Abstract
Procollagen C proteinases (pCPs) cleave type I to III procollagen C propeptides as a necessary step in assembling the major fibrous components of vertebrate extracellular matrix. The protein PCOLCE1 (procollagen C proteinase enhancer 1) is not a proteinase but can enhance the activity of pCPs ∼10-fold in vitro and has reported roles in inhibiting other proteinases and in growth control. Here we have generated mice with null alleles of the PCOLCE1 gene, Pcolce, to ascertain in vivo roles. Although Pcolce−/− mice are viable and fertile, Pcolce−/− male, but not female, long bones are more massive and have altered geometries that increase resistance to loading, compared to wild type. Mechanical testing indicated inferior material properties of Pcolce−/− male long bone, apparently compensated for by the adaptive changes in bone geometry. Male and female Pcolce−/− vertebrae both appeared to compensate for inferior material properties with thickened and more numerous trabeculae and had a uniquely altered morphology in deposited mineral. Ultrastructurally, Pcolce−/− mice had profoundly abnormal collagen fibrils in both mineralized and nonmineralized tissues. In Pcolce−/− tendon, 100% of collagen fibrils had deranged morphologies, indicating marked functional effects in this tissue. Thus, PCOLCE1 is an important determinant of bone mechanical properties and geometry and of collagen fibril morphology in mammals, and the human PCOLCE1 gene is identified as a candidate for phenotypes with defects in such attributes in humans.
Procollagen precursors of the major fibrillar collagen types I to III contain N and C propeptides that are cleaved upon secretion to yield mature triple-helical monomers that associate into fibrils. In particular, cleavage of the C propeptide has seemed an important rate-limiting step in fibrillogenesis, as monomers that retain C propeptides, unlike those which retain N propeptides, are not incorporated into fibrils in fibrillogenesis assays (15, 31). C propeptides are cleaved by procollagen C proteinase (pCP) activity provided by bone morphogenetic protein 1 (BMP1) and related metalloproteinases (15, 20). This activity is potentiated ∼10-fold by a secreted 55-kDa glycoprotein, designated the pCP enhancer 1 (PCOLCE1) (19, 33). PCOLCE1 binds specifically to the type I procollagen C propeptide and to the C telopeptide, which links the C propeptide to the triple-helical domain, and may act by inducing a conformational change that renders procollagen a fitter substrate for pCP cleavage (19, 27). PCOLCE1 comprises three major protein domains (36): two CUB domains and a C-terminal NTR domain. CUB (complement-Uegf-BMP1) domains are thought to mediate protein-protein interactions in a broad range of proteins with roles in development (8). Proteolytically processed 34- to 36-kDa forms of PCOLCE1, composed solely of the two CUB domains, are found in cell culture and retain in vitro pCP-enhancing activity and the ability to bind type I procollagen C propeptides (19, 36).
The C-terminal NTR (NETRIN) motif has homology with C-terminal domains of the netrins; complement components C3, C4, and C5; and the frizzled-related proteins and with N-terminal domains of the TIMPs (tissue inhibitors of metalloproteinases) (4). Relevant to the latter homology, the PCOLCE1 NTR domain has been identified as a non-TIMP MMP (matrix metalloproteinase) inhibitor secreted by tumor cells and, like the TIMPs, may serve to inhibit more than one proteinase (28). Thus, PCOLCE1 CUB and NTR domains may both contribute to net formation of fibrous matrix, by potentiating fibrillogenesis and inhibiting matrix-degrading proteinases, respectively. PCOLCE1 may serve additional roles, as disruption of the rat PCOLCE1 gene was reported to result in anchorage-independent growth of cultured fibroblasts (26).
A more recently isolated protein with only 43% sequence identity with PCOLCE1 but with an identical domain structure (43) has been shown to have pCP-enhancing activity levels similar to those of PCOLCE1 (33) and is designated PCOLCE2. Recently both PCOLCE1 and -2 have been found to bind the triple-helical portions of fibrillar collagens and to bind collagen fibrils in tissues (33). However, it is presently unknown how these newly described capabilities of the PCOLCEs might contribute to pCP enhancement or whether PCOLCEs, like other collagen-binding proteins, might contribute to the higher-order structure of collagen fibrils in tissues and/or interactions of such fibrils with other matrix or cell surface components. PCOLCE1 has a broad distribution of expression in soft tissues and highest expression levels in ossified bone, whereas PCOLCE2 expression is more restricted, primarily to nonossified cartilage, heart, and eye. The apparent inverse partitioning of PCOLCE1 and -2 expression to ossified and nonossified portions of developing bone, respectively, suggests different roles for the two proteins in formation of this tissue.
Here, in a direct approach towards ascertaining in vivo roles of PCOLCE1, we have generated mice with null alleles of the Pcolce gene. The Pcolce−/− phenotype demonstrates PCOLCE1 to indeed be an in vivo enhancer of procollagen processing and, in addition, a determinant of bone strength and geometry and in vivo collagen fibril morphologies in mineralized and nonmineralized tissues.
MATERIALS AND METHODS
Generation of mutant mice and MEFs.
A replacement targeting vector was generated by inserting genomic DNA fragments, isolated from a 129/SvJ mouse library (Stratagene), into vector pPNT (38). A ∼2.7-kb 5′ homology arm was inserted between pPNT NotI and XhoI sites upstream of the neo cassette, and a ∼2.6-kb 3′ homology arm was inserted between pPNT XbaI and BamHI sites downstream of the cassette. In the final targeting vector, the neo cassette replaces ∼2.5 kb of genomic sequences, including the entire second and third Pcolce exons. 129/SvJ embryonic stem (ES) cells were electroporated with NotI-linearized targeting vector and subjected to selection in G418 and ganciclovir. Four correctly targeted lines were identified by Southern blot assays of genomic DNA restricted with HindIII or BamHI and separately hybridized to 5′ or 3′ external probes, respectively. Southern blot analysis with a neo probe confirmed the absence of rearrangements or random integrations (data not shown). ES cells from three independently targeted cell lines were injected into C57BL/6 blastocysts and implanted into pseudopregnant females at the Children's Hospital Research Foundation Transgenic Facility (Cincinnati, OH). Chimeras were crossed with outbred Black Swiss mice (Taconic), and analysis of agouti offspring by Southern blotting for the targeted allele found germ line transmission from two of the independently targeted ES cell lines, leading to establishment of two separate mouse lines with Pcolce null alleles. Wild-type, heterozygous, and homozygous individuals were identified within the two lines via PCR amplification using internal neor primer 5′-GATCAGGATGATCTGGACGAAGAGC-3′ (forward) and primer 5′-AGGATGTTGCCTGGGCACCTAGTGG-3′ (reverse), corresponding to Pcolce intron IV sequences, to generate a ∼1-kb band for identification of the null allele or using the same reverse primer but forward primer 5′-CGCTGAGACTCCATCCTTAATTCGC-3′, corresponding to Pcolce intron II sequences, for generating a ∼1.35-kb band for identification of the wild-type allele. Mouse embryo fibroblasts (MEFs) were derived from 13.5-day-postconception embryos, as described previously (16).
Nucleic acid analyses.
Southern blotting was performed as described previously (37). For reverse transcription-PCR (RT-PCR), total RNA was isolated from MEFs using Trizol (Invitrogen), and reverse transcription reactions were performed as described previously (32). PCR amplifications were with Pcolce exon 1 forward primer, 5′-AACCTCCTTTCTGGGCCCATTCCT-3′ (nucleotides 61 to 84, GenBank accession no. NM_008788), and exon 6 reverse primer, 5′-GGAGCTCGTTCCCTTCAGAAGAGA-3′ (nucleotides 771 to 794). Glyceraldehyde-3-phosphate dehydrogenase-specific primers have been described elsewhere (32). PCR products were electrophoresed on agarose gels and visualized with ethidium bromide.
Protein analyses.
In Fig. 2A, 25 ng each of recombinant PCOLCE1 and PCOLCE2 (prepared as described in reference 33) was run on the same gel and transferred to the same membrane as 30 μl conditioned medium (from 10 ml total/10-cm culture dish) and the equivalent of 10% of the cell layer from the corresponding dish. To obtain samples, confluent MEFs were washed three times with phosphate-buffered saline (PBS) and incubated for 24 h in serum-free Dulbecco's modified Eagle's medium (DMEM) containing 40 μg/ml soybean trypsin inhibitor (SBTI) (Sigma). After 24 h, conditioned media were harvested, and protease inhibitors were added to final concentrations of 1 mM p-aminobenzoic acid, 1 mM N-ethylmaleimide, and 1 mM phenylmethylsulfonyl fluoride. Cells were washed with PBS, and cell layer proteins were extracted in hot sodium dodecyl sulfate (SDS)-polyacrylamide sample buffer. Proteins were resolved by SDS-polyacrylamide gel electrophoresis (PAGE), under reducing conditions on a 10% acrylamide gel, and subjected to immunoblot analysis as previously described (33, 34).
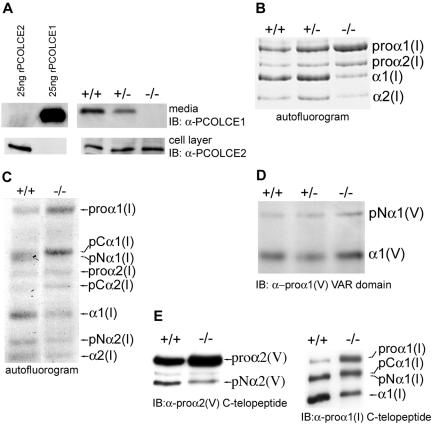
FIG. 2.
Pcolce−/− MEFs are devoid of PCOLCE1 and have reduced processing of the C propeptides of procollagen types I and V. (A) Twenty-five nanograms each of recombinant PCOLCE1 and -2 is compared by Western blotting to samples corresponding to 30 μl medium and 10% of the cell layer of 10-ml-medium/10-cm-culture-dish cultures of Pcolce+/+, Pcolce+/−, and Pcolce−/− MEFs. (B) An autofluorogram of 24-h-[3H]proline-radiolabeled material shows type I procollagen processing to be diminished in Pcolce−/− MEF medium, compared to Pcolce+/+ and Pcolce+/− MEF media. (C) An autofluorogram of pulse (1-h)/chase (9-h) [3H]proline-radiolabeled MEF medium samples, run on a long SDS-polyacrylamide gel to separate processing intermediates, shows increased and decreased amounts of pCα1(I) and pNα1(I), respectively, in a Pcolce−/− sample compared to wild type (+/+). (D and E) Western blotting shows that processing of pNα1(V) to mature α1(V) chains is unaffected (D) but that processing of pro-α2(V) to pNα2(V) chains is decreased (E, left panel) in Pcolce−/− MEFs, compared to wild type (+/+). The Western blot in the right panel of panel E compares processing of type I procollagen in the same samples used for analyzing type V procollagen processing in the left panel of panel E and in panel D.
To analyze type I collagen processing, long-term metabolic radiolabeling of confluent MEF collagens was conducted as previously described (29). Briefly, confluent MEFs were incubated for 20 h in DMEM, 10% fetal bovine serum (FBS) (HyClone), 50 μg/ml ascorbic acid, 100 μg/ml SBTI at pH 7.0. Cell cultures were then incubated for 4 h in depletion medium, consisting of DMEM, 5% dialyzed FBS, 50 μg/ml ascorbic acid, and 100 μg/ml SBTI, followed by radiolabeling in the same type of medium for 24 h with 30 μCi/ml l-[2,3-3H]proline (NEN Life Sciences). Radiolabeled collagen forms were obtained from conditioned medium and analyzed by reducing SDS-PAGE on 5% acrylamide gels and autofluorography, as previously described (35). Pulse-chase radiolabeling of collagen intermediates was performed as described above, except that depleted cells were radiolabeled for 1 h with 50 μCi/ml l-[2,3-3H] proline, at which time media were replaced with DMEM, 5% FBS, 100 μg/ml SBTI. Media were collected after a 9-h chase, and collagen forms were analyzed by autofluorography, as described above.
Preparation and analysis of type V and type I collagen forms for Western blot analysis were performed as previously described for analysis of type XI collagen intermediates (29). Briefly, confluent MEFs were grown overnight in DMEM, 10% FBS, 50 μg/ml ascorbic acid. Cells were switched to DMEM containing 1 ng/ml transforming growth factor β1 (R&D Systems), 40 μg/ml SBTI, and 50 μg/ml ascorbic acid, and 48 h later, media were collected and type V and I collagen forms were prepared as described for type I procollagen forms (above). Proteins were subjected to SDS-PAGE on reducing 5% acrylamide gels and then to immunoblot analysis using antibodies specific for the pro-α1(V) chain variable domain (39), antibody LF-67 against the pro-α1(I) C telopeptide (36) (kindly provided by Larry W. Fisher, National Institutes of Health), and antibody against the α2(V) C telopeptide (below), using previously described conditions (14, 23, 39). Rabbit polyclonal antibodies to murine pro-α2(V) C-telopeptide sequences were raised in rabbits against peptide GDIMGHYDENMPDPC, corresponding to residues 1232 to 1245 of the published mouse pro-α2(V) sequence (3) (accession number NP_031763) plus an additional cysteine for coupling to keyhole limpet hemocyanin. The pro-α2(V) antibodies were affinity purified on a column of the same peptide, as previously described (23).
Immunofluorescence.
Primary MEFs were plated in Lab Tek Chamber Slides (Nalgene Nunc) at a density of 1 × 104 cells/well; grown to 80 to 90% confluence in DMEM, 10% FBS; and then placed in DMEM, 10% FBS, 50 μg/ml ascorbic acid for 72 h. Subsequently, cells were washed for 5 min in PBS, incubated for 30 min on ice in 0.5 M acetic acid, and washed with PBS for an additional 5 min. Cells were blocked for 30 min in PBS containing 5% normal goat serum, followed by 1 h of incubation in the same blocking solution containing 1:200 dilutions of primary LF-67 rabbit antibodies against α(I) C-telopeptide sequences or against α(I) C-propeptide sequences (2) (kindly provided by Arthur Veis, Northwestern University). Following three washes of 5 min each in PBS, cells were blocked for an additional 30 min in 5% normal goat serum in PBS and then incubated in the same blocking solution containing a 1:1,000 dilution of goat anti-rabbit immunoglobulin G conjugated to Alexa 546 dye (Molecular Probes). Cells were washed three times for 5 min each in PBS and then mounted in Immu-Mount (Thermo Shandon), and coverslips were added. Images were captured using a Zeiss Axiophot II microscope.
Transmission electron microscopy.
For ultrastructural analyses of flexor digitorum longus (FDL) tendon and femur, male hind limbs were harvested; fixed for 15 min at room temperature in 4% paraformaldehyde, 2.5% glutaraldehyde, 0.1 M sodium cacodylate, pH 7.4, with 8.0 mM CaCl2 (5); and transferred to fresh fixative for an additional 105 min on ice. Hind limbs were transported on ice in 10% sucrose, 0.4% paraformaldehyde, 0.25% glutaraldehyde in 0.1 M sodium cacodylate, pH 7.4, after which FDL tendons were dissected free from surrounding tissues. For femoral analysis, left femora were freed of all soft tissue and demineralized for 3 weeks at 4°C in PBS, pH 7.2, 0.5% paraformaldehyde, 10% EDTA prior to further processing. Samples were postfixed with 1% osmium tetroxide and en bloc stained with uranyl acetate-50% ethanol. After dehydration in an ethanol series, followed by propylene oxide, tissue samples were infiltrated and embedded in a mixture of Embed 812, nadic methyl anhydride, dodecenyl succinic anhydride, and DMP-30 (Electron Microscopy Sciences). Thin sections were cut on an ultramicrotome and poststained with 2% aqueous uranyl acetate, 1% phosphotungstic acid, pH 3.2. Sections were examined at 120 kV using a Tecnai 12 transmission electron microscope equipped with a 2 K Gatan Ultrascan US1000 2 K digital camera.
Specimens for micro-computed tomography (micro-CT) analysis and four-point bending assays.
Left femora and eighth caudal vertebrae were harvested from 8-week-old Pcolce-null mice (n = 16 male, n = 15 female) and their wild-type littermates (n = 15 male, n = 15 female). Femora and vertebrae were cleaned of soft tissue at harvest and stored frozen in lactated Ringer's solution at −20°C.
Micro-CT evaluation.
Left femora and eighth caudal vertebrae were scanned and reconstructed at 18-μm voxels using a cone beam micro-CT system (GE Healthcare BioSciences, London, Ontario, Canada) to assess geometric and morphological properties. The three-dimensional data set generated by micro-CT was arranged as a series of 18-μm-thick slices, oriented along the long axis of the bone in the femora and along the superior-inferior axis of the vertebrae. A representative threshold was applied to each slice (21). Geometric analyses were performed on transverse cross sections over a 3-mm middiaphyseal segment in the femora and morphological analysis over two independent standardized volumes of trabecular bone in the proximal and distal ends of the vertebrae. Each slice of the femora data was analyzed for cross-sectional area, cortical thickness, and bending moment of inertia in the anterior-posterior direction about the medial-lateral axis of the bone. Data from each femur were averaged over the whole region of interest to provide representative values for the entire three-dimensional middiaphysis. Two standardized volumes of trabecular bone were segmented from the proximal and distal ends of the vertebral bodies and analyzed for bone volume fraction, trabecular thickness, degree of anisotropy, trabecular number, and trabecular spacing.
Mechanical properties.
Whole-bone mechanical properties were determined by loading the left femora to failure in four-point bending using a servohydraulic testing machine (858 Mini Bionix II; MTS Systems, Eden Prairie, MN) at a constant displacement rate of 0.5 mm/s. Femora were loaded in the anterior-posterior direction so that the posterior side of the bone was in tension and the anterior side was in compression. All four loading points were placed in contact with the bone by adjusting heights of the upper two points independently. Regions loaded in four-point bending corresponded to those measured by micro-CT. Displacement was monitored using a linear variable differential transducer (050 MHR; Lucas Schavitts, Hampton, VA), and load data were collected with a load cell (Sensotec, Columbus, OH) in series with the actuator. Load and displacement data were sampled at 2,000 Hz using the TestStar IIs system (version 2.4; MTS, Eden Prairie, MN). Load-displacement curves were analyzed for whole-bone yield load, ultimate load, stiffness, failure energy, and displacement ratio (ultimate displacement/yield displacement) using MATLAB software (version 7.0; The Mathworks Inc., Natick, MA). Bone tissue material properties were predicted using geometric measures from micro-CT for each bone and data from the mechanical tests. The predicted tissue elastic moduli and yield strength were calculated using standard mechanics of materials equations. These equations predict inherent material properties of the extracellular matrix, assuming a bending mode of failure.
Statistical analysis.
Multivariate general linear models and nonparametric tests were used to test for differences by genotype, with significance attributed to P < 0.05. Male and female subgroups were tested in separate models. Subsample analyses using nonparametric exact tests were employed for micro-CT evaluation. Multivariate general linear models were used to test for differences in mechanical properties. Independent sample t tests were used to analyze the predicted material properties.
RESULTS
Targeting of the Pcolce gene.
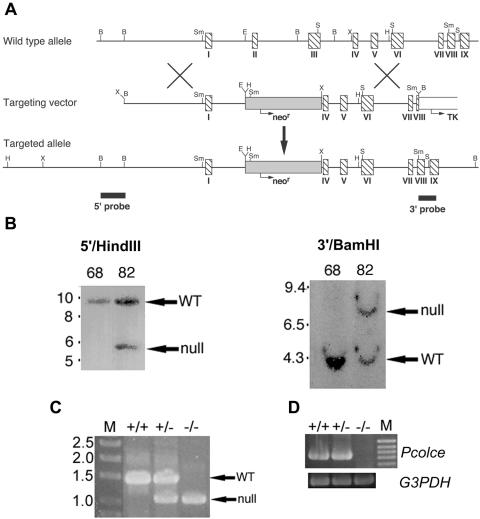
To characterize in vivo roles of PCOLCE1, mouse lines with null alleles for the PCOLCE1 gene Pcolce were created. Towards this end, a replacement targeting vector was constructed (Fig. 1A) in which a neor expression cassette replaces ∼2.5 kb of genomic sequence including the entire second and third Pcolce exons (109 and 259 bp, respectively), which together encode the more N-terminal CUB domain. Deletion of these exons not only removes all sequences encoding this domain but also creates a frameshift, such that nothing downstream of the signal peptide is translated in the original frame, and only 21 amino acids downstream of the frameshift are translated in the new frame, prior to a premature stop codon. Four correctly targeted lines were identified by Southern blot analysis of genomic DNA (Fig. 1B), followed by injection of ES cells into C57BL/6 blastocysts and implantation into pseudopregnant females. Two independently targeted ES cell lines gave rise to male chimeras that transmitted the targeted allele to progeny, upon mating to Black Swiss females, leading to establishment of two separate lines of mice with Pcolce null alleles. Genotyping within the two lines was via PCR amplification using a forward internal neor primer and a reverse primer corresponding to Pcolce intron IV sequences, to generate a ∼1-kb band corresponding to the null allele or using the same reverse primer but a forward primer corresponding to Pcolce intron II sequences to generate a ∼1.35-kb band corresponding to the wild-type allele (Fig. 1C). To assay for wild-type and mutant allele expression, wild-type, +/−, and −/− MEF RNA was subjected to RT-PCR, using a Pcolce exon 1 forward primer and an exon 6 reverse primer. Although these primers produced a clearly detectable 734-bp band, corresponding to the wild-type allele, in +/+ and +/− mouse samples (Fig. 1D), the same primers were unable to generate a 366-bp band corresponding to mutant allele mRNA from either +/− or −/− RNA (not shown). These results are consistent with the probability that mRNA from the mutant allele is rapidly degraded via nonsense-mediated decay and that the mutant Pcolce allele is thus a true null allele.
FIG. 1.
Targeted disruption of the Pcolce gene. (A) Structure of the targeting vector and Pcolce locus before and after homologous recombination. Horizontal arrows mark start sites and direction of transcription of neor (shaded box) and tk (open box) cassettes. Hatched boxes represent Pcolce exons. Black boxes represent a 554-bp 5′ external probe and a 414-bp 3′ external probe. B, BamHI; E, EcoRI; H, HindIII; S, SacI; Sm, SmaI; X, XbaI. (B) Southern blots of HindIII- or BamHI-restricted genomic DNA from a clone of untargeted ES cells (68) and from a clone of correctly targeted ES cells (82), hybridized to the 5′ or 3′ external probe, respectively. The 5′ probe detected bands of 9.5 and 5.5 kb from the wild-type (WT) and targeted alleles, respectively, while the 3′ probe detected bands of 4 and 8 kb from the wild-type and targeted alleles, respectively. Numbers at left of panels are molecular sizes in kilobases. (C) +/+, +/−, and −/− individuals were identified within the two mouse lines via PCR amplification using an internal neor primer (forward) and a reverse primer corresponding to Pcolce intron IV sequences, to generate a ∼1-kb band identifying the null allele, or using the same reverse primer but a forward primer corresponding to Pcolce intron II sequences, to generate a ∼1.35-kb band identifying the wild-type (WT) allele. M, DNA size markers marked with sizes in kilobases. (D) RT-PCR analysis of poly(A)+ RNA prepared from MEFs detected a 734-bp amplimer for +/+ and +/− MEFs but no bands for −/− MEFs. M, 100-bp marker ladder with the 1-kb marker corresponding to the topmost band visible in the figure. G3PDH, glyceraldehyde-3-phosphate dehydrogenase.
Pcolce-null mice are viable and fertile.
Heterozygotes derived from the two different lines of ES cells were separately interbred to produce homozygotes for each line, and genotypes of offspring from heterozygote-to-heterozygote matings were determined at weaning (∼3 weeks). The proportion of −/− offspring is ∼25% in both lines, showing Pcolce-null animals to be both viable and fertile, with an absence of gross abnormalities detected upon external inspection. Necropsies were performed by a veterinary pathologist on four Pcolce−/− mice, ranging in age from 1 to 1.5 years. Although myocardial degeneration with fibrosis, ventral extravasation of intervertebral disks, and mineralization of focal areas of connective tissue in limbs were noted in some mutant mice, none of these conditions were constant features in all four Pcolce−/− mice. Moreover, aspects of these conditions were also noted in some age-matched +/+ controls.
Absence of PCOLCE1 results in reduced levels of processing of C propeptides of the major fibrillar procollagens and of the pro-α2 chain of minor fibrillar collagen type V.
MEFs derived from embryos with null alleles have previously been employed in analyzing molecular/cellular effects of gene ablations (12, 29, 32, 39). Since PCOLCE1 is expressed by MEFs (24), MEFs were isolated from embryos with null Pcolce alleles, to determine effects of PCOLCE1 ablation on procollagen processing. Prior to such studies, we tested for possible MEF PCOLCE2 expression, due to possible functional overlap in cells in which PCOLCE1 and -2 are coexpressed. The possibility of such overlap would clearly be an important consideration in interpreting assays of pCP function in Pcolce-null MEFs. As can be seen in Fig. 2A, wild-type MEFs produce readily detectable amounts of both PCOLCE1 and -2. Moreover, levels of expression appeared to be relatively high, given the strength of signals from samples representing only 3% of 10 ml medium and 10% of corresponding cell layers from 10-cm plates, compared to signal strengths of 25 ng each of PCOLCE1 and -2 standards (Fig. 2A). As previously reported by Xu et al. for porcine trabecular meshwork and human fibroblasts (43), all detectable MEF PCOLCE2 was found associated with cell layers. In contrast, all detectable PCOLCE1 was in conditioned medium (Fig. 2A), and none was associated with the cell layer (not shown). Importantly, Pcolce+/− heterozygous MEFs were found to have reduced levels of secreted PCOLCE1 compared to wild type, whereas PCOLCE1 was undetectable in conditioned medium of Pcolce−/− MEFs (Fig. 2A). Thus, additional evidence is supplied that targeted Pcolce alleles in the present study are true null alleles. In contrast to results with PCOLCE1, PCOLCE2 levels were similar for cultured MEFs of all three genotypes (Fig. 2A).
PCOLCE1, initially isolated due to its observed ability to potentiate the activities of pCPs in vitro (1), is primarily described in the literature as a pCP enhancer, due to this in vitro activity. To examine whether PCOLCE1 is actually employed as a pCP enhancer by cells, we next compared procollagen processing in wild-type and Pcolce-null MEF cultures. As demonstrated in an autofluorogram of 24-h-[3H]proline-labeled material from conditioned MEF medium (Fig. 2B), processing of type I procollagen is diminished in Pcolce−/− MEF culture medium compared to wild-type and +/− MEF media, as evidenced by higher levels of procollagen and lower levels of mature α1(I) and α2(I) chains. In a separate analysis of effects of PCOLCE1 ablation on type I procollagen processing by cells, MEFs were pulsed for 1 h with [3H]proline and chased for 9 h with unlabeled medium, and samples from conditioned media were then run on a long gel to separate different processing intermediates. Processing intermediates pCα(I) (which retains N but not C propeptides) and pNα(I) (which retains C but not N propeptides) comigrate upon SDS-PAGE as a doublet, as seen for wild-type MEF medium sample (Fig. 2C). In contrast, although amounts of pCα1(I) were increased in Pcolce−/− MEF medium sample compared to wild type, detectable pNα1(I) chains were absent. This result is consistent with the conclusion that PCOLCE1 serves to specifically facilitate processing of the type I procollagen C propeptide in vivo. Underscoring this specificity, processing of the type V procollagen pNα1(V) chain, the globular N-terminal domain of which is cleaved in vitro and in vivo by BMP1 (39), is unaffected in Pcolce−/− MEF medium (Fig. 2D). The latter result is consistent with results from in vitro biochemical assays in which PCOLCE1 has been found to have no effect on processing of the N propeptide of the pro-α1(V) chain (reference 27 and data not shown). Thus, in vivo activity of PCOLCE1 enhances processing of the C propeptides of the major fibrillar collagens but not processing of the N propeptides of either the major fibrillar collagens or the minor fibrillar collagen pro-α1(V) chain. Processing of probiglycan and chordin, both of which are processed in vitro and in vivo by BMP1 (29, 32), was also unaffected in Pcolce−/− MEF medium (data not shown), consistent with results showing PCOLCE1 to have no effect on processing of either chordin (15, 27, 30) or probiglycan (data not shown) in in vitro assays.
The predominant form of type V procollagen is a heterotrimer comprising one pro-α2(V) and two pro-α1(V) chains. Although the pro-α1(V) C propeptides are cleaved by furin-like proprotein convertases, the pro-α2(V) C propeptides are cleaved by BMP1-like proteinases (39). Thus, we examined whether processing of pro-α2(V) C propeptides is affected by the absence of PCOLCE1. As can be seen (Fig. 2E, left panel), processing of the pro-α2(V) C propeptide is diminished in Pcolce−/− MEF cultures compared to wild type. The diminishment, though slight, was reproducible over the course of three independent experiments and is the first indication that PCOLCE1 can enhance the processing of substrates other than the major fibrillar procollagens.
Pcolce−/− bone phenotype.
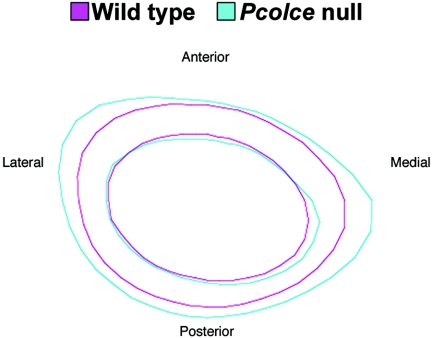
As Pcolce is expressed at particularly high levels in developing bone (33) and since type I collagen, a product of pCP activity, is the major structural protein of bone, constituting ∼80% of bone protein (22), bone was the first Pcolce−/− tissue subjected to detailed examination for abnormalities. Micro-CT and subsample analyses using nonparametric exact tests showed significant differences between wild-type and Pcolce−/− males in cross-sectional area, cortical thickness, and bending moment of inertia. Average cross-sectional representations of male cortical bone showed that excess bone apposition occurred on Pcolce−/− medial and lateral periosteum (Fig. 3), with increased bending moment (anterior-posterior), compared to wild-type male littermates. These differences were highly statistically significant. In contrast, although Pcolce−/− female femora consistently had increased cross-sectional area and cortical thickness compared to wild-type littermate female femora, these differences were not statistically significant with the numbers of mice used. As suggested by the disparate results in the separate female and male analyses, the effect of genotype on the parameters of interest differs by sex. This sex difference is statistically significant for cross-sectional area (P < 0.0010), cortical thickness (P=0.02), and bending moment y (P = 0.001).
FIG. 3.
Average cross-sectional representations of male cortical bone. Cone beam micro-CT scanning showed male Pcolce−/− null (blue, n = 16) medial and lateral femoral periostea to have excess bone apposition, with increased bending moment (medial-lateral) compared to wild-type male littermates (magenta, n = 15).
Multivariate general linear models of four-point bending data were used to test for differences in whole-bone mechanical properties by genotype. Results revealed that male Pcolce−/− mutants had significantly greater yield load and stiffness than did littermate controls, although there was no difference between the two groups in displacement ratio (Table 1).
TABLE 1.
Mechanical analyses by gender and genotypea
| Gender | Genotype | Yield load (N) | Stiffness (N/mm) | Displacement ratio |
|---|---|---|---|---|
| Female | Pcolce−/− | 20.9 (4.9) | 214 (35) | 3.00 (1.3) |
| WTb | 20.9 (5.5) | 207 (40) | 2.88 (1.3) | |
| Male | Pcolce−/− | 25.9 (6.6)* | 256 (65)* | 2.62 (1.0) |
| WT | 21.0 (5.8) | 204 (43) | 2.76 (1.1) |
Data are given as means (standard deviations). *, significant (P ≤ 0.05).
WT, wild type.
Predicted modulus and yield strength were calculated from the mechanical testing and geometrical properties using standard mechanics of materials equations. Independent sample t tests showed a significant difference in elastic modulus by genotype in males but no significant difference in predicted strength by genotype in males or females (Table 2). Male Pcolce−/− mutant femora had a lower predicted modulus than did male wild-type femur, thus suggesting inferior material properties of the Pcolce-null bone. These results suggest that the changed geometry, leading to increased bending moment, and the increased cross-sectional area of Pcolce−/− male femora were sufficient compensation by the organism for the inferior material properties of these bones, such that the bone maintained its overall strength compared to those of wild-type male littermate controls.
TABLE 2.
Predicted material properties by gender and genotypea
| Gender | Genotype | Modulus (MPa) | Yield strength (MPa) |
|---|---|---|---|
| Female | Pcolce−/− | 6,278 (703) | 114 (22) |
| WTb | 6,866 (923) | 123 (19) | |
| Male | Pcolce−/− | 5,461 (972)* | 107 (9.9) |
| WT | 6,758 (941) | 113 (33) |
Data are given as means (standard deviations). *, significant (P ≤ 0.05).
WT, wild type.
With respect to trabecular tissue in vertebral bodies, subsample analyses using nonparametric exact tests showed significant differences in bone volume fraction, trabecular thickness, and degree of anisotropy (degree of spatial orientation) by genotype. Both male and female Pcolce−/− mice had a larger bone volume fraction and thicker, disoriented trabeculae compared to littermate controls (Table 3). Similar results were observed in the proximal and distal regions of the vertebrae.
TABLE 3.
Distal vertebral geometric analyses by gender and genotypea
| Gender | Genotype | BVF | TbTh (mm) | DA |
|---|---|---|---|---|
| Female | Pcolce−/− | 0.418 (0.04)* | 0.074 (0.01)* | 1.87 (0.12)* |
| WT | 0.366 (0.08) | 0.063 (0.01) | 1.99 (0.11) | |
| Male | Pcolce−/− | 0.519 (0.05)* | 0.071 (0.00)* | 1.76 (0.11)* |
| WT | 0.457 (0.06) | 0.063 (0.01) | 1.97 (0.10) |
Data are given as means (standard deviations). *, significant (P ≤ 0.05). Abbreviations: BVF, bone volume fraction; TbTh, trabecular thickness; DA, degree of anisotropy; WT, wild type.
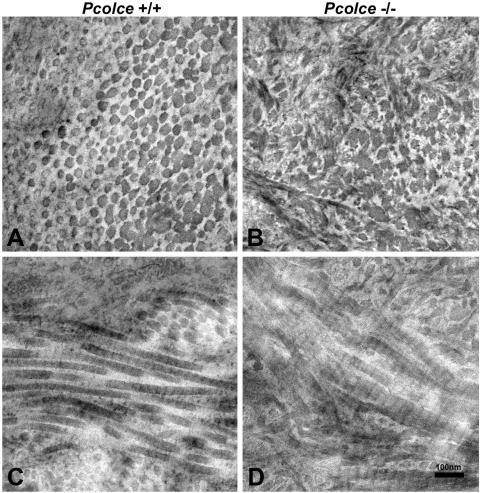
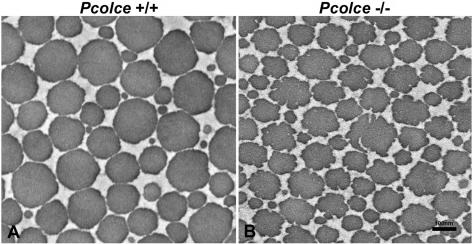
The ultrastructure of Pcolce−/− male femora was next compared to that of wild-type male control femora by transmission electron microscopy. As can be seen in Fig. 4, the collagen fibrils of male Pcolce−/− femur show aberrant, fluted, or scalloped cross sections (Fig. 4B), unlike the far more circular outlines of collagen fibrils in wild-type male control femur (Fig. 4A). Longitudinal views (Fig. 4C and D) show Pcolce−/− femur collagen fibrils to have irregular, broadened profiles, with some branching, and to be somewhat reminiscent of longitudinal views of the “hieroglyphic” fibrils of Ehlers-Danlos syndrome type VIIC patients (9).
FIG. 4.
Transmission electron micrographs of collagen fibril morphology in 4-week-postnatal Pcolce−/− and wild-type (Pcolce+/+) left femora. Unlike wild-type fibril cross sections (A), all mutant collagen fibrils have irregular scalloped profiles in cross section (B), whereas longitudinal views of collagen fibrils (C and D) show Pcolce−/− fibrils to have irregular, broadened profiles, with some branching.
Pcolce−/− soft tissue phenotype.
Type I collagen is also a major structural protein in most soft tissues. In tendon, characterized by densely packed collagen fibrils organized as fibers parallel to the tendon axis, it is the major structural component and is responsible for the mechanical attributes of this tissue. As is evident from transmission electron micrographs comparing 30-day-postnatal Pcolce−/− and wild-type tendon (Fig. 5A and B), collagen fibrils in this tissue are also markedly affected by the absence of PCOLCE1, with Pcolce−/− fibrils showing fluted/scalloped cross sections, in contrast to the smooth, circular cross sections of wild-type tendon fibers. It is also notable that 100% of collagen fibrils in Pcolce−/− tendon have abnormal cross sections, suggesting pronounced effects on the mechanical properties and function of Pcolce-null tendon.
FIG. 5.
Transmission electron micrographs of collagen fibril morphology in 30-day-postnatal Pcolce−/− (A) and wild-type littermate (B) FDL tendon cross sections. All mutant collagen fibrils have an irregular, scalloped appearance in contrast to the smooth circular cross sections of the wild-type tendon fibrils.
Analyses towards understanding the mechanical basis of the effects of PCOLCE1 ablation on tissue collagen fibrils.
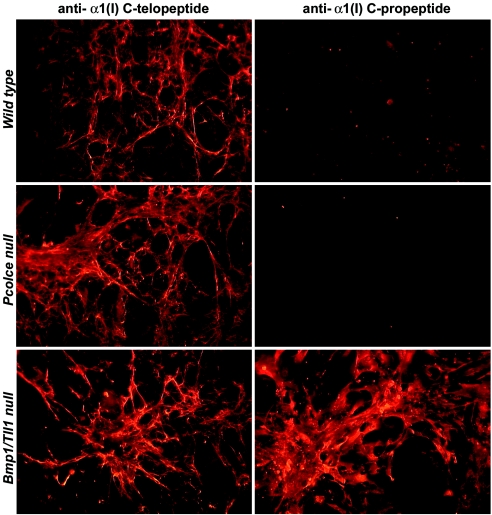
It is difficult to explain the various changes in collagen fibril morphology in Pcolce−/− bone and tendon on the basis of changes in kinetics of the cleavage of the major fibrillar procollagen C propeptides alone. Previously, it has been suggested that abnormal collagen fibrils in embryos null for genes that encode pCPs can result from incorporation of type I collagen pC monomers (which retain C propeptides) into growing fibrils (29, 35). Since ablation of a pCP enhancer might also result in the incorporation of type I collagen pC forms into nascent fibrils, type I collagen fibrils deposited by wild-type and Pcolce-null MEFs were assayed for possible retention of C propeptides and compared to those deposited by MEFs from embryos doubly homozygous null for the Bmp1 gene, which generates alternatively spliced RNAs for the two pCP metalloproteinases BMP1 and mTLD, and the Tll1 gene, which encodes the related pCP mTLL1. As can be seen in Fig. 6, antibody directed against the pro-α1(I) chain C propeptide (2) clearly reacted with collagen fibrils in Bmp1/Tll1 doubly null MEF cultures but did not react with collagen fibrils in either wild-type or Pcolce−/− MEF cultures. Thus, although these data support the contention that aberrant Bmp1/Tll1 doubly null collagen fibrils contain monomers with retained C propeptides, the same does not appear to be the case for Pcolce−/− collagen fibrils.
FIG. 6.
Collagen fibrils of Bmp1/Tll1 doubly null, but not Pcolce-null, MEFs contain readily detectable levels of type I procollagen pro-α1(I) C propeptides. Wild-type, Pcolce-null, and Bmp1/Tll1 doubly null MEF cell layers were subjected to immunofluorescence staining, employing antibodies directed against the C-telopeptide (11) or C-propeptide (2) region of the pro-α1(I) chain.
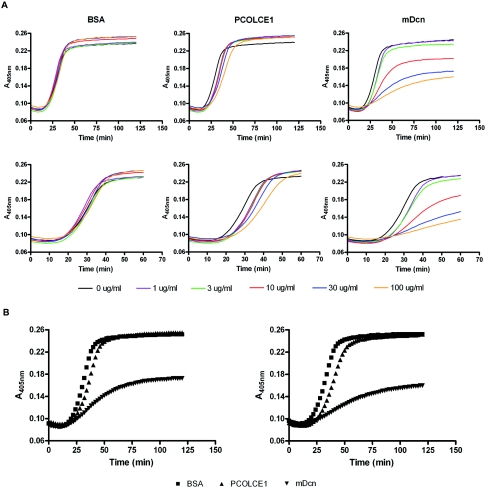
The fluted cross-sectional profiles of Pcolce−/− tendon collagen fibrils are somewhat reminiscent of collagen fibril morphologies encountered in the same tissue in mice with null alleles for genes encoding small leucine-rich proteoglycans (SLRPs), such as decorin, lumican, and fibromodulin (10, 18). Such phenotypes, together with analyses via in vitro fibrillogenesis assays (12, 40, 41), have indicated that at least some SLRPs regulate fibrillogenesis directly, via interactions with collagen monomers. To determine whether PCOLCE1 might also regulate fibrillogenesis by interacting with collagen monomers via mechanisms similar to those of SLRPs, increasing amounts of PCOLCE1 were added to in vitro type I collagen fibrillogenesis assays. As can be seen in Fig. 7A, PCOLCE1, like bovine serum albumin (BSA), had no appreciable effect on the final turbidity of the reaction (thought to reflect the lateral growth of fibrils) and only an apparently small effect on the reaction rate. In contrast, the mature SLRP decorin (mDcn; decorin minus the propeptide) is seen to markedly reduce rates of fibrillogenesis and final turbidity levels, as previously reported (12, 40), in a concentration-dependent manner. Shapes of fibrillogenesis curves in the presence of 30 μg/ml and 100 μg/ml BSA, PCOLCE1, and decorin are superimposed for comparison in Fig. 7B. Shapes of curves were reproducible for BSA, PCOLCE1, and decorin over the course of five experiments. Thus, unlike SLRPs, PCOLCE1 does not appear to have a marked effect on fibrillogenesis via direct interactions with collagen monomers. This is consistent with the results of Hulmes et al. (17), who previously found a protein fragment containing the two PCOLCE1 CUB domains to have no appreciable effect on morphology of collagen fibrils formed in vitro.
FIG. 7.
Testing for a possible role for PCOLCE1 in regulating type I collagen fibrillogenesis. (A) Different concentrations of PCOLCE1 are compared to matched concentrations of mature decorin (mDcn; decorin minus the propeptide) (positive control) and BSA (negative control) for possible effects in a fibrillogenesis assay of type I collagen, in which turbidity (measured at 450 nm) is primarily a measure of lateral growth of fibrils (13), and duration of the lag phase at the beginning of the assay, during which there is no detectable change in turbidity, is thought to involve primarily linear elongation of thin fibrils (13). (B) Type I collagen fibrillogenesis curves in the presence of 30 μg/ml (left panel) or 100 μg/ml (right panel) BSA, PCOLCE1, and mDcn are superimposed for comparison. Collagen was 0.5 mg/ml in all assays.
DISCUSSION
Here, we have generated and characterized mice with null alleles for the Pcolce gene, to define in vivo roles for the protein PCOLCE1. Absence of PCOLCE1 resulted in a site-specific, gender-specific phenotype that affected the material properties and geometry of bone. Pcolce−/− males appeared to compensate for weaker cortical bone material by increasing the size and modifying the cross-sectional shape of long bones, such as femora. This increase and modification in geometrical properties drove an increase in structural mechanical integrity. Male Pcolce−/− femora are thus somewhat reminiscent of the femora of Mov13+/− mice, which have one null allele for the α1(I) chain of type I collagen and serve as a model for osteogenesis imperfecta type I (OI type I) (7). The similarity is that Mov13+/− mice, like Pcolce−/− males, compensate for inferior cortical bone material properties by generating more massive bone with altered geometry, thus maximizing the bending strength/moment of inertia (6). Interestingly, although Pcolce−/− female femora tended to have increased cross-sectional area and cortical thickness compared to wild-type littermate female femora, these differences were not statistically significant with the numbers of mice used. The gender effect in cortical bone may be physiologic in nature and due to availability of hormones and/or cytokines. However, an exact determination for the sex difference in cortical bone cannot be made from the studies done thus far.
In contrast to cortical bone, the absence of PCOLCE1 appeared to cause identical phenomena in male and female Pcolce−/− trabecular bone. Pcolce-null mice increased their trabecular bone volume fraction by thickening trabeculae and by adding more trabecular struts (data not shown). As in the cortical bone of femora, the thickening and increased number of struts in Pcolce−/− trabecular bone may represent an attempt to compensate for weaker material properties of the bone. Interestingly, histograms of the gray-scale voxel values from micro-CT (calibrated through the use of a standard phantom during scanning) suggested that the mutant trabecular bone might have an alteration in the morphology of the mineral being deposited. Thus, we are left with the exciting and quite novel possibility that an alteration in the mineral-collagen interaction is driving the reduced material properties of Pcolce-null bone.
Further investigation at the tissue and ultrastructural level is needed in order to verify the material properties of Pcolce−/− bone and to characterize the molecular network of the collagen and mineral crystals. However, Pcolce-null bone is clearly unlike Mov13+/− bone. Although Mov13+/− and male Pcolce−/− cortical bone both have inferior material properties compared to wild type, Pcolce−/− cortical bone, unlike Mov13+/− bone (shown to be representative of OI type I cortical bone), is not “brittle.” Thus, although Mov13+/− femora shatter by almost simultaneous breakage at both cortices at maximum load (7) and demonstrate little resistance to crack propagation, male Pcolce−/− femora, like those of the wild type, typically fractured initially at a single cortical surface (the one in tension), with propagation of the fracture along the medullary canal (data not shown) and continued energy absorption until complete failure. This difference indicates that the Pcolce−/− bone phenotype is not easily explainable by invoking the haploinsufficiency of type I collagen deposition that underlies the Mov13+/−, OI type I phenotype.
If Mov13+/− mice are models for OI type I, then what human condition is mirrored by the Pcolce−/− phenotype? Studies designed to determine the material properties of Pcolce-null bone, which may help suggest a human correlate of the phenotype, and studies of the Pcolce-null molecular network of collagen and mineral, which will provide insights into the mechanism whereby PCOLCE1 influences collagen-mineral interactions, are currently under way.
The fluted/scalloped appearance of bone and tendon collagen fibrils in Pcolce-null mice is reminiscent of features found in the tendons of mice with null alleles for some SLRP genes. However, an important difference is that, unlike SLRP-deficient mice, which generally acquire the fibril phenotype later in development (during periods of fibril growth from fusion of intermediates), Pcolce−/− mice demonstrate the fibril phenotype throughout development. The latter finding suggests that the Pcolce−/− phenotype results from dysfunctional regulation of an early step in collagen fibrillogenesis. We also demonstrate here that PCOLCE1 does not markedly modulate fibrillogenesis in a direct manner similar to that of SLRPs. Thus, since neither the absence of SLRP-type regulatory activity on fibrillogenesis nor a mere diminution of collagen deposition seems adequate for explaining the profound and varied effects on collagen fibril morphology and the unique bone phenotype in Pcolce−/− mice, some other mechanism(s) must be sought. An interesting possibility is that dysregulation of type V procollagen processing, as demonstrated here by the decreased processing rate of pro-α2(V) C propeptides in the absence of PCOLCE1, contributes to the abnormal fibrillogenesis seen in Pcolce−/− tissues. Previous studies have shown the low-abundance collagen type V to regulate the lateral growth of heterotypic fibrils in which it is incorporated with the much more abundant collagen type I (25). Moreover, the developmental evidence herein for a PCOLCE1 regulatory role that occurs at an early step in collagen fibrillogenesis conforms with previous evidence that type V collagen can also regulate type I collagen fibrillogenesis early in development, via a collagen fibril nucleation event (42). Another possible mechanism contributing to the Pcolce−/− phenotype is suggested by the ability of PCOLCE1 to bind assembled collagen fibrils in tissues (33) and involves the possibility that PCOLCE1 associated with such fibrils may normally affect their structure and/or stability and/or interactions with other extracellular matrix components. Such possibilities remain to be explored.
The Pcolce−/− tendon phenotype differs from that of mice null for SLRP genes in that 100% of Pcolce−/− tendon fibrils have aberrant profiles. Future studies will explore the functional consequences of these aberrant collagen fibrils for Pcolce−/− tendon and will also explore the possibility of joint laxity, due to the possibility of similar aberrant fibrils in ligamentous attachments. Such studies should provide further insights into how such a phenotype might present clinically for humans with abnormalities of the orthologous PCOLCE1 gene.
It remains unknown to what extent effects of PCOLCE1 ablation in Pcolce-null mice and MEFs may be masked by functional redundancy provided by PCOLCE2. PCOLCE1 and -2 show similar levels of pCP-enhancing activity in vitro, against both procollagen types I and II, and the two proteins are coexpressed in MEFs and heart and other tissues (33). However, the two proteins appear to have nonoverlapping expression domains in bone and cartilage, respectively (33), and although PCOLCE1 and -2 are both secreted (33), PCOLCE2 is retained by cell layers, whereas PCOLCE1 is not (33, 43). The latter difference is likely to influence modes of activity and is relevant to questions regarding extents of functional overlap and to the question of whether PCOLCE2 is capable of diffusing over sufficient distances to complement loss of PCOLCE1 in tissues where the latter, but not the former, is ordinarily expressed. Such questions will be addressed via generating and characterizing mice with disrupted alleles for the PCOLCE2 gene Pcolce2. Moreover, intercrossing of mice with Pcolce and Pcolce2 null alleles will produce progeny, some of which are doubly homozygous null for both genes, thereby removing any possible functional redundancy/overlap and allowing determination of the full range of in vivo functions of this class of mammalian gene products.
Acknowledgments
We thank Art Veis (Northwestern University Medical School) and Larry Fisher (National Institute of Dental and Craniofacial Research) for generously providing antibodies to type I procollagen and Biao Zuo for expert technical assistance with transmission electron microscopy.
This work was supported by grants GM63471, GM71679, and AR47746 (D.S.G.); AR44745 (D.E.B.); and AR042919 (M.H.) and the University of Michigan Musculoskeletal Core Center grant AR46024 (S.A.G.), all provided by the National Institutes of Health.
REFERENCES
- 1.Adar, R., E. Kessler, and B. Goldberg. 1986. Evidence for a protein that enhances the activity of type I procollagen C-proteinase. Collagen Relat. Res. 6:267-277. [DOI] [PubMed] [Google Scholar]
- 2.Alvares, K., F. Siddiqui, J. Malone, and A. Veis. 1999. Assembly of the type 1 procollagen molecule: selectivity of the interactions between the α1(I) and α2(I)-carboxyl propeptides. Biochemistry 38:5401-5411. [DOI] [PubMed] [Google Scholar]
- 3.Andrikopoulos, K., H. R. Suzuki, M. Solursh, and F. Ramirez. 1992. Localization of pro-alpha 2(V) collagen transcripts in the tissues of the developing mouse embryo. Dev. Dyn. 195:113-120. [DOI] [PubMed] [Google Scholar]
- 4.Bányai, L., and L. Patthy. 1999. The NTR module: domains of netrins, secreted frizzled related proteins, and type I procollagen C-proteinase enhancer protein are homologous with tissue inhibitors of metalloproteases. Protein Sci. 8:1636-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birk, D. E., and R. L. Trelstad. 1984. Extracellular compartments in matrix morphogenesis: collagen fibril, bundle, and lamellar formation by corneal fibroblasts. J. Cell Biol. 99:2024-2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonadio, J., K. J. Jepsen, M. K. Mansoura, R. Jaenisch, J. L. Kuhn, and S. A. Goldstein. 1993. A murine skeletal adaptation that significantly increases cortical bone mechanical properties. J. Clin. Investig. 92:1697-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonadio, J., T. L. Saunders, E. Tsai, S. A. Goldstein, J. Morris-Wiman, L. Brinkley, D. F. Dolan, R. A. Altschuler, J. E. Hawkins, Jr., J. F. Bateman, T. Mascara, and R. Jaenisch. 1990. Transgenic mouse model of the mild dominant form of osteogenesis imperfecta. Proc. Natl. Acad. Sci. USA 87:7145-7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bork, P., and G. Beckmann. 1993. The CUB domain: a widespread module in developmentally regulated proteins. J. Mol. Biol. 231:539-545. [DOI] [PubMed] [Google Scholar]
- 9.Byers, P. H. 1995. Disorders of collagen biosynthesis and structure, p. 4029-4077. In C. R. Scriver, A. L. Beaudet, W. S. Sly, and D. Valle (ed.), The metabolic and molecular bases of inherited disease. McGraw-Hill, New York, N.Y.
- 10.Danielson, K. G., H. Baribault, D. F. Holmes, H. Graham, K. E. Kadler, and R. V. Iozzo. 1997. Targeted disruption of decorin leads to abnormal collagen fibril morphology and skin fragility. J. Cell Biol. 136:729-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher, L. W., J. T. Stubbs III, and M. F. Young. 1995. Antisera and cDNA probes to human and certain animal model bone matrix noncollagenous proteins. Acta Orthop. Scand. Suppl. 266:61-65. [PubMed] [Google Scholar]
- 12.Ge, G., N.-S. Seo, X. Liang, D. R. Hopkins, M. Höök, and D. S. Greenspan. 2004. Bone morphogenetic protein-1/Tolloid-related metalloproteinases process osteoglycin and enhance its ability to regulate collagen fibrillogenesis. J. Biol. Chem. 279:41626-41633. [DOI] [PubMed] [Google Scholar]
- 13.Gelman, R. A., B. R. Williams, and K. A. Piez. 1979. Collagen fibril formation. Evidence for a multistep process. J. Biol. Chem. 254:180-186. [PubMed] [Google Scholar]
- 14.Gopalakrishnan, B., W.-M. Wang, and D. S. Greenspan. 2004. Biosynthetic processing of the pro-α1(V)pro-α2(V)pro-α3(V) procollagen heterotrimer. J. Biol. Chem. 279:30904-30912. [DOI] [PubMed] [Google Scholar]
- 15.Greenspan, D. S. 2005. Biosynthetic processing of collagen molecules. Top. Curr. Chem. 247:149-183. [Google Scholar]
- 16.Hogan, B. L. M., R. Beddington, F. Constantini, and E. Lacy. 1994. Manipulating the mouse embryo: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Hulmes, D. J. S., A. P. Mould, and E. Kessler. 1997. The CUB domains of procollagen C-proteinase enhancer control collagen assembly solely by their effect on procollagen C-proteinase/bone morphogenetic protein-1. Matrix Biol. 16:41-45. [DOI] [PubMed] [Google Scholar]
- 18.Jepson, K. J., F. Wu, J. H. Peragallo, J. Paul, L. Roberts, Y. Ezura, A. Oldberg, D. E. Birk, and S. Chakravarti. 2002. A syndrome of joint laxity and impaired tendon integrity in lumican- and fibromodulin-deficient mice. J. Biol. Chem. 277:35532-35540. [DOI] [PubMed] [Google Scholar]
- 19.Kessler, E., and R. Adar. 1989. Type I procollagen C-proteinase from mouse fibroblasts. Purification and demonstration of a 55-kDa enhancer glycoprotein. Eur. J. Biochem. 186:115-121. [DOI] [PubMed] [Google Scholar]
- 20.Kessler, E., K. Takahara, L. Biniaminov, M. Brusel, and D. S. Greenspan. 1996. Bone morphogenetic protein-1: the type I procollagen C-proteinase. Science 271:360-362. [DOI] [PubMed] [Google Scholar]
- 21.Kuhn, J. L., S. A. Goldstein, L. A. Feldkamp, R. W. Goulet, and G. Jesion. 1990. Evaluation of a microcomputed tomography system to study trabecular bone structure. J. Orthop. Res. 8:833-842. [DOI] [PubMed] [Google Scholar]
- 22.Kuivaniemi, H., G. Tromp, and D. J. Prockop. 1991. Mutations in collagen genes: causes of rare and some common diseases in humans. FASEB J. 5:2052-2060. [DOI] [PubMed] [Google Scholar]
- 23.Lee, S., D. E. Solow-Cordero, E. Kessler, K. Takahara, and D. S. Greenspan. 1997. Transforming growth factor-β regulation of bone morphogenetic proteoin-1/procollagen C-proteinase and related proteins in fibrogenic cells and keratinocytes. J. Biol. Chem. 272:19059-19066. [DOI] [PubMed] [Google Scholar]
- 24.Lee, S.-T., E. Kessler, and D. S. Greenspan. 1990. Analysis of site-directed mutations in human pro-α2(I) collagen which block cleavage by the C-proteinase. J. Biol. Chem. 265:21992-21996. [PubMed] [Google Scholar]
- 25.Linsenmayer, T. F., E. Gibney, F. Igoe, M. K. Gordon, J. M. Fitch, L. I. Fessler, and D. E. Birk. 1993. Type V collagen: molecular structure and fibrillar organization of the chicken α1(V) NH2-terminal domain, a putative regulator of corneal fibrillogenesis. J. Cell Biol. 121:1181-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masuda, M., H. Igarashi, M. Kano, and H. Yoshikura. 1998. Effects of procollagen C-proteinase enhancer protein on the growth of cultured rat fibroblasts revealed by an excisable retroviral vector. Cell Growth Differ. 9:381-391. [PubMed] [Google Scholar]
- 27.Moali, C., B. Font, F. Ruggiero, D. Eichenberger, P. Rousselle, V. Francois, A. Oldberg, L. Bruckner-Tuderman, and D. J. S. Hulmes. 2005. Substrate-specific modulation of a multisubstrate proteinase. J. Biol. Chem. 280:24188-24194. [DOI] [PubMed] [Google Scholar]
- 28.Mott, J. D., C. L. Thomas, M. T. Rosenbach, K. Takahara, D. S. Greenspan, and M. J. Banda. 2000. Post-translational proteolytic processing of procollagen C-terminal proteinase enhancer releases a metalloproteinase inhibitor. J. Biol. Chem. 275:1384-1390. [DOI] [PubMed] [Google Scholar]
- 29.Pappano, W. N., B. M. Steiglitz, I. C. Scott, D. R. Keene, and D. S. Greenspan. 2003. Use of Bmp1/Tll1 doubly homozygous null mice and proteomics to identify and validate in vivo substrates of BMP-1/Tolloid-like metalloproteinases. Mol. Cell. Biol. 23:4428-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petropoulou, V., L. Garrigue-Antar, and K. E. Kadler. 2005. Identification of the minimum domain structure of bone morphogenetic protein-1 (BMP-1) for chordinase activity: chordinase activity is not enhanced by procollagen C-proteinase enhancer-1 (PCPE-1). J. Biol. Chem. 280:22616-22623. [DOI] [PubMed] [Google Scholar]
- 31.Prockop, D. J., and D. J. S. Hulmes. 1994. Assembly of collagen fibrils de novo from soluble precursors: polymerization and copolymerization of procollagen, pN-collagen, and mutated collagens, p. 47-90. In P. D. Yurchenco, D. E. Birk, and R. P. Mecham (ed.), Extracellular matrix assembly and structure. Academic Press, New York, N.Y.
- 32.Scott, I. C., Y. Imamura, W. N. Pappano, J. M. Troedel, A. D. Recklies, P. J. Roughley, and D. S. Greenspan. 2000. Bone morphogenetic protein-1 processes probiglycan. J. Biol. Chem. 275:30504-30511. [DOI] [PubMed] [Google Scholar]
- 33.Steiglitz, B. M., D. R. Keene, and D. S. Greenspan. 2002. PCOLCE2 encodes a functional procollagen C-proteinase enhancer (PCPE2) that binds collagen and differs in distribution of expression, and post-translational modification from the previously described PCPE1. J. Biol. Chem. 277:49820-49830. [DOI] [PubMed] [Google Scholar]
- 34.Steiglitz, B. M., M. Ayala, K. Narayanan, A. George, and D. S. Greenspan. 2004. Bone morphogenetic protein-1/Tolloid-like proteinases process dentin matrix protein-1. J. Biol. Chem. 279:980-986. [DOI] [PubMed] [Google Scholar]
- 35.Suzuki, N., P. A. Labosky, Y. Furuta, L. Hargett, R. Dunn, A. B. Fogo, K. Takahara, D. M. P. Peters, D. S. Greenspan, and B. L. M. Hogan. 1996. Failure of ventral body wall closure in mouse embryos lacking a procollagen C-proteinase encoded by Bmp1, a mammalian gene related to Drosophila tolloid. Development 122:3587-3595. [DOI] [PubMed] [Google Scholar]
- 36.Takahara, K., E. Kessler, L. Biniaminov, M. Brusel, R. L. Eddy, S. Jani-Sait, T. B. Shows, and D. S. Greenspan. 1994. Type I procollagen COOH-terminal proteinase enhancer protein: identification, primary structure, and chromosomal localization of the cognate human gene (PCOLCE). J. Biol. Chem. 269:26280-26285. [PubMed] [Google Scholar]
- 37.Toriello, H. V., T. W. Glover, K. Takahara, P. H. Byers, D. E. Miller, J. V. Higgins, and D. S. Greenspan. 1996. A translocation interrupts the COL5A1 gene in a patient with Ehlers-Danlos syndrome and hypomelanosis of Ito. Nat. Genet. 13:361-365. [DOI] [PubMed] [Google Scholar]
- 38.Tybulewicz, V. L. J., C. E. Crawford, P. K. Jackson, R. T. Bronson, and R. C. Mulligan. 1991. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell 65:1153-1163. [DOI] [PubMed] [Google Scholar]
- 39.Unsöld, C., W. N. Pappano, Y. Imamura, B. M. Steiglitz, and D. S. Greenspan. 2002. Biosynthetic processing of the pro-α1(V)2pro-α2(V) collagen heterotrimer by bone morphogenetic protein-1 and furin-like proprotein convertases. J. Biol. Chem. 277:5596-5602. [DOI] [PubMed] [Google Scholar]
- 40.Vogel, K. G., and J. A. Trotter. 1987. The effect of proteoglycans on the morphology of collagen fibrils formed in vitro. Collagen Relat. Res. 7:105-114. [DOI] [PubMed] [Google Scholar]
- 41.Vogel, K. G., M. Paulsson, and D. Heinegård. 1984. Specific inhibition of type I and type II collagen fibrillogenesis by the small proteoglycan of tendon. Biochem. J. 223:587-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wenstrup, R. J., J. B. Florer, E. W. Brunskill, S. M. Bell, I. Chervoneva, and D. E. Birk. 2004. Type V collagen controls the initiation of collagen fibril assembly. J. Biol. Chem. 279:53331-53337. [DOI] [PubMed] [Google Scholar]
- 43.Xu, H., T. S. Acott, and M. K. Wirtz. 2000. Identification and expression of a novel type I procollagen C-proteinase enhancer protein gene from the glaucoma candidate region on 3q21-q24. Genomics 66:264-273. [DOI] [PubMed] [Google Scholar]