells expressing vector alone or MyrAkt. In contrast to cells expressing vector alone, cells expressing MyrAkt exhibited higher polysome-to-ribosome ratios (Fig. 3C), probably due to a higher proliferation index and transformed phenotype. Consistent with previous results (10), considerable amounts of YB-1 were detected throughout the polysome region and in the postpolysomal zone in both cell lines (Fig. 3D). However, in MyrAkt-expressing cells, significantly more YB-1 was detected in the top fractions of the gradient, which was predicted to contain unbound proteins (Fig. 3D, lanes 1 and 2). Of further note, the upper band, which is sometimes visible in YB-1 preparations from NIH 3T3 cells (Fig. 1C and D, 2E, and 4E) and likely represents the phosphorylated YB-1 form, is highly enriched in these fractions. Distribution of other mRNA-binding proteins, such as poly(A)-binding protein, which is predominantly associated with polysomal mRNAs (4), and translational repressor TIAR (22), were not changed, indicating that dissociation of a proportion of YB-1 from mRNA complexes in the presence of activated Akt may be functionally important. Interestingly, Akt was also detected in postpolysomal fractions (Fig. 3D, lanes 3 to 7), suggesting that Akt by itself may be associated with postpolysomal mRNPs.
FIG. 4.
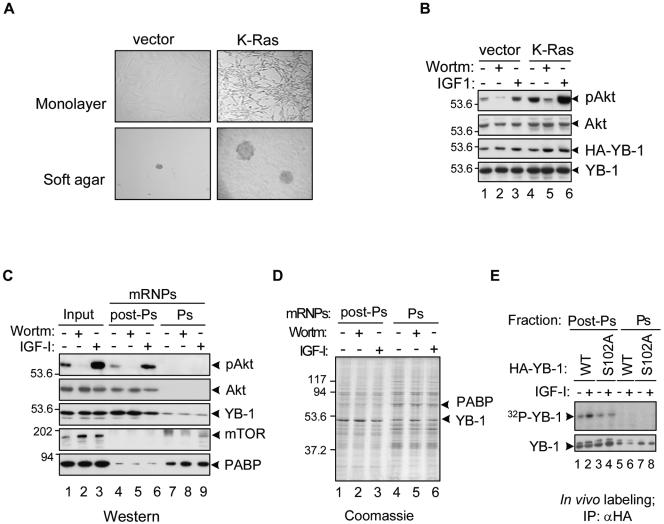
Akt is a component of postpolysomal mRNPs that may mediate YB-1 phosphorylation when it is activated. (A and B) Nontransformed (vector) and K-Ras-transformed NIH 3T3 cells were grown in monolayers or on soft agar plates and analyzed by light microscopy (A). Whole-cell extracts were also analyzed by IB using the corresponding antibodies (B). Wortm, wortmannin; pAkt, phosphorylated Akt. The positions of molecular mass markers (in kilodaltons) are shown to the left of the gel. (C) K-Ras-transformed NIH 3T3 cells were treated with 0.1 μM wortmannin (Wortm) or 50 ng/ml IGF-I for 2 h (+). Five percent of cytosolic cell extracts (input) or 20% of postpolysomal (post-Ps) or polysomal (Ps) mRNP preparations were used for IB. PABP, poly(A)-binding protein. (D) Coomassie blue staining of the mRNP preparations analyzed in panel C. (E) NIH 3T3 cells were transiently transfected with constructs expressing the HA-tagged wild-type (WT) or S102A mutant YB-1 protein. Following 24 h posttransfection, cells were treated with IGF-I (+) and simultaneously labeled with [32P]orthophosphate for 2 h. Postpolysomal or polysomal fractions were then utilized for IP with anti-HA antibodies (αHA). YB-1 proteins were detected by autoradiography (top) or IB using anti-YB-1 antibodies (bottom).
To directly address whether Akt might be present in mRNP complexes, we chose to utilize K-Ras-transformed NIH 3T3 cells in which Akt activity is physiologically increased (23) rather than cells overexpressing an artificial MyrAkt construct. In contrast to cells expressing vector alone, activated K-Ras cells display transformed spindle-shape morphology, form colonies in soft agar (Fig. 4A), and possess substantially elevated levels of phospho-Akt (Fig. 4B). These features make K-Ras-transformed cells a useful model to study the effects of Akt activation on YB-1 functions. Consistent with sucrose gradient fractionation results, substantial amounts of both total and phospho-Akt were present in preparations of postpolysomal mRNPs (Fig. 4C). In contrast, residual amounts of mTOR, if any, were detected exclusively in polysomal mRNP preparations (Fig. 4C). It is important to note that the preparations described in this study represent mRNP and not large protein-protein complexes, since they were purified using oligo(dT) cellulose chromatography, which is based upon adsorption of the mRNA component; treatment with RNase A was shown to abolish YB-1 binding to oligo(dT) cellulose (10). Besides, these mRNP complexes exhibit characteristic protein composition (4, 29), with YB-1 being the only major protein within postpolysomal mRNPs and with both YB-1 and poly(A)-binding protein present in polysomal mRNPs (Fig. 4D). We next tested whether phosphorylation of YB-1 associated with postpolysomal or polysomal mRNPs may be induced by Akt activation. The HA-tagged wild-type or S102A mutant YB-1 proteins were recovered from the corresponding fractions after in vivo orthophosphate labeling of NIH 3T3 cells. As seen in Fig. 4E, following IGF-I treatment, the levels of phosphorylated wild-type YB-1 but not the S102A mutant YB-1 protein were strongly induced in postpolysomal fractions. Together, these data indicate that similar to YB-1, Akt is a component of translationally inactive postpolysomal mRNPs that upon activation may mediate YB-1 phosphorylation.
YB-1 phosphorylation by Akt specifically reduces its interaction with the capped 5′ terminus.
In order to directly determine whether Akt-mediated phosphorylation may affect the RNA-binding ability of YB-1, we performed gel retardation assays using the wild-type or S102A YB-1 proteins phosphorylated by the activated or inactive Akt forms in the presence of cold ATP. As expected, increasing amounts of YB-1 caused a decrease in the mobility of uncapped chloramphenicol acetyltransferase (CAT) mRNA used in this assay (Fig. 5A). Similar results were reported for β-actin (26) or α-globin (36) mRNAs, consistent with the notion that YB-1 binding to mRNA in vitro is strong but rather nonspecific (12, 24). No considerable difference between the phosphorylated and nonphosphorylated forms of wild-type YB-1 or the S102A mutant YB-1 protein was observed (Fig. 5A, compare top and bottom gels), indicating that phosphorylation by Akt does not interfere with the RNA-binding ability of YB-1. We have shown previously that in addition to general RNA-binding activity, YB-1 is also capable of displacing eIF4E from the cap structure, thereby providing selective stabilization of capped mRNAs (13). We thus examined whether interaction of YB-1 with cap might be regulated by Akt phosphorylation. Recent reports suggested utilization of m7GTP-Sepharose as a tool to analyze YB-1 binding to the cap structure (1, 3). In our hands, however, binding of endogenous YB-1 to m7GTP-Sepharose was extremely weak and not comparable to that of eIF4E (V. Evdokimova, unpublished observations). In addition, the recombinant YB-1 protein bound to m7GTP-Sepharose was not competed by the cap analogue, suggesting that association of YB-1 with the mRNA cap structure may occur only within the context of mRNA. We thus took advantage of a previously established system to monitor UV cross-linking of YB-1 to the 32P-cap-labeled mRNA (13). As seen in Fig. 5B, both the wild-type and S102A mutant YB-1 proteins mock phosphorylated by the inactive form of Akt were efficiently cross-linked to the cap-labeled luciferase (LUC) mRNA (lanes 5 and 6) but not to the same 32P-5′-triphosphate-labeled mRNA (lanes 8 and 9 and lanes 11 and 12). Remarkably, phosphorylation of the wild-type but not mutant YB-1 by activated Akt significantly reduced its cross-linking to the cap-labeled mRNA (Fig. 5B, compare lanes 2 and 3), indicating that Akt may modulate the ability of YB-1 to associate directly with or adjacent to the cap mRNA structure.
To further examine the physiological consequences of YB-1 phosphorylation by Akt, we tested the effect of phosphorylated versus nonphosphorylated YB-1 on the translational activity of the capped bicistronic mRNA CAT-PVI IRES-LUC. Increasing concentrations of YB-1 mock phosphorylated by the inactive form of Akt caused a dose-dependent decrease of CAT protein levels, whose expression is cap dependent (Fig. 5C, lanes 2 to 7). However, translation of LUC mRNA, which is driven by poliovirus (PVI) internal ribosome entry site (IRES), remained largely unaffected, suggesting that nonphosphorylated YB-1 primarily inhibits cap-dependent translation. By contrast, YB-1 phosphorylated by activated Akt inhibited LUC but not CAT expression (Fig. 5C, compare lanes 2 to 7 to lanes 9 to 14; see also Fig. 5D). Using YB-1 phosphorylated by activated versus inactive forms of Akt in the presence of [γ-32P]ATP, we confirmed that YB-1 maintained its phosphorylation status over 60 min of incubation in rabbit reticulocyte cell-free translation system (Fig. 5E), and therefore, differential effects of the YB-1 forms on cap-dependent and IRES-dependent translation were indeed attributable to the YB-1 phosphorylation status. To exclude the possibility that the YB-1 forms might alter stability of the reporter mRNA, we analyzed the time course of its degradation in a cell-free translation system. As seen in Fig. 5F, degradation kinetics of the reporter mRNA were similar in the presence of phosphorylated or mock-phosphorylated YB-1. Slightly higher stability of the reporter mRNA in the presence of mock-phosphorylated YB-1 may reflect its higher affinity to the capped 5′ terminus of the mRNA and consequently its ability to prevent 5′→3′ exonucleolytic degradation (13), but it cannot explain differential effects of the YB-1 forms on the translational efficiency of the reporter mRNA. Considering that the general mRNA-binding activity of YB-1 was not significantly changed by phosphorylation (Fig. 5A), the most plausible explanation for these results is that phosphorylation by Akt reduces YB-1 association with the cap structure and leads to its redistribution along the other parts of the mRNA molecule. This may increase concentration of YB-1 on the IRES structure, thereby inhibiting IRES-dependent translation. We therefore argue that phosphorylation by Akt specifically reduces YB-1 ability to bind to the capped 5′ terminus of mRNA and to inhibit cap-dependent translation.
Microarray analysis of mRNAs associated with YB-1.
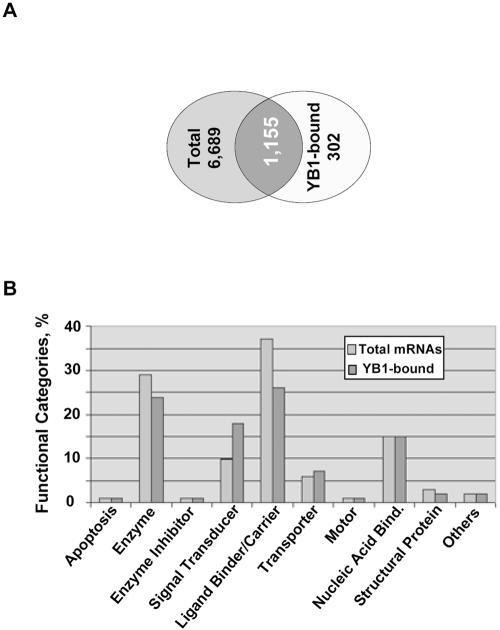
We utilized Affymetrix 22,626-gene murine microarray chips to examine RNAs present in total cell extracts or in immunoprecipitated YB-1-RNP complexes. The latter approach is called ribonomics and is based upon identification of mRNAs bound to and regulated by the protein of interest (43). YB-1-bound RNAs were immunoprecipitated from K-Ras-transformed NIH 3T3 cells ectopically expressing HA-YB-1 using anti-HA antibodies immobilized on protein G-Sepharose beads. To control for nonspecific binding, RNAs from the control cells transduced with vector alone were isolated on anti-HA antibody beads in parallel. Normalized profiles of mRNAs bound to YB-1 were then generated by subtracting the genes that were nonspecifically bound to the beads in the absence of HA-YB-1. We first investigated differences in total cellular mRNA populations and those associated with YB-1. About 40% of all the genes on the array were hybridized using the inputted total cellular RNA (Fig. 6A). Approximately 18% of the total messages represented on the arrays were identified in YB-1 complexes (Fig. 6A). Among the YB-1-associated messages, over 300 transcripts were not detectable in total RNA preparations (for complete lists of identified mRNAs, see ftp://genomecore-chla.usc.edu/public/AKT-YB1/), suggesting that specific mRNA subsets may be selectively enriched within YB-1 complexes. Specifically, low-abundance messages encoding cell growth and maintenance molecules constituted one-third of the unique YB-1-bound transcripts (data not shown). Additional evidence for YB-1 mRNA binding selectivity was obtained by analyzing functional distribution of total and YB-1-bound transcripts. While most of the functional gene categories were quite similarly represented between the two profiles, we observed a much higher proportion of signal transduction-related probe sets among the YB-1-bound fraction than in total RNA profiles (Fig. 6B). Furthermore, large sets of transcripts identified in YB-1 complexes appear to code for proteins that belong to the same regulatory pathway. These include 7 members of the ATP-binding cassette transporter family that are known to confer multidrug resistance in cancer cells as well as 5 components of the insulin signaling pathway, 13 members of mitogen-activated protein kinase cascade, 7 cyclins and cyclin-dependent kinases, a number of proteins involved in translational control, etc. (Table 2). Besides, more than 200 transcripts found in the complex with YB-1 encode growth factors, receptors, and associated proteins, including fibroblast growth factor 9, fibroblast growth factor receptor 5, granulocyte-macrophage colony-stimulating factor (GM-CSF), IGF-I, IGF-binding proteins 4 and 5, platelet-derived growth factor 4 (PDGF-4), PDGF receptor α (PDGFR-α) and β, TGF-α and -β, TGFβi4, and vascular endothelial growth factor B (VEGF-B) and C. These findings are consistent with other studies demonstrating specific binding of YB-1 to certain cytokine and growth factor mRNAs including IL-2, GM-CSF, and VEGF (6-8). Although the above interactions still need to be confirmed, these findings suggest a possibility that translational activity and/or stability of many transcripts is regulated by a common mechanism.
FIG. 6.
Functional distribution of total and YB-1-bound transcripts derived from K-Ras-transformed NIH 3T3 cells. (A) Venn diagram showing overlap between total and YB-1-bound transcripts. (B) The major functional categories represented in total and YB-1-bound mRNA populations. Bind., binding.
TABLE 2.
List of representative YB-1-bound transcripts associated with cell growth and stress responsesa
| Accession no. | Gene name | Accession no. | Gene name | |
|---|---|---|---|---|
| BC026496.1 | ATP-binding cassette, subfamily A (MDR), member 8A | AF160966.1 | Heat shock transcription factor 4 | |
| AK014703.1 | Insulin-degrading enzyme | |||
| NM_023732.1 | ATP-binding cassette, subfamily B (MDR), member 6 | BC012409.1 | Insulin-like growth factor 1 | |
| BB787243 | Insulin-like growth factor-binding protein 4 | |||
| AW537380 | ATP-binding cassette, subfamily B (MDR), member 7 | NM_027397.1 | Insulin-related protein 2 | |
| NM_009951.1 | Insulin-like growth factor 2, binding protein 1 | |||
| NM_021022.2 | ATP-binding cassette, subfamily B (MDR), member 11 | BC002081.1 | Jun oncogene, c-jun | |
| NM_015771.1 | Large tumor suppressor 2 | |||
| BG071908 | ATP-binding cassette, subfamily C (MRP), member 1 | NM_008520.1 | Latent transforming growth factor beta-binding protein 3 | |
| NM_013806.1 | ATP-binding cassette, subfamily C (MRP), member 2 | BB554226 | Latent transforming growth factor beta-binding protein 4 | |
| BC011273.1 | ATP-binding cassette, subfamily D (ALD), member 1 | NM_008606.1 | Matrix metalloproteinase 11 | |
| AV296905 | Metastasis-associated 1-like 1 | |||
| BM120925 | BCL2-like 11 (apoptosis facilitator) | NM_054082.1 | Metastasis-associated 3 | |
| NM_007545.1 | BH3-interacting (BCL2 family) domain, apoptosis agonist | BC024514.1 | Mitogen-activated protein kinase 9 | |
| BC021640.1 | Mitogen-activated protein kinase 12 | |||
| BC026138.1 | Calpain 1 | BC012235.1 | Mitogen-activated protein kinase 14 | |
| AK013497.1 | Calpain 10 | NM_011946.1 | Mitogen-activated protein kinase kinase kinase 2 | |
| BB752393 | Caspase 7 | U11548.1 | Mitogen-activated protein kinase kinase kinase 4 | |
| BB815299 | Caspase 9 | BB734681 | Mitogen-activated protein kinase kinase kinase 5 | |
| NM_008587.1 | c-mer proto-oncogene, mertk | AW547374 | Mitogen-activated protein kinase kinase kinase 7 | |
| M21149.1 | Colony-stimulating factor 1 (macrophage) | AF262046.1 | Mitogen-activated protein kinase 8-interacting protein 3 | |
| NM_007631.1 | Cyclin D1 | |||
| NM_007633.1 | Cyclin E1 | M57683.1 | Platelet-derived growth factor receptor alpha (PDGFRα) | |
| BC003958.1 | Cyclin G-associated kinase | |||
| BI112766 | Cyclin L2 | AA499047 | Platelet-derived growth factor receptor beta (PDGFRβ) | |
| BG070401 | Cyclin M2 | |||
| NM_009833.1 | Cyclin T1 | NM_019932.1 | Platelet factor 4 | |
| BC025046.1 | Cyclin-dependent kinase 8 | BE948730 | Prostaglandin E receptor 1 (subtype EP1) | |
| AY027937.1 | Damage-specific DNA-binding protein 2 | NM_009025.1 | RAS p21 protein activator 3 | |
| AF370126.1 | Delta/notch-like EGF-related receptor | BG868120 | Rous sarcoma virus oncogene | |
| BC005522.1 | Down-regulated in metastasis | BI662291 | Serum response factor | |
| AK017841.1 | E2F transcription factor 1 | NM_022332.1 | Suppression of tumorigenicity 7 | |
| X06746.1 | Early growth response 2 | NM_009423.1 | TNF receptor-associated factor 4b | |
| AK013765.1 | Endothelial cell growth factor 1 (platelet-derived) | NM_011633.1 | TNF receptor-associated factor 5 | |
| NM_007906.1 | eIF1 alpha 2 | AK015764.1 | Transcriptional repressor NAC 1 | |
| NM_010122.1 | eIF2B | BB277931 | Transducer of ERBB2, 2 | |
| AY033898.1 | eIF2 alpha kinase 1 | M92420.1 | Transforming growth factor alpha (TGF-α) | |
| NM_010123.1 | eIF3 | NM_009369.1 | Transforming growth factor beta induced | |
| AK013033.1 | eIF4E-binding protein 1 | AU016382 | Transforming growth factor beta induced 4 (TGFβi4) | |
| AK012082.1 | eIF4E nuclear import factor 1 | |||
| NM_007986.1 | Fibroblast activation protein | NM_009310.1 | Tumor-associated antigen 1 | |
| NM_013518.1 | Fibroblast growth factor 9 | NM_054042.1 | Tumor endothelial marker 1 precursor | |
| AF127140.1 | Fibroblast growth factor receptor 4 | BB233088 | Tumor necrosis factor alpha induced | |
| AK010420.1 | Growth arrest and DNA damage inducible 45 beta (GADD45β) | NM_023680.1 | Tumor necrosis factor receptor superfamily, member 22 | |
| M33324.1 | Growth hormone receptor | U48800.1 | Vascular endothelial growth factor B (VEGF-B) | |
| BC013716.1 | Heat shock factor 1 | NM_009506.1 | Vascular endothelial growth factor C (VEGF-C) |
The full list of the genes is available at ftp://genomecore-chla.usc.edu/public/AKT-YB1/.
TNF, tumor necrosis factor.
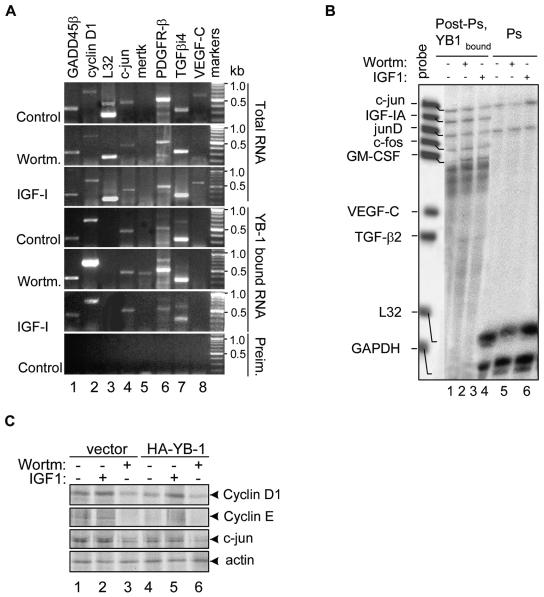
To verify the identities of selected candidate mRNAs in YB-1 complexes and to delineate potential effects of PI3K-Akt signaling, we performed semiquantitative RT-PCR analysis of RNA preparations derived from K-Ras-NIH 3T3 cells challenged with IGF-I or wortmannin. Having established that phosphorylation by Akt may reduce YB-1 binding to the capped 5′ terminus of the mRNA and its inhibitory effect on cap-dependent translation, we choose to analyze the postpolysomal YB-1-bound mRNAs in order to determine whether association of translationally inactive transcripts with YB-1 might be affected by Akt activation. RNAs were thus recovered directly from postpolysomal fractions or after IP with anti-YB-1 or preimmune rabbit antibodies. For these analyses, we chose several representative growth- and stress-related mRNAs, such as GADD45β, cyclin D1, c-jun, mertk, PDGFR-β, TGFβi4, and VEGF-C (Table 2). Since none of the transcripts encoding ribosomal proteins was found in the complex with YB-1, we utilized L32 mRNA to control for unspecific binding. Not surprisingly considering the short treatment times and the semiquantitative nature of RT-PCR, the levels of each tested transcript recovered from total postpolysomal RNA populations were not significantly altered in response to wortmannin or IGF-I (Fig. 7A, three top gels). However, binding of certain transcripts to YB-1 was dramatically changed by the above treatments. For example, mertk mRNA, which was undetectable in total RNA preparations, was highly enriched in the pool of YB-1-bound transcripts after exposure to wortmannin (Fig. 7A, lane 5). Similarly, levels of cyclin D1 and PDGFR-β mRNAs were markedly elevated in YB-1 complexes compared to total postpolysomal RNA preparations, especially in response to wortmannin (Fig. 7A, lanes 2 and 6). Stimulation with IGF-I reduced association of YB-1 with many transcripts (Fig. 7A). Of note, L32 mRNA was readily detected in the preparation of total postpolysomal RNAs but not in the pool of YB-1-bound transcripts (Fig. 7A, lane 3). For unclear reasons, we failed to detect VEGF-C mRNA in the complex with YB-1. None of the mRNAs tested was detected when preimmune antibodies were used as bait for RNA isolation (Fig. 7A, bottom gel), further indicating the selectivity of YB-1 binding to certain mRNA species.
FIG. 7.
Binding of different mRNAs to YB-1 is dynamic and depends on growth conditions. (A) Semiquantitative RT-PCR analysis of selected candidate mRNAs derived from postpolysomal RNA preparations, total or YB-1 bound RNAs. K-Ras-NIH 3T3 cells treated with 0.1 μM wortmannin (Wortm.) or 50 ng/ml IGF-I for 2 h were utilized in this assay. Similar results were produced in three independent experiments using independently isolated YB-1-bound and total postpolysomal RNAs; the representative data are shown. The positions of molecular size markers (in kilobases) are shown to the right of the gels. GADD45β, growth arrest- and DNA damage-induced 45β; Preim., preimmune rabbit antibodies. (B) Multiprobe RNase protection assay of mRNAs recovered from postpolysomal (Post-Ps.) complexes with YB-1 or from polysomal (Ps) fractions of K-Ras-transformed cells treated with wortmannin (Wortm) or IGF-I (+), as described above for panel A. A customized growth factor template set was used in this assay. Note that undigested probe derived from the plasmid template is always larger than the actual RNase-digested product. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (C) Effect of YB-1 overexpression on translation efficiencies of certain candidate mRNAs. K-Ras-NIH 3T3 cells expressing vector alone or HA-YB-1 were treated with IGF-I or wortmannin as described above for panel A and metabolically labeled with [35S]methionine for the last 60 min. Cytosolic cell extracts were utilized for IPs using mixtures of cyclin D1 and cyclin E antibodies or c-jun and actin antibodies immobilized on protein A/G-Sepharose beads to ensure equal recovery of the proteins from the immunoprecipitates. Immunoprecipitated proteins were then detected by SDS-10% PAGE and autoradiography.
To simultaneously screen for changes in the mRNA profiles in response to IGF-I or wortmannin, YB-1-bound postpolysomal mRNAs or mRNAs derived from polysomal fractions were analyzed by multiprobe RNase protection assay using template sets specific for a panel of growth-related mRNA targets (Fig. 7B). We also employed housekeeping mRNAs, L32 and GAPDH, as controls. In agreement with RT-PCR data, L32 mRNA was not detected in the complex with YB-1; however, both L32 and GAPDH mRNAs were highly enriched in polysomal fractions (Fig. 7B, lanes 4 to 6). In contrast, several growth-related mRNAs, such as IGF-IA, c-fos, and GM-CSF, were detected in association with YB-1 in the fraction of postpolysomal mRNPs but not in polysomes (Fig. 7B, lanes 1 to 3). Others, such as c-jun and junD, were present in both RNA populations. Treatment with IGF-I diminished the association of c-jun mRNA with YB-1 and increased its levels in polysomes (Fig. 7B, compare lanes 2 and 3 to lanes 5 and 6). The levels of IGF-IA, junD, and c-fos mRNAs associated with YB-1 were also somewhat decreased in response to IGF-I; however, these messages were not visible in polysomal fractions, possibly due to sensitivity limitations of this approach. Overall, these data indicate that many growth-related mRNAs are kept silent in the complex with YB-1, and at least some of them may be released for translation upon induction of PI3K-Akt signaling.
In order to directly determine whether YB-1 phosphorylation may enhance the translation efficiencies of the target mRNAs, we performed metabolic [35S]methionine labeling of K-Ras-NIH 3T3 cells ectopically expressing YB-1 or vector alone. Cells were left untreated or treated with IGF-I or wortmannin for 2 h, followed by IP of the candidate proteins. Due to the short treatment times, this strategy allowed us to monitor the effects of the above compounds on translation, excluding their potential influence on transcription of the corresponding genes. As seen in Fig. 7C, the expression levels of cyclin D1, cyclin E, and c-jun were reduced in YB-1-overexpressing cells than in cells expressing vector alone (compare lanes 1 and 4). However, stimulation with IGF-I rescued cyclin D1 protein expression, in agreement with the notion that activation of Akt signaling releases YB-1-bound cyclin D1 transcripts for translation. The ability of IGF-I to rescue the other transcripts was less evident, suggesting additional complexity for this process. The effect was specific for the YB-1-bound mRNA targets, since the expression levels of β-actin were unaffected in either cell line under the same conditions. Therefore, an increase in YB-1 levels results in translational repression of specific mRNA subsets, whereas activation of Akt signaling releases them for translation. These data are consistent with the idea that translational activation of certain mRNAs is a direct consequence of YB-1 phosphorylation by Akt.
DISCUSSION
This study provides compelling evidence that YB-1 associates with the serine/threonine kinase Akt in vitro and in vivo. We found that Akt mediates phosphorylation of YB-1 in vitro at Ser-102 and that the phosphorylation of the wild-type YB-1 but not a Ser-102-to-Ala mutant YB-1 protein was induced by IGF-I in quiescent NIH 3T3 cells. Akt-mediated YB-1 phosphorylation did not alter the general RNA-binding ability of YB-1 but did reduce its cap-binding capability (Fig. 5A and B). Consistent with this notion, phosphorylated YB-1 was less capable of repressing cap-dependent translation but exhibited a stronger inhibitory effect towards IRES-dependent translation (Fig. 5C). The total levels of YB-1 within inactive mRNPs did not seem to be changed significantly, as we did not observe a massive overall dissociation of YB-1 from inactive mRNPs in response to IGF-I stimulation or oncogenic transformation with K-Ras or MyrAkt. Akt phosphorylation likely redirects the repressor activity of YB-1, thereby altering mRNA subsets bound to YB-1. Indeed, YB-1 interactions with the majority of mRNAs tested in this study were dynamic and dependent on PI3K-Akt signaling. The levels of many transcripts, including cyclin D1, c-jun, mertk, and PDGFR-β mRNAs, were elevated in the postpolysomal complexes with YB-1 from wortmannin-treated cells and reduced following IGF-I stimulation (Fig. 7A and B), supporting the idea that phosphorylation of YB-1 is sufficient to activate translation of stored mRNA species. These observations correlated well with a reduction of the expression of the corresponding proteins in YB-1-overexpressing cells, which was reversed by IGF-I stimulation (Fig. 7C).
A role for Akt and Ras signaling in oncogenic activation via differential recruitment of preexisting mRNAs to polysomes has recently emerged as an important mechanism for regulating expression of growth-related and oncogenic proteins (31, 32). This is also substantiated by our finding that Akt by itself was detected within translationally inactive mRNPs. Moreover, a total of 93 transcripts identified in the YB-1 complexes in our study (Table 3) were among the 343 messages previously suggested to be kept silent and rapidly activated by Ras/Akt signaling (32).
TABLE 3.
List of common genes found among the YB-1-bound transcripts and the transcripts that were previously shown to be recruited to polysomes in response to activation of Ras and Akt signalinga
| Accession no. | Gene name |
|---|---|
| AF332087.1 | Activin A receptor, type 1 |
| BG230208 | Adaptor protein complex AP-1, gamma 1 subunit |
| NM_013799.1 | Arginine-tRNA-protein transferase 1 |
| BC011273.1 | ATP-binding cassette, subfamily D (ALD), member 1 |
| AK012821.1 | Branched-chain aminotransferase 1, cytosolic |
| NM_013468.1 | Cardiac responsive adriamycin protein |
| BB815299 | Caspase 9 |
| AW539140 | Chromodomain helicase DNA-binding protein 1 |
| AF177196.1 | Deiodinase, iodothyronine, type II |
| BE630687 | Deoxycytidine kinase |
| BB165431 | DNA methyltransferase (cytosine-5) 1 |
| BG066325 | Dynamin 2 |
| AK006149.1 | Endosulfine alpha |
| AF127033.1 | Fatty acid synthase |
| AF007248.1 | Fibrillin 1 |
| BF228318 | Fibulin 2 |
| BB759833 | forkhead box C1 |
| AK010239.1 | frizzled homolog 7 (Drosophila melanogaster) |
| AF043219.1 | General transcription factor II, I |
| L23312.1 | Huntington disease gene homolog |
| AF013486.1 | Interferon (alpha and beta) receptor 2 |
| NM_008446.1 | Kinesin family member 4 |
| NM_008453.1 | Kruppel-like factor 3 (basic) |
| BB201735 | LIM and SH3 protein 1 |
| D00622.1 | Low-density lipoprotein receptor-related protein |
| NM_008569.1 | Meiotic check point regulator |
| C80350 | Minichromosome maintenance deficient (Saccharomyces cerevisiae) |
| U11548.1 | Mitogen-activated protein kinase kinase kinase 4 |
| NM_018871.1 | 3-Monooxgenase/tryptophan activation protein |
| X15052.1 | Neural cell adhesion molecule 1 |
| AI987929 | N-myc downstream regulated 1 |
| NM_008681.1 | N-myc downstream regulated-like |
| BF785921 | Nuclear factor I/C |
| NM_008173.1 | Nuclear receptor subfamily 3, group C, member 1 |
| NM_016861.1 | PDZ and LIM domain 1 (elfin) |
| NM_008903.1 | Phosphatidic acid phosphatase 2a |
| U52193.1 | Phosphatidylinositol 3-kinase, alpha polypeptide |
| U01023.1 | Phosphoribosylglycinamide formyltransferase |
| NM_009088.1 | RNA polymerase 1 to 4 |
| BF468377 | SAC2 (supressor of actin mutations 2, homolog)-like |
| BM236195 | Selectin, endothelial cell, ligand |
| AK012411.1 | Serine (or cysteine) proteinase inhibitor (clade F), member 1 |
| NM_009195.1 | Solute carrier family 12, member 4 |
| NM_011368.2 | Src homology 2 domain-containing transforming protein C1 |
| NM_011638.1 | Transferrin receptor |
| NM_009369.1 | Transforming growth factor beta induced |
| BF139489 | Tumor differentially expressed 1 |
| AI788797 | Utrophin |
This list was generated by the alignment with the published gene set of 343 mRNAs (see supplemental data for reference 31 [http://www.molecule.org/cgi/content/full/12/4/889/DC1]).
Although the exact requirements for mRNA recognition remain to be defined, it is clear that YB-1 exhibits selectivity towards specific mRNA subsets. A significant number of mRNAs detected in YB-1 complexes are low-abundance messages coding for regulatory proteins involved in cell growth and survival and stress responses. Of further note, we did not observe abundant housekeeping transcripts, such as those coding for L32 or glyceraldehyde-3-phosphate dehydrogenase in the pool of YB-1-bound mRNA. This is despite the fact that L32 mRNA belongs to a family of mRNAs containing 5′-end oligopyrimidine tract and is known to be redistributed from untranslated postpolysomal particles into polysomes after mitogenic activation of quiescent cells (21). One possibility is that YB-1 may directly recognize highly structured GC-rich 5′ untranslated regions characteristic of many growth-related mRNAs (25). Another possibility is that YB-1 simply binds to messages that are physically excluded from translation due to their inability to compete for translation initiation factors, especially eIF4E, which is the least abundant and thus, the rate-limiting factor in the binding of ribosomes to the mRNA (38). The latter possibility is supported by our previous findings that YB-1 binding to the cap structure is alleviated by sequestration of eIF4E with 4E-BP1 or by adding cap analogue (13). We therefore propose that certain messages, whose translation is associated with a higher requirement for cap-dependent initiation, are being bound to YB-1 and kept in a silent state. Storage of these messages can be especially important considering their low abundance, short half-life when being translated, and key role in regulating cell growth. This pool of stored mRNAs may be utilized in response to growth stimuli, thereby allowing cells to respond rapidly to changing extracellular environments. It should also be noted that some of these transcripts, such as c-jun and junD, were also present in polysomal fractions (Fig. 7B), indicating that YB-1 may maintain a “backup” pool of messages which can be utilized by translational machinery as the pool of their polysomal counterparts declines.
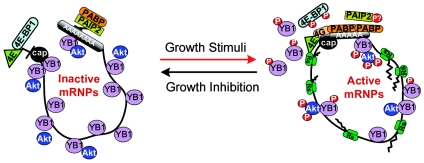
The above data support a model whereby phosphorylation by Akt renders YB-1 less competitive for binding to the capped 5′ end of mRNAs, such that it could be more easily displaced by the translation initiation complex driven by eIF4E (for a model, see Fig. 8). In this manner, activation of mRNA species silenced by YB-1 may require both YB-1 phosphorylation to reduce its affinity to the cap and sufficient availability of the active eIF4E to subsequently recruit the YB-1-bound transcripts to polysomes. This mechanism ensures that the mRNA is released to polysomes only under favorable translational conditions. Otherwise, it is kept silent and stable in the complex with YB-1. This would necessitate a strongly coordinated process that is dependent on a balance between the YB-1 level, Akt activity, and availability of eIF4E and perhaps other components of the translational machinery. Elevation of YB-1 above physiological levels therefore might be predicted to block PI3K-Akt signaling due to (i) exceeding the ability of Akt to phosphorylate the excess of YB-1 and/or Akt inactivation by sequestering it into nonfunctional complexes with YB-1, and (ii) silencing of growth-related messages whose translational activity is poor and highly dependent on eIF4E. Indeed, it has been demonstrated that YB-1 overexpression specifically overcomes PI3K- and Akt-induced oncogenesis by inhibiting protein synthesis (1, 2). Although up-regulation of YB-1 has been detected in many human cancers (for a review, see reference 24), this may represent a preventive cellular mechanism directed to overcome neoplastic growth. Deregulation of this mechanism by hyperactivation of Akt, eIF4E, or other means may contribute to uncontrolled proliferation in cancer cells. This possibility awaits further experimental confirmation.
FIG. 8.
Model for translational regulation via Akt-mediated YB-1 phosphorylation. Under normal growth conditions, competition between mRNAs for available translational components excludes many of them from translation and causes their accumulation in the pool of inactive postpolysomal mRNPs. Growth-related mRNAs are primarily affected, owing to their highly structured 5′ untranslated regions and the small amount of active eIF4E in the cell. A great number of these messages are kept silent and stable in the complex with YB-1, which supposedly functions as a gatekeeper blocking access of eIF4E and degradation enzymes to the mRNA species (13). Upon activation, Akt phosphorylates YB-1 and reduces its affinity to the capped 5′ terminus of mRNA. Akt signaling also leads to phosphorylation of 4E-BP1-inhibitory protein, thereby increasing levels of active eIF4E. In turn, eIF4E is capable of displacing phosphorylated YB-1 and releasing the YB-1-inhibited mRNAs to polysomes. PABP, poly(A)-binding protein; PAIP2, PABP-interacting protein 2; P, phosphate group; Rs, ribosome.
Acknowledgments
We thank Nissim Hay (University of Illinois at Chicago) for Rat1a cells expressing MyrAkt, Betty Shaub and Xuan Chen (Children's Hospital, Los Angeles) for generation of Affymetrix data, and Nataliya Melnyk and Michelle Pollard (University of British Columbia) for help with generation of cell lines and excellent technical assistance.
This study was supported by the National Cancer Institute of Canada and by a Translational Research Grant from the Children's Oncology Group (to P.H.B.S.). This work was also funded by the Johal Program in Pediatric Oncology Basic and Translational Research at the Child and Family Research Institute (to P.H.B.S. and V.E.).
REFERENCES
- 1.Bader, A. G., K. A. Felts, N. Jiang, H. W. Chang, and P. K. Vogt. 2003. Y box-binding protein 1 induces resistance to oncogenic transformation by the phosphatidylinositol 3-kinase pathway. Proc. Natl. Acad. Sci. USA 100:12384-12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bader, A. G., and P. K. Vogt. 2004. An essential role for protein synthesis in oncogenic cellular transformation. Oncogene 23:3145-3150. [DOI] [PubMed] [Google Scholar]
- 3.Bader, A. G., and P. K. Vogt. 2005. Inhibition of protein synthesis by Y box-binding protein 1 blocks oncogenic cell transformation. Mol. Cell. Biol. 25:2095-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blobel, G. 1973. A protein of molecular weight 78,000 bound to the polyadenylate region of eukaryotic messenger RNAs. Proc. Natl. Acad. Sci. USA 70:924-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunn, G. J., P. Fadden, T. A. Haystead, and J. C. J. Lawrence. 1997. The mammalian target of rapamycin phosphorylates sites having a (Ser/Thr)-Pro motif and is activated by antibodies to a region near its COOH terminus. J. Biol. Chem. 272:32547-32550. [DOI] [PubMed] [Google Scholar]
- 6.Capowski, E. E., S. Esnault, S. Bhattacharya, and J. S. Malter. 2001. Y box-binding factor promotes eosinophil survival by stabilizing granulocyte-macrophage colony-stimulating factor mRNA. J. Immunol. 167:5970-5976. [DOI] [PubMed] [Google Scholar]
- 7.Chen, C.-Y., R. Gherzi, J. S. Andersen, G. Gaietta, K. Jurchott, H.-D. Royer, M. Mann, and M. Karin. 2000. Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes Dev. 14:1236-1248. [PMC free article] [PubMed] [Google Scholar]
- 8.Coles, L. S., M. A. Bartley, A. Bert, J. Hunter, S. Polyak, P. Diamond, M. A. Vadas, and G. J. Goodall. 2004. A multi-protein complex containing cold shock domain (Y-box) and polypyrimidine tract binding proteins forms on the vascular endothelial growth factor mRNA. Potential role in mRNA stabilization. Eur. J. Biochem. 271:648-660. [DOI] [PubMed] [Google Scholar]
- 9.Darnbrough, C. H., and P. J. Ford. 1981. Identification in Xenopus laevis of a class of oocyte-specific proteins bound to messenger RNA. Eur. J. Biochem. 113:415-424. [DOI] [PubMed] [Google Scholar]
- 10.Davydova, E. K., V. M. Evdokimova, L. P. Ovchinnikov, and J. W. B. Hershey. 1997. Overexpression in COS cells of p50, the major core protein associated with mRNA, results in translation inhibition. Nucleic Acids Res. 25:2911-2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Didier, D. K., J. Schiffenbauer, S. L. Woulfe, M. Zacheis, and B. D. Schwartz. 1988. Characterization of the cDNA encoding a protein binding to the major histocompatibility complex class II Y box. Proc. Natl. Acad. Sci. USA 85:7322-7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evdokimova, V. M., and L. P. Ovchinnikov. 1999. Translational regulation by Y-box transcription factor: involvement of the major mRNA-associated protein, p50. Int. J. Biochem. Cell. Biol. 31:139-149. [DOI] [PubMed] [Google Scholar]
- 13.Evdokimova, V. M., P. Ruzanov, H. Imataka, B. Raught, Y. Svitkin, L. P. Ovchinnikov, and N. Sonenberg. 2001. The major mRNA-associated protein YB-1 is a potent 5′ cap-dependent mRNA stabilizer. EMBO J. 20:5491-5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evdokimova, V. M., A. S. Sitikov, P. N. Simonenko, O. A. Lazarev, K. S. Vasilenko, V. A. Ustinov, C.-L. Wei, J. W. B. Hershey, and L. P. Ovchinnikov. 1995. The major protein of messenger ribonucleoprotein particles in somatic cells is a member of the Y-box binding transcription factor family. J. Biol. Chem. 270:3186-3192. [DOI] [PubMed] [Google Scholar]
- 15.Gingras, A.-C., S. P. Gygi, B. Raught, R. D. Polakiewicz, R. T. Abraham, M. F. Hoekstra, R. Aebersold, and N. Sonenberg. 1999. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 13:1422-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gingras, A. C., B. Raught, and N. Sonenberg. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913-963. [DOI] [PubMed] [Google Scholar]
- 17.Hay, N., and N. Sonenberg. 2004. Upstream and downstream of mTOR. Genes Dev. 18:1926-1945. [DOI] [PubMed] [Google Scholar]
- 18.Holland, E. C. 2004. Regulation of translation and cancer. Cell Cycle 3:452-455. [PubMed] [Google Scholar]
- 19.James, A. C., J. G. Veitch, A. R. Zareh, and T. Triche. 2004. Sensitivity and specificity of five abundance estimators for high-density oligonucleotide microarrays. Bioinformatics 20:1060-1065. [DOI] [PubMed] [Google Scholar]
- 20.Jurchott, K., S. Bergmann, U. Stein, W. Walther, M. Janz, I. Manni, G. Piaggio, E. Fietze, M. Dietel, and H. D. Royer. 2003. YB-1 as a cell cycle-regulated transcription factor facilitating cyclin A and cyclin B1 gene expression. J. Biol. Chem. 278:27988-27996. [DOI] [PubMed] [Google Scholar]
- 21.Kaspar, R. L., T. Kakegawa, H. Cranston, D. R. Morris, and M. W. White. 1992. A regulatory cis element and a specific binding factor involved in the mitogenic control of murine ribosomal protein L32 translation. J. Biol. Chem. 267:508-514. [PubMed] [Google Scholar]
- 22.Kedersha, N. L., M. Gupta, W. Li, I. Miller, and P. Anderson. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 147:1431-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khwaja, A., P. Rodriguez-Viciana, S. Wennstrom, P. H. Warne, and J. Downward. 1997. Matrix adhesion and Ras transformation both activate a phosphoinositide 3-OH kinase and protein kinase B/Akt cellular survival pathway. EMBO J. 16:2783-2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohno, K., H. Izumi, T. Uchiumi, M. Ashizuka, and M. Kuwano. 2003. The pleiotropic functions of the Y-box-binding protein, YB-1. BioEssays 25:691-698. [DOI] [PubMed] [Google Scholar]
- 25.Kozak, M. 1991. An analysis of vertebrate mRNA sequences: intimations of translational control. J. Cell Biol. 115:887-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsumoto, K., K. J. Tanaka, and M. Tsujimoto. 2005. An acidic protein, YBAP1, mediates the release of YB-1 from mRNA and relieves the translational repression activity of YB-1. Mol. Cell. Biol. 25:1779-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsumoto, K., and A. Wolffe. 1998. Gene regulation by Y-box proteins: coupling control of transcription and translation. Trends Cell Biol. 8:318-323. [DOI] [PubMed] [Google Scholar]
- 28.Minich, W. B., N. L. Korneyeva, and L. P. Ovchinnikov. 1989. Translational active mRNPs from rabbit reticulocytes are qualitatively different from free mRNA in their translatability in cell-free system. FEBS Lett. 257:257-259. [DOI] [PubMed] [Google Scholar]
- 29.Minich, W. B., and L. P. Ovchinnikov. 1992. Role of cytoplasmic mRNP proteins in translation. Biochimie 74:477-483. [DOI] [PubMed] [Google Scholar]
- 30.Nekrasov, M. P., M. P. Ivshina, K. G. Chernov, E. A. Kovrigina, V. M. Evdokimova, A. A. Thomas, J. W. Hershey, and L. P. Ovchinnikov. 2003. The mRNA-binding protein YB-1 (p50) prevents association of the eukaryotic initiation factor eIF4G with mRNA and inhibits protein synthesis at the initiation stage. J. Biol. Chem. 278:13936-13943. [DOI] [PubMed] [Google Scholar]
- 31.Rajasekhar, V. K., A. Viale, N. D. Socci, M. Wiedmann, X. Hu, and E. C. Holland. 2003. Oncogenic Ras and Akt signaling contribute to glioblastoma formation by differential recruitment of existing mRNAs to polysomes. Mol. Cell 12:889-901. [DOI] [PubMed] [Google Scholar]
- 32.Rajasekhar, V. K., and E. C. Holland. 2004. Postgenomic global analysis of translational control induced by oncogenic signaling. Oncogene 23:3248-3264. [DOI] [PubMed] [Google Scholar]
- 33.Ranjan, M., S. R. Tafuri, and A. P. Wolffe. 1993. Masking mRNA from translation in somatic cells. Genes Dev. 7:1725-1736. [DOI] [PubMed] [Google Scholar]
- 34.Richter, J. D., and L. D. Smith. 1984. Reversible inhibition of translation by Xenopus oocyte-specific proteins. Nature 309:378-380. [DOI] [PubMed] [Google Scholar]
- 35.Skabkin, M. A., V. Evdokimova, A. M. Thomas, and L. Ovchinnikov. 2001. The major messenger ribonucleoprotein particle protein p50 (YB-1) promotes nucleic acid strand annealing. J. Biol. Chem. 276:44841-44847. [DOI] [PubMed] [Google Scholar]
- 36.Skabkin, M. A., O. I. Kiselyova, K. G. Chernov, A. V. Sorokin, E. V. Dubrovin, I. V. Yaminsky, V. D. Vasiliev, and L. P. Ovchinnikov. 2004. Structural organization of mRNA complexes with major core mRNP protein YB-1. Nucleic Acids Res. 32:5621-5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sommerville, J., and M. Ladomery. 1996. Masking of mRNA by Y-box proteins. FASEB J. 10:435-443. [DOI] [PubMed] [Google Scholar]
- 38.Sonenberg, N., and A. C. Gingras. 1998. The mRNA 5′ cap-binding protein eIF4E and control of cell growth. Curr. Opin. Cell Biol. 10:268-275. [DOI] [PubMed] [Google Scholar]
- 39.Sorokin, A. V., A. A. Selyutina, M. A. Skabkin, S. G. Guryanov, I. V. Nazimov, C. Richard, J. Th'ng, J. Yau, P. H. B. Sorensen, L. P. Ovchinnikov, and V. Evdokimova. 2005. Proteasome-mediated cleavage of the Y-box-binding protein 1 is linked to DNA-damage stress response. EMBO J. 24:3602-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stenina, O. I., K. M. Shaneyfelt, and P. E. DiCorleto. 2001. Thrombin induces the release of the Y-box protein dbpB from mRNA: a mechanism of transcriptional activation. Proc. Natl. Acad. Sci. USA 98:7277-7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sutherland, B. W., J. Kucab, J. Wu, C. Lee, M. C. Cheang, E. Yorida, D. Turbin, S. Dedhar, C. Nelson, M. Pollak, H. Leighton Grimes, K. Miller, S. Badve, D. Huntsman, C. Blake-Gilks, M. Chen, C. J. Pallen, and S. E. Dunn. 2005. Akt phosphorylates the Y-box binding protein 1 at Ser102 located in the cold shock domain and affects the anchorage-independent growth of breast cancer cells. Oncogene 24:4281-4292. [DOI] [PubMed] [Google Scholar]
- 42.Tafuri, S. R., and A. P. Wolffe. 1990. Xenopus Y-box transcription factors: molecular cloning, functional analysis and developmental regulation. Proc. Natl. Acad. Sci. USA 87:9028-9032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tenenbaum, S. A., C. C. Carson, P. J. Lager, and J. D. Keene. 2000. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc. Natl. Acad. Sci. USA 97:14085-14090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toker, A., and A. C. Newton. 2000. Akt/protein kinase B is regulated by autophosphorylation at the hypothetical PDK-2 site. J. Biol. Chem. 275:8271-8274. [DOI] [PubMed] [Google Scholar]
- 45.Wolffe, A. 1994. Structural and functional properties of the evolutionarily ancient Y-box family of nucleic acid binding proteins. BioEssays 16:245-251. [DOI] [PubMed] [Google Scholar]