Abstract
PC5 belongs to the proprotein convertase family and activates precursor proteins by cleavage at basic sites during their transit through the secretory pathway and/or at the cell surface. These precursors include prohormones, proreceptors, growth factors, adhesion molecules, and viral glycoproteins. The Pcsk5 gene encodes two alternatively spliced isoforms, the soluble PC5A and transmembrane PC5B. We have carefully analyzed the expression of PC5 in the mouse during development and in adulthood by in situ hybridization, as well as in mouse tissues and various cell lines by quantitative reverse transcription-PCR. The data show that adrenal cortex and intestine are the richest sources of PC5A and PC5B, respectively. To better define the specific physiological roles of PC5, we have generated a mouse Pcsk5Δ4-deficient allele missing exon 4 that encodes the catalytic Asp173. While Δ4/+ heterozygotes were healthy and fertile, genotyping of progeny obtained from Δ4/+ interbreeding indicated that Δ4/Δ4 embryos died between embryonic days 4.5 and 7.5. These data demonstrate that Pcsk5 is an essential gene.
A family of subtilisin-like proteases is responsible for the processing of precursor proteins that transit through the secretory pathway of mammalian cells. This family comprises nine members: PC1/3, PC2, furin, PC4, PACE4, PC5/6, PC7, SKI-1/S1P, and NARC-1/PCSK9 (29, 30, 32). The first seven proprotein convertases (PCs) are more closely related to each other than to SKI-1 and NARC-1. They belong to the kexin subfamily (SKI-1 and NARC-1 belong to the pyrolysin and proteinase K subfamilies, respectively) and share a cleavage specificity for basic motifs of the type (K/R)-(X)n-(K/R)↓, in which n = 0, 2, 4, or 6. The basic residues present at the C terminus of the smaller product are usually trimmed out by a carboxypeptidase that generates an active molecule, although, in some cases, additional posttranslational modifications may be required (for a review, see reference 13). PCs themselves are synthesized as precursors. Autocatalytic cleavage at one or two intramolecular sites allows the activation of the zymogen by removal of its N-terminal prosegment. The latter acts as an intramolecular chaperone for folding of the enzyme and as a transient in trans inhibitor. PC1/3 and PC2 are present in neuroendocrine tissues, where they contribute to activate hormones or neuropeptides. PC4 is restricted to gonads. Although widespread, the other basic motif-specific PCs exhibit different and contrasting expression patterns (31). In cell lines, coexpression of candidate precursors with these PCs revealed that, in many cases, substrates preferentially processed by PC5 are also efficiently cleaved by furin, PC7, and to a lesser extent by PACE4. This is the case in some α chains of integrins (1); the neural adhesion molecule L1 (17); transforming growth factor β-like proteins (10, 36), including Müllerian inhibiting substance (22); the receptor protein tyrosine phosphatase RPTPμ (5); membrane-bound metalloproteinases such as ADAM-17 (34); and possibly the membrane type-1 matrix metalloproteinase MT1-MMP (39). This ex vivo redundancy probably reflects the ability of these enzymes to compensate for each other in vivo in a tissue-specific manner. In this context, the knockout of these enzymes is very informative. Furin expression is essential during development since its knockout leads to embryonic lethality around embryonic day 11 (E11) (28). However, its liver-specific knockout had no effect on the viability of mice, and in this organ, several candidates for substrates of furin were still well processed (27).
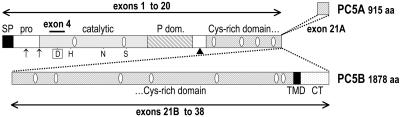
The mouse PC5 gene, Pcsk5, encodes two isoforms generated by alternative splicing, PC5A and PC5B (Fig. 1). They share their first 20 exons, which encode the signal peptide, prosegment, catalytic domain, and N-terminal region of the Cys-rich domain (CRD). A specific additional exon (21A) encodes the last 38 residues of PC5A, while exons 21B to 38 encode the last 1,000 residues of PC5B. PC5A and its closest family member, PACE4, share similar C-terminal CRDs. The latter were recently shown (26) to be responsible for the interaction of these enzymes with cell surface heparan sulfate proteoglycans (HSPGs) via tissue inhibitors of metalloproteases (TIMPs). This agrees with the cell surface localization of most PC5 candidate substrates.
FIG. 1.
Isoforms PC5A and PC5B and their processing. Exons 1 to 20 of the mouse Pcsk5 gene encode the signal peptide (SP; amino acids [aa] 1 to 34), prosegment (aa 35 to 116), catalytic domain (aa 117 to 458), P domain (aa 459 to 600), and N-terminal part of the Cys-rich domain (aa 638 to 877). Exon 21A, specific to PC5A, encodes its last 38 residues, while the 18 additional exons of PC5B (21B to 38) extend the Cys-rich domain up to a transmembrane domain (TMD; aa 1769 to 1789) followed by an 88-aa-long cytosolic tail (CT). The residues of the catalytic triad comprise Asp173 (boxed D), encoded by exon 4; His214 (H); and Ser388 (S). The position of the oxyanion hole Asn315 (N) is also shown. Arrows in the prosegment indicate autocatalytic cleavage sites, and the arrowhead points to the intracellular cleavage site, generating an ∼68-kDa PC5A-ΔC form. White ellipses emphasize the putative N-glycosylation sites (Asn 227, 383, 667, 754, 804, 854, 951, 1016, 1220, 1317, 1523, 1711, and 1733).
To date, only furin (28) and SKI-1/S1P (40) have been shown to be essential genes. Knockout of PC1 (42), PC2 (14), PC4 (21), PACE4 (6), or PC7 (37) genes resulted in viable mice to various degrees, with no or relatively mild phenotypes. To discriminate between substrates specific to PC5 or common to PC5 and other PCs, we set up a strategy aimed at deleting a 4.5-kb genomic fragment containing exon 4 that encodes the catalytic Asp173. Our data show that the absence of catalytically functional PC5 leads to early embryonic death, indicating that, like the genes for furin (Pcsk3) and SKI-1 (Mbtps1), Pcsk5 is essential.
MATERIALS AND METHODS
ISH study.
In situ hybridization (ISH) was performed as follows. Ten- to 12-μm-thick cryosections were prepared from embryos at different stages of development or from adult mice (whole-body sections), fixed in 4% formaldehyde, and hybridized as previously described (20) with mouse sense (negative control) and antisense cRNA probes. The latter probes (807 bases) correspond to the mouse PC5 coding region for residues 80 to 348 and were synthesized using 35S-UTP and 35S-CTP (>1,000 Ci/mmol; Amersham Biosiences, Piscataway, NJ).
Quantitative analysis of the PC5 transcripts in cell lines and tissues.
Cells, which were washed three times with phosphate-buffered saline (PBS), and fresh mouse tissues were directly incubated with Trizol reagent (Invitrogen, Burlington, Ontario, Canada). Total RNA was extracted as recommended by the manufacturer. RNA integrity, cDNA synthesis, and quantitative PCR (QPCR) were performed as previously described (11). For each tissue or cell line, one to three RNA samples were analyzed at least twice in triplicates. Specific primers sitting on different exons were used for the simultaneous amplification of the normalizing cDNA for ribosomal protein S16 (11) and the cDNA for either PC5A+B, PC5A, or PC5B messengers: 5′-ACTCTTCAGAGGGTGGCTA versus 5′-GCTGGAACAGTTCTTGAATC and 5′-TGACCACTCTTCAGAGAATGGATAC versus 5′-GAGATACCCACTAGGGCAGC for human and mouse PC5A+B, respectively; 5′-AGGATTCAAGAACTGTTCCA versus 5′-AGCATACAGAAGCCTCCTT for mouse PC5A; and 5′-GCAATGCCTCCCACTCCC versus 5′-TGCTCGTAAAACTCAGCCTCC for mouse PC5B.
Construction of the targeting vector.
The 12-kb Ecl136II-XhoI genomic region comprising exon 4 was subcloned in three steps in the 38LoxPNeo vector containing a PGK-neo cassette framed with loxP sites (IncyteGenomics). First, the 5′ arm, a 4,061-bp Ecl136II fragment, was subcloned upstream of the loxP1 site in the vector digested with EcoRI and filled (Ecl136II recognizes the same site as SacI and generates blunt ends). Second, a 4,427-bp EcoRV fragment was inserted downstream of the loxP1 site in a filled HindIII site, leading thus to the replacement of 66 bp of genomic DNA that lay between the Ecl136II and EcoRV sites by the 96-bp loxP1 segment. Finally, the 3′ arm, a 3,454-bp EcoRV-filled XhoI fragment, was subcloned downstream of the loxP2 site in a blunt SfiI site. Digestion of the targeting vector with NotI (5′ region of the polylinker) and KpnI (1,100 bp upstream of the XhoI site in the 3′ arm) generated a 12,869-bp insert that was gel purified and transfected into R1 embryonic stem (ES) cells, (129/Sv × 129/Sv-CP)F1 (23).
Creation of a PC5-null allele.
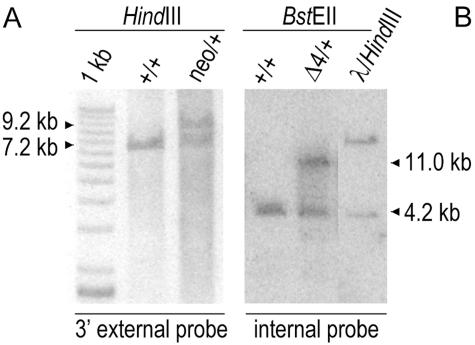
ES cells were selected for G418 resistance, and about 600 clones were digested by HindIII and analyzed by Southern blotting using an XhoI-HindIII 507-bp probe that lies downstream of the 3′ arm. Six positive clones were identified by the presence of 9.2- and 7.2-kb bands corresponding to Pcsk5neo and wild-type alleles, respectively. After expansion and further analysis, all of the positive clones presented multiple insertions at the Pcsk5 locus. However, the analysis of one of them, 5B10, revealed correct 5′ and 3′ insertion sites of the tandem repeats by using external 5′ and 3′ probes and digestion with various restriction enzymes. This ES clone was thus injected into C57BL/6 blastocysts to produce chimeras that successfully transmitted the neo allele when mated to C57BL/6 mice. Heterozygous mice were then crossed with cytomegalovirus (CMV)-Cre-expressing transgenic mice (12), leading to the excision of the exon 4 (Pcsk5Δ4 allele).
Probes and PCR for ES characterization and genotyping mice.
The 5′ and 3′ external probes were EcoRI-EcoNI (652 bp) and XhoI-HindIII (507 bp) fragments, both located outside of the region included in the targeting vector. Two internal probes were used either located in the neomycin resistance gene or downstream of the neo cassette insertion site (482 bp starting from the EcoRV site). For PCR genotyping of mice, specific pairs of primers for wild type (WT) (5′-AGCCTGTGGGCACATGTACGTT versus 5′AGAGCCCTGAGACCTCAATGAGAA) and Δ4 (5′TTGGTGAGAATTCCGATCATATTCAA versus 5′TGCACTTTGCCCATTCCAGA) alleles were used.
Animals.
PC5 wild-type and heterozygous mice in the C57BL/6 background (mimimum of five successive backcrosses) were housed in a 12-h dark cycle and fed a standard laboratory diet (Charles River rodent chow 5075; Purina Mills, St. Louis, MO). Mice were genotyped by PCR analysis of tail DNA. The institutional Animal Care and Ethics Committee approved all procedures described below.
PCR genotyping of embryos.
PC5Δ4/+ females were superstimulated with 5 IU of pregnant mare serum gonadotropin (PMSG) and superovulated with 5 IU of human chorionic gonadotropin (hCG) 47 h later (23). They were then mated to PC5Δ4/+ males, and embryos were either collected the next morning at E0.5 in the ampulla of the oviduct or dissected at E7.5. After a brief treatment with ∼300 mg/ml hyaluronidase, E0.5 embryos were washed in potassium simplex optimized medium (KSOM) containing 20 mM HEPES, pH 7.4, transferred in 20-μl droplets of KSOM (25 to 30 embryos/droplet) overlaid with mineral oil, and incubated for 5 days at 37°C (3). Each blastocyst was recovered in 5 μl of PBS and incubated for 1 h at 95°C. Because of the small amount of DNA available, genotypes were analyzed by a nested PCR using 1 μl of matrix and the primers used for genotyping (see above) and, for nesting, the following primers: 5′-CACTTGGCTACAAATATGCGAAC versus 5′-TTTCAGCATTCTAGGTTTCCAGA and 5′-GGTCCCTCGAAGAGGTTCA versus 5′-CTCACAGGAAACACAGGCACT for detection of the WT and Δ4 alleles, respectively.
PC5Δ4-expressing vector.
Deletion of the sequence corresponding to exon 4 in a pIRES2-enhanced green fluorescent protein (EGFP) recombinant vector containing a mouse PC5A cDNA (pIR-mPC5A-V5) was achieved by a two-step PCR (15). Two sense and antisense primers reconstituting the exon 3-to-exon 5 junction (5′-GTATGTGGTACATGGATGCTCTGGCAAGTTGCG and 5′-GCAACTTGCCAGAGCATCCATGTACCACATACTTGGCC) and two distal primers, located upstream of a unique EcoRI site in the polylinker (sense, 5′-CGGACTCAGATCTCGAGC) and downstream of a unique BsmBI in the mPC5A sequence (antisense, 5′-CACATCGCAACTTGCCATGTACCACATACTTGGCC), led to the final amplification of a 1,180-bp fragment that was subcloned in a pDRIVE vector (QIAGEN, Mississauga, Ontario, Canada). The recombinant pDRIVE was then double digested with EcoRI and BsmBI to generate a 1,121-bp fragment that substituted for the original one containing exon 4 in pIR-mPC5A-V5.
Cell culture, transfections, Western blotting, and antibodies.
Human embryonic kidney HEK293 cells were grown in Dulbecco's modified Eagle's medium containing 10% heat-inactivated fetal calf serum (Canadian Life Technologies, Burlington, Ontario, Canada) at 37°C in 5% CO2. Cells (1 × 106 plated on 35-mm plates) were transfected with plain or recombinant vectors (0.6 μg) using Effectene (QIAGEN, Mississauga, Ontario, Canada). The medium was replaced 12 h after transfection by a serum-free medium, and cells were grown for an additional 24 h before being harvested for protein extraction in 1× radioimmunoprecipitation assay (RIPA) buffer. Media were also collected for Western blot analysis and activity measurements. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to nitrocellulose filters (Hybond-ECL; Amersham Biosciences). After blocking with 5% dry milk in Tris-buffered saline containing 0.05% Tween 20, filters were incubated with anti-V5-horseradish peroxidase conjugate (InVitrogen, Carlsbad, CA) at a dilution of 1:5,000. The antigen-antibody complexes were revealed using an enhanced chemiluminescence kit (ECL Plus; Amersham Biosciences). For immunoprecipitations, media or protein extracts were diluted and adjusted at 0.5× RIPA buffer, incubated overnight at 4°C with an anti-prosegment of PC5 (ppPC5) (25) at 1/100, and further incubated 2 h with 20 μl of protein A/G-agarose (Santa-Cruz Biotechnology, Santa Cruz, CA). The beads were then washed twice with 1× RIPA buffer and a third time with PBS before being boiled in loading buffer.
In vitro PC5 activity.
Supernatants (60 μl) of HEK293 cells transfected with the empty or PC5A- or PC5Δ4-expressing vectors were incubated in presence of a mixture of 50 μM fluorogenic substrate pyruvic acid-RTKR-7-amido-4-methylcomarine, 2 mM CaCl2, 50 mM Tris (pH 7), and 100 μM β-mercaptoethanol in a total volume of 100 μl after a 20-min preincubation of the 40-μl mixture. Fluorescence emission was detected at 460 nm, as described previously (25).
RESULTS
Tissue distribution of PC5 during development and in adulthood.
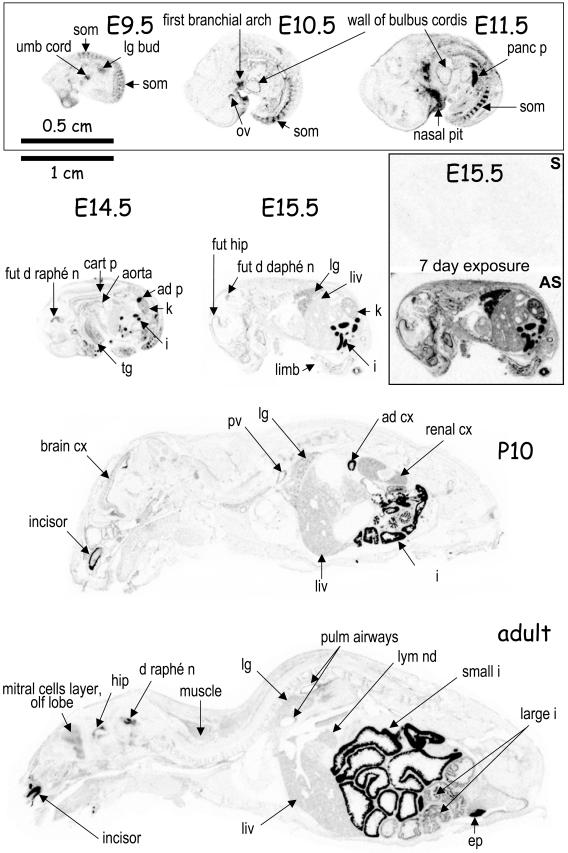
Since the present work deals with inactivation of the Pcsk5 gene, we have characterized by ISH and/or QPCR the expression of mouse PC5 during development and in adulthood (Fig. 2). The first stage that we were able to examine by ISH was E9.5, where labeling was observed in the somites, bulb of umbilical cord, and lung bud. At E10.5 to E11.5, the first bronchial arch, wall of bulbus cordis, and the nasal pit were also labeled, as well as the pancreatic primordium at E11.5. The adrenal primordium, visible at E14.5, is already one of the richest expression sites of PC5/6. By E15.5, the PC5 pattern of expression becomes similar to that of the adult, with strong labeling in small intestine, kidney, and lung. In the developing brain, the future hippocampus (CA3 region) and dorsal raphe nucleus are well labeled (E14.5 to E15.5). The strong labeling of incisors, already observed at E19.5 (data not shown), is maintained at P10 and in the adult. After birth, at P4 (data not shown) and at P10, the adrenal cortex is strongly labeled. Note also that several layers of the brain cortex are PC5 positive at P10. Finally, PC5 is rich in vasculature (P10 and adult), including the aorta, which is labeled as early as E14.5.
FIG. 2.
PC5 expression during mouse development and in adulhood. Distribution of PC5 transcripts was obtained by ISH of cryosections with a 32P-labeled specific probe. Embryos at E9.5, E10.5, and E11.5 are twice enlarged compared to the others. In the E15.5 inset, a longer exposure of sections hybridized with either the control sense (S) or antisense (AS) probe is shown. The main sites of expression are indicated as follows: ad, adrenal; ad p, adrenal primordium; cart p, cartilage primordium; cx, cortex; ep, epididymis; k, kidney; fut d raphé n, future dorsal raphe nucleus; hip, hippocampus; i, intestine; liv, liver; lg, lung; lym nd, lymphatic node; ov, otic vesicle; panc p, pancreas primordium; pv, portal vein; pulm, pulmonary; som, somite; tg, tongue; umb cord, umbilical cord.
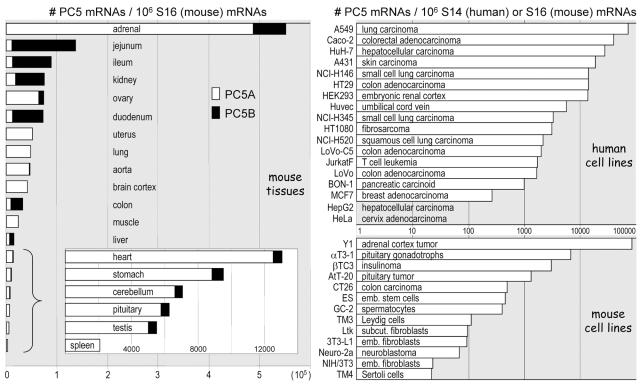
Many of these data were confirmed and extended by QPCR estimation of the mRNA expression of each PC5 isoform in adult mouse tissues (Fig. 3). Among the tissues examined, the adrenal was the richest source of PC5, with four- to sixfold-higher levels than the small intestine. Interestingly, with the exception of the liver that contains equal levels of each isoform, the tissues can be distributed in two groups. The first one, in which PC5A dominates (88 to 100%), comprises the adrenal, ovary, uterus, lung, aorta, brain cortex, muscle, heart, stomach, cerebellum, pituitary, testis, and spleen. The other group is characterized by elevated levels of PC5B (74 to 92%) and comprises the jejunum, ileum, kidney, duodenum, and colon. Finally, we estimated PC5 levels in various human and mouse cell lines. While there is no strict correlation between PC5 levels in cell lines and the tissues from which they originate, it is notable that the most highly expressing line, Y1, is derived from a tumor of the adrenal cortex. Two cell lines, HepG2 and HeLa, seem totally devoid of PC5 expression and may thus constitute valuable tools to analyze the requirement of PC5 activity for the processing of candidate substrates.
FIG. 3.
PC5 expression in tissues and cell lines. Using QPCR, mRNAs of isoforms A (white bars) and B (black bars) of PC5 were independently quantified in tissues (primers in exons 20/21 and 37/38, respectively), while a global estimation of A and B isoforms (primers in exons 19/20) was performed for cell lines. For the last six tissues in which PC5 levels were low, an inset was added showing an enlargement of the levels of PC5A and PC5B. Note that a linear scale was used for tissues (left panel) and a logarithmic one for cell lines (right panel).
Production of PC5-null mice.
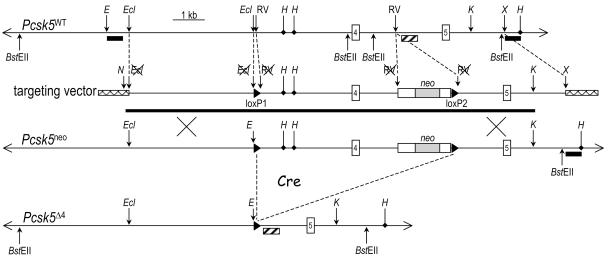
A Pcsk5 allele was modified in ES cells by homologous recombination using a targeting vector containing about 12 kb of genomic DNA, in which the exon 4 is framed with two loxP sites, the 3′ one being adjacent to a cassette allowing expression of neomycin phosphotransferase under the control of the PGK promoter (Fig. 4). We chose to target exon 4 since it contains one of the three catalytic residues, the Asp173 that is essential for enzyme activity. Following electroporation of ES cells, screening of about 600 G418-resistant ES clones led to the identification of six positive clones. Using a 3′ external probe, they were characterized by the presence of 7.2- and 9.2-kb bands corresponding to wild-type and Pcsk5neo alleles, respectively (Fig. 5A). Unfortunately, further analysis with various internal probes revealed multiple insertions (xneo) in all six clones. Only one of them, 5B10, had its tandem repeats correctly inserted when checked by using internal as well as 5′ and 3′ external probes, following digestion with various restriction enzymes (data not shown). This ES clone was thus injected into C57BL/6 blastocysts to produce chimeras that successfully transmitted the Pcsk5xneo allele when mated to C57BL/6 mice. However, intercrossing of xneo/+ mice generated no xneo/xneo mice, indicating that the presence of the multiple insertions at the Pcsk5 locus led to embryonic lethality. Out of 107 mice, 62% and 42% exhibited the genotypes xneo/+ and +/+, respectively. We thus decided to completely excise these multiple insertions by two successive crosses with CMV-Cre transgenic mice (12), leading to the deletion of ∼4.5 kb comprising exon 4 (Pcsk5Δ4 allele), as revealed by digestion of genomic DNA with BstEII that cleaves outside of the region corresponding to the transfected insert (Fig. 5B). Accordingly, this resulted in a nonconditional knockout of the Pcsk5 gene. In order to eliminate the possibility of rearrangements elsewhere in the genome, we backcrossed Δ4/+ mice to C57BL/6 mice for a minimum of 5 generations.
FIG. 4.
Pcsk5 knockout strategy. Three genomic DNA fragments covering 12 kb of the Pcsk5 locus were subcloned into the targeting vector, in which the exon 4 is flanked with two loxP sites (open triangles) and a PGK-neo cassette. ES cells were transfected with the NotI-KpnI insert (solid line). The expected neo allele is shown. Mating of heterozygotes bearing a Pcsk5neo recombinant allele with transgenic mice expressing the Cre recombinase generated progeny in which Cre was expressed and excised the sequences between the loxP sites. The generated Pcsk5Δ4-inactivated allele misses ∼4.5 kb of genomic DNA, including exon 4. E, EcoRI; RV, EcoRV; Ecl, Ecl136II; H, HindIII; K, KpnI; N, NotI; X, XhoI.
FIG. 5.
Genomic analysis of 5B10-derived mice. Genomic DNA (6 μg) from C57BL/6 (+/+), 5B10-derived (neo/+), and Δ4/+ heterozygotes issued from mating of 5B10-derived mice to Cre-expressing ones was digested and analyzed by Southern blotting. Using a 3′ external probe and HindIII digestion, wild-type and neo recombinant alleles generate 7.2- and 9.2-kb fragments, respectively. Using an internal probe and BstEII digestion, wild-type and Δ4 alleles generate 4.2- and 11-kb fragments, respectively. The lanes marked “1 kb” and “λ/HindIII” contain specific ladders.
The Δ4/Δ4 genotype is embryonic lethal.
No homozygous mice were obtained from Δ4/+ intercrosses. Out of 204 mice, 65% were Δ4/+ and 35% were +/+, as expected if the Δ4/Δ4 genotype leads to embryonic death, and no bias was observed between male and female progeny (Table 1, data for 3 weeks). To determine the stage of lethality, heterozygous males were mated with stimulated females and the 35 embryos isolated at E14.5 were genotyped: 77% were Δ4/+ and 23% were +/+, indicating that lethality had occurred before this developmental stage. In a second step, 36 embryos were carefully dissected at E7.5 and their genotyping also revealed the absence of any Δ4/Δ4 embryos. Finally, zygotes were collected in the ampulla of the oviduct at E0.5 and cultured for 5 days at 37°C to obtain blastocysts. Their genotyping revealed a Mendelian distribution of the +/+, Δ4/+, and Δ4/Δ4 genotypes (Table 1), indicating that oocyte fertilization and cell division in the early embryo were efficiently achieved, independent of the PC5 genotype. Altogether, our data suggest that embryonic death likely occurs between implantation (∼E4.5) and E7.5.
TABLE 1.
Genotyping of Δ4/+ × Δ4/+ progeny
| Genotype | No. (%) of Δ4/+ intercross progeny
|
|||
|---|---|---|---|---|
| Cultured at E0.5 | E7.5 | E14.5 | 3 wk | |
| +/+ | 13 (24) | 9 (25) | 8 (23) | 72 (35) |
| Δ4/+ | 27 (49) | 27 (75) | 27 (77) | 132 (65) |
| Δ4/Δ4 | 15 (27) | 0 (0) | 0 (0) | 0 (0) |
| Total | 55 (100) | 36 (100) | 35 (100) | 204 (100) |
Analysis of Δ4/+ PC5 mRNA and protein.
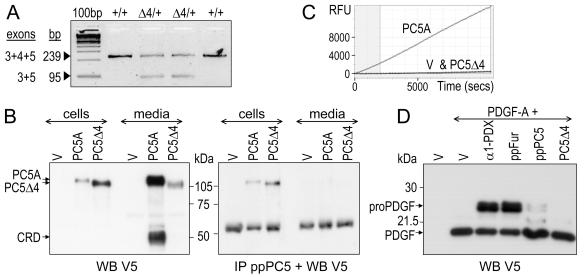
To follow the fate of the PC5 mRNA, QPCR analysis was performed on RNA isolated from either +/+ or Δ4/+ livers. Primers hybridizing to exons 3 and 5 were used since connection of neighboring exons was reported to occur upon exon deletion (4, 14). Indeed, the presence of a lower band (95 bp) in Δ4/+ samples and sequencing of the corresponding DNA revealed that exons 3 and 5 are fused in the mRNA deriving from the recombinant allele (Fig. 6A). In addition, quantitative analysis showed that the Δ4 mRNA is as well produced as the wild-type one (data not shown), indicating that heterozygotes have 50% of mRNA coding for active PC5 compared to that in wild-type mice. Since exons 3 and 5 share the same reading frame, exon 4 deletion may lead to the synthesis of a truncated PC5Δ4 protein lacking 48 residues (from His140 to Tyr187). To check the cellular fate of PC5Δ4, a recombinant pIRES2-EGFP vector containing a cDNA encoding mouse PC5A C-terminally tagged with V5 was modified to mimic the exon 4 deletion (−144 bp). In transfected HEK293 cells and their media, PC5Δ4 showed a slightly lower apparent molecular mass than PC5A, compatible with the loss of 48 residues (∼5 kDa; Fig. 6B). Microsequencing revealed that proPC5A is the major intracellular species (25). Since similar ratios of PC5A to PC5Δ4 (experiment done three times) were revealed by V5 Western blotting, without or with prior immunoprecipitation with a PC5 prosegment-specific antibody, we can deduce that proPC5Δ4 is predominant in the cell. In the medium of wild-type PC5A, in addition to the ∼110-kDa PC5A, an ∼50-kDa C-terminal product (V5 positive) was observed, representing the cleaved CRD as previously reported (26). Taking into account the 10-fold-higher loading for the media than for the cell lysates, we estimated that while most of PC5A was secreted, only ∼5% of PC5Δ4 was found in the medium. Although the same quantity of DNA was used for transfection, the total amount of PC5Δ4 detected was much lower than that of PC5A, suggesting that PC5Δ4 was largely degraded. Interestingly, the secreted fraction of PC5Δ4, which migrated faster than PC5A, underwent zymogen processing since otherwise proPC5Δ4 would have been ∼4 kDa heavier than PC5A. This indicates that proPC5Δ4 can exit from the endoplasmic reticulum and be processed in trans. The inactivity of PC5Δ4 in HEK293 media, although expected from the loss of one of the catalytic residues, wasassessed using the fluorogenic substrate Pyr-RTKR-AMC. PC5Δ4 activity was not detected since it was found equivalent to that of control medium (empty vector), which was at least 30-fold lower than that obtained from PC5Α (Fig. 6C).
FIG. 6.
PC5Δ4 expression and activity. (A) QPCR analysis of PC5 mRNA from +/+ and Δ4/+ livers using primers located in exons 3/5 revealed a unique 239 bp product in +/+ liver and an additional 95-bp one in Δ4/+ liver resulting from an altered splicing connecting exons 3 and 5. (B) HEK293 cells were transfected with the pIRES2-EGFP vector empty (V) or expressing wild-type PC5A or PC5Δ4, both C-terminally V5 tagged. Cell extracts (20 μg) and media (40 μl out of 1 ml) were analyzed by anti-V5 Western blotting (WB) prior (left panel) or after (right panel) immunoprecipitation (IP) with a prosegment antibody (anti-ppPC5). (C) Online PC5 activity in relative fluorescence units (RFU) was measured by incubation of 60 μl of medium with the fluorogenic substrate Pyr-RTKR-AMC. Note the absence of activity in PC5Δ4 and control V media. (D) HEK293 cells were cotransfected with 0.1 μg of a vector expressing PDGF-A carrying a C-terminal V5 tag and 0.9 μg of a vector expressing either no product (V), α1-PDX, the prosegment of furin (ppFur), the prosegment of PC5 (ppPC5), or PC5Δ4. Media (2 μl out of 1 ml) were analyzed by Western blotting using V5 antibodies.
Embryonic lethality is expected to result from the loss of PC5 activity. However, since a truncated form of PC5 is produced from the Δ4 allele, the possible in trans inhibitory effect of PC5Δ4 on the activity of the other PCs, namely furin, whose knockout is also embryonic lethal, was assessed. For this, the extent of cleavage of a V5-tagged pro-PDGF-A, which is efficiently processed by furin in HEK293 cells (33), was estimated in the absence or in presence of overexpressed α1-PDX, the prosegment of furin, or the prosegment of PC5 or PC5Δ4 (Fig. 6D). No inhibitory effect of PC5Δ4 could be detected while the PC inhibitors, α1-PDX and the furin prosegment, efficiently inhibited proPDGF-A processing. In addition, only a very mild inhibitory effect of the PC5 prosegment was detected. Finally, even under overexpression conditions, PC5Δ4 is not inhibitory.
DISCUSSION
In the present work, we carefully documented PC5 mRNA expression during mouse development, in adulthood, and in human and mouse cell lines (Fig. 2 and 3). The data agree with and extend those obtained in rats, which show that PC5 distribution is distinct and often complementary to those of furin and PACE4 (8, 9, 19, 31, 41). PC5 is richest in the adrenal cortex, small intestine, kidney, ovary, uterus, lung, aorta, and brain cortex. Our data revealed that, except in the liver where both isoforms are equally expressed, PC5A is the major isoform in most tissues (87 to 100%), and only the intestine and kidney show a predominance of PC5B (74 to 92%). The two isoforms (Fig. 1) were shown to have distinct properties: (i) they do not show similar efficiencies toward various substrates in cell lines (2, 16, 18), and (ii) the transmembrane PC5B is restricted to the constitutive secretory pathway (7) and cycles between the Golgi and the cell surface (38), while the soluble PC5A is sorted to both regulated and constitutive secretory pathways (7). However, PC5A can also be associated with the plasma membrane via a ternary complex involving its CRD, TIMPs, and HSPGs (26). Thus, the distinct tissue distribution of the two isoforms may not be fortuitous but rather may reflect isoform-specific functions.
In order to assess the in vivo functions of PC5, we developed a knockout strategy that led to the inactivation of both isoforms involving deletion of the common exon 4 encoding the catalytic Asp173 (Fig. 4). While heterozygote mice harboring a Pcsk5Δ4 allele (+/Δ4) are healthy and fertile, homozygosity leads to early embryonic lethality that occurs between the blastocyst implantation stage (E4.5) and E7.5 (Table 1), demonstrating that Pcsk5 is an essential gene.
Deletion of exon 4 resulted in a truncated mRNA in which exons 3 and 5 are fused in frame (Fig. 6A). Cellular overexpression of a cDNA mimicking the exon 4 deletion resulted in low levels of PC5Δ4. In addition, the protein was mostly found in the cell as a zymogen, whereas only a small fraction (<5%) was processed in trans and secreted (Fig. 6B). Thus, the level of the inactive PC5Δ4 protein produced from the Pcsk5Δ4 allele is expected to be negligible. Indeed, the available antibodies directed against PC5 did not detect endogenous PC5Δ4 in heterozygotes (data not shown). Furthermore, media obtained from HEK293 cells overexpressing PC5Δ4 revealed no in vitro activity as compared to that of the wild type (Fig. 6C), and there was no inhibitory effect on the ex vivo processing of pro-platelet-derived growth factor A (proPDGF-A) by endogenous PCs (Fig. 6D). Taken together, these data demonstrate that the residual expression of PC5Δ4 is not responsible for the observed lethality. Similarly, PC2 inactivation through disruption of exon 3 also led to an alternative splicing producing proPC2Δ3 that remained as an inactive zymogen in vivo (14). Finally, using the Flp/loxP system, the recent production of a conditional PC5 knockout in which exon 1 was targeted led, in the presence of a CMV-Cre transgene, to the absence of PC5 transcripts and resulted in early lethality of Δ1/Δ1 embryos (Essalmani et al., unpublished data).
Using an antisense morpholino oligonucleotide (24), it was shown that uterine expression of PC5 is required for decidualization at the implantation site in the mouse, but no information is available concerning the requirement for embryonic expression of PC5 at the same stage. Since the litter size does not differ between +/+ and +/Δ4 females (data not shown), we can rule out that the embryonic lethality observed upon intercrosses of +/Δ4 mice is due to a maternal PC5 deficit. This indicates that early de novo synthesis of PC5 in either embryonic or extraembryonic cells is essential. Whether embryonic expression of PC5 is essential for implantation or at later stages remains to be defined. It has been mentioned in a review article (35), but without experimental details, that PC5B inactivation led to embryonic death at E10.5 to E11.5, i.e., at least 3 days after that observed herein upon PC5A+B inactivation. This suggests that PC5B is essential, but the lethality observed in its absence may be delayed by PC5A. Is PC5A also essential? If not, then the key role(s) played by PC5B may reside in extraembryonic giant trophoblasts (41) or unidentified early progenitor cells. In that context, the extensive expression at E9 of PC5, but not furin and PACE4, in the rat materno-embryonic junction, whose trophoblast giant cells derive from the embryo, is compelling (41). Finally, the elucidation of the critical cognate substrate(s) will require the identification of the exact stage and cell types associated with the observed lethality. Future studies will determine if the essential role of PC5 observed during early development also applies to later stages. The available conditional knockout will allow the induction of PC5 inactivation at different stages as well as in specific tissues, thereby leading to the identification of the substrates and physiological roles of PC5.
Acknowledgments
We are very grateful to Qinzhang Zhu for his excellent technical support in handling ES cells and to Claudia Toulouse for expert animal husbandry. We thank David Lohnes for his generous gift of CMV-Cre transgenic mice. We also thank Daniel Dufort for sharing his expertise in dissecting embryos, Antonella Pasquato for in vitro PC5 activity measurements, and Jean Vacher for a critical reading of the manuscript.
This work was entirely supported by CIHR grant MGP-44363 to N.G.S.
REFERENCES
- 1.Bergeron, E., A. Basak, E. Decroly, and N. G. Seidah. 2003. Processing of alpha4 integrin by the proprotein convertases: histidine at position P6 regulates cleavage. Biochem. J. 373:475-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergeron, E., M. J. Vincent, L. Wickham, J. Hamelin, A. Basak, S. T. Nichol, M. Chretien, and N. G. Seidah. 2005. Implication of proprotein convertases in the processing and spread of severe acute respiratory syndrome coronavirus. Biochem. Biophys. Res. Commun. 326:554-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biggers, J. D., L. K. McGinnis, and M. Raffin. 2000. Amino acids and preimplantation development of the mouse in protein-free potassium simplex optimized medium. Biol. Reprod. 63:281-293. [DOI] [PubMed] [Google Scholar]
- 4.Bruneau, B. G., G. Nemer, J. P. Schmitt, F. Charron, L. Robitaille, S. Caron, D. A. Conner, M. Gessler, M. Nemer, C. E. Seidman, and J. G. Seidman. 2001. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell 106:709-721. [DOI] [PubMed] [Google Scholar]
- 5.Campan, M., M. Yoshizumi, N. G. Seidah, M. E. Lee, C. Bianchi, and E. Haber. 1996. Increased proteolytic processing of protein tyrosine phosphatase mu in confluent vascular endothelial cells: the role of PC5, a member of the subtilisin family. Biochemistry 35:3797-3802. [DOI] [PubMed] [Google Scholar]
- 6.Constam, D. B., and E. J. Robertson. 2000. SPC4/PACE4 regulates a TGFbeta signaling network during axis formation. Genes Dev. 14:1146-1155. [PMC free article] [PubMed] [Google Scholar]
- 7.De, B., I. M. Marcinkiewicz, D. Malide, C. Lazure, K. Nakayama, M. Bendayan, and N. G. Seidah. 1996. The isoforms of proprotein convertase PC5 are sorted to different subcellular compartments. J. Cell Biol. 135:1261-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong, W., M. Marcinkiewicz, D. Vieau, M. Chretien, N. G. Seidah, and R. Day. 1995. Distinct mRNA expression of the highly homologous convertases PC5 and PACE4 in the rat brain and pituitary. J. Neurosci. 15:1778-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong, W., B. Seidel, M. Marcinkiewicz, M. Chretien, N. G. Seidah, and R. Day. 1997. Cellular localization of the prohormone convertases in the hypothalamic paraventricular and supraoptic nuclei: selective regulation of PC1 in corticotrophin-releasing hormone parvocellular neurons mediated by glucocorticoids. J. Neurosci. 17:563-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubois, C. M., F. Blanchette, M. H. Laprise, R. Leduc, F. Grondin, and N. G. Seidah. 2001. Evidence that furin is an authentic transforming growth factor-beta1-converting enzyme. Am. J. Pathol. 158:305-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubuc, G., A. Chamberland, H. Wassef, J. Davignon, N. G. Seidah, L. Bernier, and A. Prat. 2004. Statins upregulate PCSK9, the gene encoding the proprotein convertase neural apoptosis-regulated convertase-1 implicated in familial hypercholesterolemia. Arterioscler. Thromb. Vasc. Biol. 24:1454-1459. (First published 3 June 2004; doi: 10.1161/01.ATV.0000134621.14315.43.) [DOI] [PubMed] [Google Scholar]
- 12.Dupe, V., M. Davenne, J. Brocard, P. Dolle, M. Mark, A. Dierich, P. Chambon, and F. M. Rijli. 1997. In vivo functional analysis of the Hoxa-1 3′ retinoic acid response element (3′RARE). Development 124:399-410. [DOI] [PubMed] [Google Scholar]
- 13.Fricker, L. D., and E. H. Leiter. 1999. Peptides, enzymes and obesity: new insights from a ‘dead’ enzyme. Trends Biochem. Sci. 24:390-393. [DOI] [PubMed] [Google Scholar]
- 14.Furuta, M., H. Yano, A. Zhou, Y. Rouille, J. J. Holst, R. Carroll, M. Ravazzola, L. Orci, H. Furuta, and D. F. Steiner. 1997. Defective prohormone processing and altered pancreatic islet morphology in mice lacking active SPC2. Proc. Natl. Acad. Sci. USA 94:6646-6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hospital, V., E. Nishi, M. Klagsbrun, P. Cohen, N. G. Seidah, and A. Prat. 2002. The metalloendopeptidase nardilysin (NRDc) is potently inhibited by heparin-binding EGF-like growth factor (HB-EGF). Biochem. J. 367:229-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin, W., I. V. Fuki, N. G. Seidah, S. Benjannet, J. M. Glick, and D. J. Rader. 2005. Proprotein covertases are responsible for proteolysis and inactivation of endothelial lipase. J. Biol. Chem. 280:36551-36559. [DOI] [PubMed] [Google Scholar]
- 17.Kalus, I., B. Schnegelsberg, N. G. Seidah, R. Kleene, and M. Schachner. 2003. The proprotein convertase PC5A and a metalloprotease are involved in the proteolytic processing of the neural adhesion molecule L1. J. Biol. Chem. 278:10381-10388. (First published 15 January 2003; doi: 10.1074/jbc.M208351200.) [DOI] [PubMed] [Google Scholar]
- 18.Lissitzky, J. C., J. Luis, J. S. Munzer, S. Benjannet, F. Parat, M. Chretien, J. Marvaldi, and N. G. Seidah. 2000. Endoproteolytic processing of integrin pro-alpha subunits involves the redundant function of furin and proprotein convertase (PC) 5A, but not paired basic amino acid converting enzyme (PACE) 4, PC5B or PC7. Biochem. J. 346:133-138. [PMC free article] [PubMed] [Google Scholar]
- 19.Lusson, J., D. Vieau, J. Hamelin, R. Day, M. Chretien, and N. G. Seidah. 1993. cDNA structure of the mouse and rat subtilisin/kexin-like PC5: a candidate proprotein convertase expressed in endocrine and nonendocrine cells. Proc. Natl. Acad. Sci. USA 90:6691-6695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marcinkiewicz, M., J. Marcinkiewicz, A. Chen, F. Leclaire, M. Chretien, and P. Richardson. 1999. Nerve growth factor and proprotein convertases furin and PC7 in transected sciatic nerves and in nerve segments cultured in conditioned media: their presence in Schwann cells, macrophages, and smooth muscle cells. J. Comp. Neurol. 403:471-485. [DOI] [PubMed] [Google Scholar]
- 21.Mbikay, M., H. Tadros, N. Ishida, C. P. Lerner, E. De Lamirande, A. Chen, M. El Alfy, Y. Clermont, N. G. Seidah, M. Chretien, C. Gagnon, and E. M. Simpson. 1997. Impaired fertility in mice deficient for the testicular germ-cell protease PC4. Proc. Natl. Acad. Sci. USA 94:6842-6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nachtigal, M. W., and H. A. Ingraham. 1996. Bioactivation of Mullerian inhibiting substance during gonadal development by a kex2/subtilisin-like endoprotease. Proc. Natl. Acad. Sci. USA 93:7711-7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagy, A., M. Gersenstein, K. Vintersten, and R. Behringer. 2003. Manipulating the mouse embryo. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Nie, G., Y. Li, M. Wang, Y. X. Liu, J. K. Findlay, and L. A. Salamonsen. 2005. Inhibiting uterine PC6 blocks embryo implantation: an obligatory role for a proprotein convertase in fertility. Biol. Reprod. 72:1029-1036. [DOI] [PubMed] [Google Scholar]
- 25.Nour, N., A. Basak, M. Chretien, and N. G. Seidah. 2003. Structure-function analysis of the prosegment of the proprotein convertase PC5A. J. Biol. Chem. 278:2886-2895. [DOI] [PubMed] [Google Scholar]
- 26.Nour, N., G. Mayer, J. S. Mort, A. Salvas, M. Mbikay, C. J. Morrison, C. M. Overall, and N. G. Seidah. 2005. The cysteine-rich domain of the secreted proprotein convertases PC5A and PACE4 functions as a cell surface anchor and interacts with tissue inhibitors of metalloproteinases. Mol. Biol. Cell 16:5215-5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roebroek, A. J., N. A. Taylor, E. Louagie, I. Pauli, L. Smeijers, A. Snellinx, A. Lauwers, W. J. Van De Ven, D. Hartmann, and J. W. Creemers. 2004. Limited redundancy of the proprotein convertase furin in mouse liver. J. Biol. Chem. 279:53442-53450. [DOI] [PubMed] [Google Scholar]
- 28.Roebroek, A. J., L. Umans, I. G. Pauli, E. J. Robertson, F. van Leuven, W. J. Van De Ven, and D. B. Constam. 1998. Failure of ventral closure and axial rotation in embryos lacking the proprotein convertase furin. Development 125:4863-4876. [DOI] [PubMed] [Google Scholar]
- 29.Seidah, N. G., S. Benjannet, L. Wickham, J. Marcinkiewicz, S. B. Jasmin, S. Stifani, A. Basak, A. Prat, and M. Chretien. 2003. The secretory proprotein convertase neural apoptosis-regulated convertase 1 (NARC-1): liver regeneration and neuronal differentiation. Proc. Natl. Acad. Sci. USA 100:928-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seidah, N. G., and M. Chretien. 1999. Proprotein and prohormone convertases: a family of subtilases generating diverse bioactive polypeptides. Brain Res. 848:45-62. [DOI] [PubMed] [Google Scholar]
- 31.Seidah, N. G., M. Chretien, and R. Day. 1994. The family of subtilisin/kexin like pro-protein and pro-hormone convertases: divergent or shared functions. Biochimie 76:197-209. [DOI] [PubMed] [Google Scholar]
- 32.Seidah, N. G., and A. Prat. 2002. Precursor convertases in the secretory pathway, cytosol and extracellular milieu. Essays Biochem. 38:79-94. [DOI] [PubMed] [Google Scholar]
- 33.Siegfried, G., A. M. Khatib, S. Benjannet, M. Chretien, and N. G. Seidah. 2003. The proteolytic processing of pro-platelet-derived growth factor-A at RRKR(86) by members of the proprotein convertase family is functionally correlated to platelet-derived growth factor-A-induced functions and tumorigenicity. Cancer Res. 63:1458-1463. [PubMed] [Google Scholar]
- 34.Srour, N., A. Lebel, S. McMahon, I. Fournier, M. Fugere, R. Day, and C. M. Dubois. 2003. TACE/ADAM-17 maturation and activation of sheddase activity require proprotein convertase activity. FEBS Lett. 554:275-283. [DOI] [PubMed] [Google Scholar]
- 35.Taylor, N. A., W. J. Van De Ven, and J. W. Creemers. 2003. Curbing activation: proprotein convertases in homeostasis and pathology. FASEB J. 17:1215-1227. [DOI] [PubMed] [Google Scholar]
- 36.Ulloa, L., J. W. Creemers, S. Roy, S. Liu, J. Mason, and S. Tabibzadeh. 2001. Lefty proteins exhibit unique processing and activate the MAPK pathway. J. Biol. Chem. 276:21387-21396. [DOI] [PubMed] [Google Scholar]
- 37.Villeneuve, P., S. Feliciangeli, G. Croissandeau, N. G. Seidah, M. Mbikay, P. Kitabgi, and A. Beaudet. 2002. Altered processing of the neurotensin/neuromedin N precursor in PC2 knock down mice: a biochemical and immunohistochemical study. J. Neurochem. 82:783-793. [DOI] [PubMed] [Google Scholar]
- 38.Xiang, Y., S. S. Molloy, L. Thomas, and G. Thomas. 2000. The PC6B cytoplasmic domain contains two acidic clusters that direct sorting to distinct trans-Golgi network/endosomal compartments. Mol. Biol. Cell 11:1257-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yana, I., and S. J. Weiss. 2000. Regulation of membrane type-1 matrix metalloproteinase activation by proprotein convertases. Mol. Biol. Cell 11:2387-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang, J., J. L. Goldstein, R. E. Hammer, Y. A. Moon, M. S. Brown, and J. D. Horton. 2001. Decreased lipid synthesis in livers of mice with disrupted Site-1 protease gene. Proc. Natl. Acad. Sci. USA 98:13607-13612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng, M., N. G. Seidah, and J. E. Pintar. 1997. The developmental expression in the rat CNS and peripheral tissues of proteases PC5 and PACE4 mRNAs: comparison with other proprotein processing enzymes. Dev. Biol. 181:268-283. [DOI] [PubMed] [Google Scholar]
- 42.Zhu, X., A. Zhou, A. Dey, C. Norrbom, R. Carroll, C. Zhang, V. Laurent, I. Lindberg, R. Ugleholdt, J. J. Holst, and D. F. Steiner. 2002. Disruption of PC1/3 expression in mice causes dwarfism and multiple neuroendocrine peptide processing defects. Proc. Natl. Acad. Sci. USA 99:10293-10298. [DOI] [PMC free article] [PubMed] [Google Scholar]