Abstract
The Notch receptor is part of a highly conserved signalling system of central importance to animal development. Its ANK (ankyrin) domain is required for Notch-mediated signal transduction. The crystal structure of the human Notch 1 ANK domain was solved by molecular replacement at 1.9 Å (1 Å=0.1 nm) resolution, and it shows that the features identified in the Drosophila homologue are conserved. The domain has six of the seven ANK repeats predicted from sequence. The putative first repeat, which has only part of the consensus and a long insertion, is disordered in both molecules in the asymmetric unit, possibly due to the absence of the RAM (RBPJκ-associated molecule) region N-terminal to it. The exposed hydrophobic core is involved in intermolecular interactions in the crystal. Evolutionary trace analysis identified several residues that map to the hairpins of the structure and may be of functional importance. Based on the Notch 1 ANK structure and analysis of homologous Notch ANK sequences, we predict two possible binding sites on the domain: one on the concave surface of repeat 2 and the other below the hairpins of repeats 6–7.
Keywords: ankyrin, binding site, crystal structure, Notch, signalling
Abbreviations: ANK, ankyrin; CNS, Crystallography and NMR System; CSL, CBF1/RBPJκ; Su(H), Lag-1; IκB, inhibitory κB; MAM, Mastermind; NICD, Notch intracellular domain; PDB, Protein Data Bank; RAM, RBPJκ-associated molecule; RMSD, root mean square deviation
INTRODUCTION
The Notch receptor is the central element of a signalling system that is involved in a wide range of events during animal development [1]. It is primarily involved in binary cell fate decisions in which the Notch signalling event directs the cell to adopt a particular cell fate, while blocking differentiation towards an alternative fate [1]. The Notch receptor is a Type I transmembrane receptor consisting of a large extracellular domain, containing 36 EGF (epidermal growth factor) and three LNR (Lin12-Notch) repeats, and an NICD (Notch intracellular domain), which includes a region comprising seven ANK (ankyrin) repeats. Functionally significant mutations that map to the ANK repeats show that it is essential for signalling [2–6].
There are four mammalian Notch homologues (Notch 1–4). Their ANK domains share significant sequence identity (identity relative to the human Notch 1 is 78% for Notch 2, 77% for Notch 3 and 53% for Notch 4). The overall structure for the Notch receptor is not known, although structures for some regions of the human Notch 1 extracellular domain have been reported [7,8], as well as the structure of the Drosophila Notch ANK domain [9].
Ligands for Notch are members of the DSL (Delta, Serrate, Lag-2) family of transmembrane proteins. Ligand binding to the extracellular domain results in two sequential cleavages of Notch. The second, mediated by γ-secretase, releases NICD, which can enter the nucleus where it interacts with members of the CSL [CBF1/RBPJκ, Su(H), Lag-1] family of transcription factors and participates in transcription activation [10]. For this reason, Notch is sometimes referred to as a membrane-tethered transcription factor [11].
NICD is instrumental in the activation of Notch target genes. The transcriptional regulator CSL is a constitutive repressor of Notch target genes through its association with transcriptional co-repressors. NICD displaces these co-repressors and forms a transcription-activating complex with CSL [12–15]. The binding of NICD to CSL recruits other proteins to the complex, in particular MAM (MAML1, Mastermind) [16–19].
Genetic analysis reveals that NICD has other functional sites in addition to the ANK repeats. N-terminal to the ANK repeats is the RAM (RBPJκ-associated molecule) region, which is required for binding of NICD to CSL [13]. This region is likely to be unfolded in the native state as judged by CD spectra [19]. C-terminal to the ANK repeats, there is a polyglutamine sequence and a PEST (Pro-Glu-Ser-Thr) motif, as well as a presumed transactivating domain [20]. Like the RAM region, this part of NICD is likely to be unfolded (M. Ehebauer, unpublished work). The ANK domain is therefore the only part of NICD for which a protein fold can probably be defined, although other parts of the sequence may be ordered when they are complexed with other signalling proteins.
ANK domains are generally responsible for mediating protein–protein interactions [21,22]. Genetic analysis of Notch suggests that its ANK domain is functionally important and it has been shown to interact with several proteins. Among them are MAM [16], Deltex [23,24], p300 [25,26], PCAF [p300/CREB (cAMP-response-element-binding protein)-binding-protein-associated factor] and GCN5 [27]. It is also known to contribute to the interaction with CSL [14,15], although there is no evidence that it interacts directly with CSL. Even though interactions between the histone acetyltransferase p300 and NICD have been reported it has been suggested that it is MAM that is responsible for recruiting this potential co-activator to NICD [28].
ANK repeats are found in a great many proteins [21,22] and although considerable sequence variability exists between them, they have a conserved secondary and tertiary structure. A repeat consists of 33 amino acids arranged in two antiparallel α-helices connected by a short loop. The helices of one repeat are connected to the next repeat by a structure that resembles a β-hairpin. These hairpins are oriented perpendicular to the helices, giving the domain an L shape in cross-section. This arrangement of repeats produces a groove along the long axis of the domain, which is frequently the main binding interface [22]. Binding partners commonly have contacts between the β-hairpin tips and/or the surface of the inner helices [22]. Intradomain interactions involving ANK domains have also been described. In these structures, the ANK domain is closely bound by another domain of the same protein, although the ANK groove is left exposed, at least in part [29,30]. Terminal repeats are often truncated and, having more polar residues, often deviate from the consensus ANK repeat sequence [22]. The polar residues that replace conserved hydrophobic residues in these repeats protect the hydrophobic core of the domain. Absence of the terminal capping repeats may destabilize the domain as a whole. Deletion of the terminal capping seventh repeat in the Notch ANK domain indicated that this repeat contributes significantly to the overall stability of this domain [31,32].
In the present study, we describe the structure of the ANK domain of human Notch 1. We show that the putative first repeat is disordered in both molecules in the asymmetric unit, possibly due to the absence of the N-terminal RAM region. This structure shows that features identified in the Drosophila homologue are conserved in mammalian orthologues.
MATERIALS AND METHODS
Bioinformatics
Amino acid sequences of Notch ANK domain homologues were aligned using ClustalW [33]. Secondary structure for the human Notch 1 ANK domain was predicted using JPred [34]. Evolutionary trace analysis was used to identify possible residues that are of structural and functional importance. For this purpose, a multiple sequence alignment containing 23 homologues of Notch ANK domains was analysed using TraceSuite II [35]. Only residues that had the highest evolutionary time cut off were considered important and were mapped to the structure of the human Notch 1 ANK domain. To identify more accurately residues that may form part of binding sites, the method of Chelliah et al. [36] was used. This method distinguishes those restraints placed on protein structure from additional restraints that arise due to function. The putative first repeat was not included in this analysis, because it is disordered in the structure reported here.
Protein expression, purification and crystallization
The ANK domain of human Notch 1 was expressed in Rosetta (DE3) Escherichia coli cells using the pET41a (+) vector from Novagen. The construct used for expression encoded the Notch 1 residues Met1873–Asp2115 (in the present paper, labelled Met1–Asp243). It was cloned into the vector using the restriction digest of a PCR product produced using 5′-GATCATTAATATGGACGTCAATGTCCGCGG-3′ as sense primer and 5′-GTCACTCGAGGTCCAGCAGCCTCACGATGTCG-3′ as antisense primer. The PCR product was digested with AseI and AvaI and ligated into the vector's NdeI and XhoI sites. The protein was C-terminally His-tagged with eight histidine residues. Due to the manner of cloning, there was a two-amino-acid insertion (Leu-Glu) between the protein's coding sequence and the tag. Cells transformed with the ANK domain expression vector were grown at 37 °C in a shake incubator until the culture had reached mid-exponential phase. The cells were then induced with 1 mM IPTG (isopropyl β-D-thiogalactoside) for 3 h. After induction, the culture was centrifuged at 2500 g for 10 min, and the resultant cell pellet was resuspended in 20% sucrose, 50 mM Tris/HCl (pH 8.0), 150 mM NaCl and 5 mM DTT (dithiothreitol) to which recombinant lysozyme was added to a final concentration of 1 mg/ml and also Roche Complete™ protease inhibitor, as directed by the manufacturer. The suspension was incubated on ice for 1 h and was then sonicated to fully disrupt the cell membranes and shear the genomic DNA. The sonicate was centrifuged at 17200 g for 15 min to produce cleared lysate.
The cleared lysate was loaded on to a Ni-NTA (Ni2+-nitrilotriacetate) column (Qiagen) for affinity purification. Protein was incubated with the affinity matrix for 1 h before collection of the flow-through. The column was washed twice using 50 mM Tris/HCl (pH 8.0), 300 mM NaCl and 20 mM imidazole and was then eluted using 50 mM Tris/HCl (pH 8.0), 300 mM NaCl and 250 mM imidazole. The affinity-purified ANK domain was then loaded on to a HiTrap Q-HP anion-exchange column (Phenomenex) pre-equilibrated with buffer A: 50 mM Tris/HCl (pH 8.0) and 50 mM NaCl. Protein was eluted using a gradient of 0–100% of buffer B: 50 mM Tris/HCl (pH 8.0) and 1 M NaCl. Collected fractions were loaded on to a Superdex 200 HR 10/30 size-exclusion column (Phenomenex) and eluted using 50 mM Tris/HCl (pH 8.0) and 50 mM NaCl.
The pure protein was usually concentrated to 20 mg/ml and crystallized by the hanging-drop vapour diffusion method using 0.9 M (NH4)2HPO4, 0.2 M NaCl and 0.1 M imidazole (pH ∼8.5) as precipitant. Crystallization was performed at room temperature (25 °C). Crystals were flash-cooled in liquid nitrogen using a cryo-protectant solution composed of 30% (v/v) glycerol, 0.9 M (NH4)2HPO4, 0.2 M NaCl and 0.1 M imidazole (pH ∼8.5).
Data collection, data processing, structure solution and refinement
Diffraction data of a single cryo-cooled crystal were collected at station 14.1 of the Daresbury Synchrotron Radiation Source, Warrington, U.K. A data set was collected by the rotation method with a 1° oscillation per frame at a wavelength of 1.488 Å (1 Å=0.1 nm). The set of images was indexed, scaled and integrated using the programs DENZO and SCALEPACK [37]. The data-processing statistics are shown in Table 1.
Table 1. Data collection and refinement statistics.
Values in parentheses are for the highest resolution shell. Rsym=Σh|Ih−〈I〉/ΣhIh, where Ih is the intensity of reflection h and 〈I〉 is the mean intensity of all symmetry related reflections. Rcryst=Σ||Fobs|−|Fcalc||/Σ|Fobs|, where Fobs and Fcalc are observed and calculated structure factor amplitudes. Rfree as for Rcryst using a random subset of data excluded from the refinement, 5% of the total dataset was used. Estimated co-ordinate error based on R value was calculated using Refmac [42]. Ramachandran plot analysis was calculated using PROCHECK [43].
| Parameter | Value |
|---|---|
| Data collection | |
| X-ray source | Daresbury Synchrotron Radiation Source station 14.1 |
| Detector | CCD ADSC unsupported-q4 |
| Space group | P65 |
| Unit cell | a=b=96.84 Å, c=108.99 Å |
| α=β=90°, γ=120° | |
| Resolution | 30–1.90 Å (1.94–1.90 Å) |
| Completeness | 98.5% (98.6%) |
| Rsym | 7.9 (38.5) |
| 〈I/σ〉 | 12.4 |
| Percentage with I/σ>3 | 81.2% (42.8%) |
| Redundancy | 3.6 |
| Number of unique reflections | 44926 |
| Wilson plot B-factor | 26.9 Å2 |
| Refinement | |
| Rcryst | 16.1% |
| Rfree | 19.3% |
| Number of reflections | |
| Working | 3138 |
| Test | 108 |
| Molecules per asymmetric unit | 2 |
| Number of non-hydrogen atoms | 3483 |
| Model quality | |
| Estimated co-ordinate error | 0.109 Å |
| RMSD bonds | 0.018 Å |
| RMSD angles | 1.43° |
| Overall mean B-factor | 26.8 Å2 |
| Ramachandran plot analysis | |
| Number of residues in | |
| Most favoured regions | 90.7% |
| Additionally allowed regions | 9.3% |
| Disallowed regions | 0% |
The VM coefficient [38] calculation (VM=2.4 Å3·Da−1) indicates the probable presence of two molecules in the asymmetric unit, designated chain A and B. The structure was solved by molecular replacement using the program AMoRe [39] with the co-ordinates of the Drosophila Notch ANK domain as a probe {Protein Data Bank (PDB) code 1OT8 [9]}. One round of rigid-body refinement, simulated-annealing and B-factor refinement was performed with CNS (Crystallography and NMR System) [40]. The σA-weighted 2Fo−Fc and Fo−Fc electron-density maps were calculated by CNS and visually inspected to allow rebuilding and refitting using XtalView [41]. Much of the first ANK repeat (first 51 residues in chain A and 54 in chain B) of both molecules in the asymmetric unit was not included, as no electron density was evident. Additional density was observed at the C-terminus of chain A, where a leucine residue was added (Leu244 chain A). This leucine is part of the insert between the ANK domain proper and the His-tag used for purification. Three more rounds of simulated annealing and B-factor refinement were performed, followed by three rounds of refinement using REFMAC5 [42]. Water molecules were introduced progressively throughout the refinement process. No electron density for the side chains of residues Gln52, Thr53, Arg55 and Arg69 in chain A was observed in the final density maps. Likewise, no density for the side chains of Arg55 and Lys178 in chain B was observed. In the final model, these residues were modelled as alanine. These residues are either part of the disordered first ANK repeat or, in the case of Arg69 and Lys178, solvent-exposed. The stereochemistry was validated using PROCHECK [43] and WHAT_CHECK [44]. Surface potentials were calculated using APBS (Adaptive Poisson–Boltzmann Solver) [45].
RESULTS AND DISCUSSION
Sequence analysis
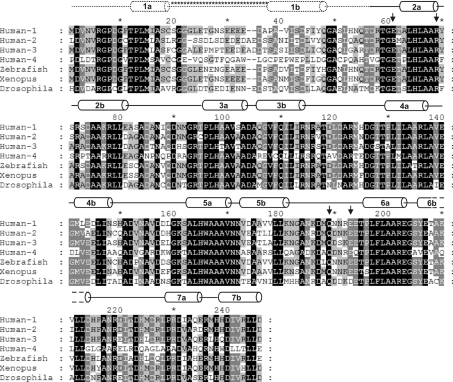
The ANK domain is the most conserved region of NICD and was predicted to have six repeats. Recent analyses have identified an additional seventh repeat [9,19,31,46]. Five of the ANK repeats in human Notch 1 conform to the ANK consensus sequence defined by Mosavi et al. [22]. The terminal repeats of Notch 1 ANK diverge significantly from this consensus. The first repeat starts with the ANK motif sequence GXTPL(M)XAXXXG, which is the consensus sequence of the first helix of an ANK repeat (X denotes any amino acid; the M in parenthesis is usually an H in the consensus sequence). This ANK motif is conserved among Notch homologues (Figure 1). The second half of the first repeat does not conform to the ANK consensus and is the least conserved region of the domain (Figure 1). The secondary-structure prediction algorithm JPred predicted two helices for the first repeat, one for the conserved N-terminus and a second helix 16 residues C-terminal to the first (Figure 1). The latter helix does not conform to the ANK consensus. Indeed, excluding the conserved N-terminal region, the first repeat does not resemble an ANK repeat. The seventh repeat also deviates from the ANK consensus, although to a lesser extent. Polar residues have replaced some conserved hydrophobic residues in this repeat.
Figure 1. Sequence alignment of Notch ANK domains.
Multiple sequence alignment of the ANK domain of human Notch 1–4, zebrafish, Xenopus and Drosophila. The secondary structure assignment of human Notch 1 is shown above the sequences. Helices are labelled according to their location in repeats 1–7. The putative first repeat (helix 1a and 1b) predicted by JPred [34], which was not evident in the electron density, is shown by dotted lines. The long insert predicted for this repeat is indicated with asterisks (*). Residues in black boxes are absolutely conserved; those in grey boxes are conserved in several of the homologues. Black arrows indicate residues that are predicted to be part of binding sites (see the Results and discussion section). The alignment was produced using ClustalW [33] and GeneDoc (http://www.psc.edu/biomed/genedoc).
Overall structure
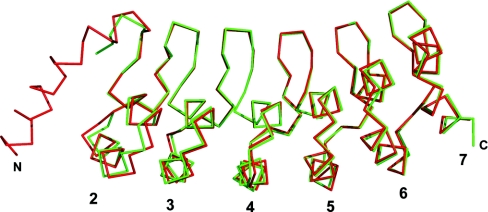
The Notch 1 ANK protein crystallized with a His8-tag at the C-terminus. No electron density for the His8-tag could be observed, although one non-native leucine residue of the insertion between the domain and the tag was evident at the C-terminus of chain A. The final model has an Rcryst of 16.1% and an Rfree of 19.3%; the refinement statistics and an assessment of the model geometry are given in Table 1. There are few crystallographic contacts between chains A and B. All of them involve residues in the terminal repeats. Although there has been some debate about the formation of ANK dimers, the crystallographic dimer is unlikely to be physiological [9]. The two chains are very similar with a RMSD (root mean square deviation) of the main-chain atoms of 0.359 Å. Overall, the structure reported here is comparable to the Drosophila ANK domain (main-chain atom RMSD=0.654 Å for residues 50–237 of the Drosophila structure and 56–243 of the structure reported in the present paper). The two structures are shown superimposed in Figure 2.
Figure 2. Superposition of the Drosophila Notch and the human Notch 1 ANK structures.
Shown in red is the Cα trace of chain A of the Drosophila Notch ANK structure (PDB code 1OT8; [9]) superimposed on the green Cα trace of chain A of the human Notch 1 ANK domain. The Drosophila structure has one additional helix at the N-terminus. The Figure was created using PyMOL (http://pymol.sourceforge.net).
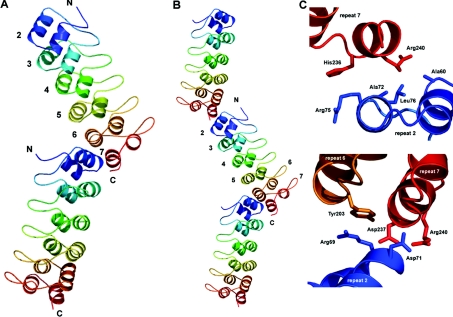
The structure of the human Notch 1 ANK domain has the expected ANK fold, although the third repeat is 34 residues in length instead of the canonical 33 (Figure 3A) (PDB accession code 1YYH). There is no density for the N-terminal first repeat, which is probably disordered. It has been suggested that this region is only ever partially folded, because the unfolding transition of Notch ANK variants, in which analogous alanine residues in each repeat were replaced with glycine residues, indicated that substitutions in the first repeat do not severely affect domain stability [32]. The prediction of an unusually long loop (16 residues) (Figure 1) with a large number of charged or polar amino acids may be one reason for the observed unfolding of this repeat. It is possible that part of the first repeat folds loosely against the second repeat, allowing the long charged loop and the rest of the repeat to be solvent-exposed, accounting for the partially unfolded nature reported for this repeat [32].
Figure 3. Crystal structure of the human Notch 1 ANK domain.
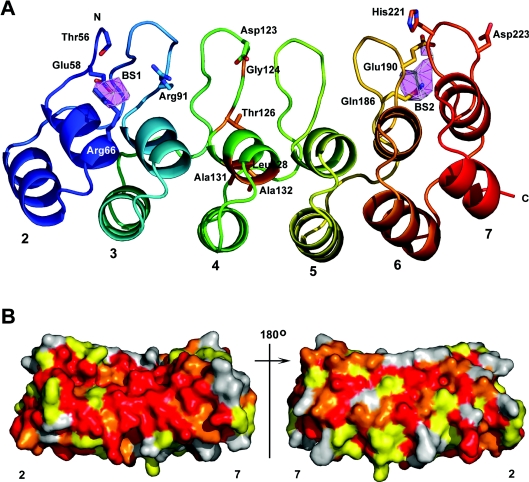
(A) Both molecules in the asymmetric unit are shown. Repeats 2–7 are individually coloured starting with blue at the N-terminus and ending with red at the C-terminus. (B) Notch ANK domains pack end-to-end in the crystal lattice. A symmetry-related ANK domain is shown with both ANK chains in the asymmetric unit. (C) Residues involved in the formation of crystal contacts between successive domains in the crystal. Figures are rotated from their original position in (B) to better show the contacts. Residues involved in forming the contacts were identified using the protein–protein interaction server (http://www.biochem.ucl.ac.uk/bsm/PP/server/). The Figure was created using PyMOL.
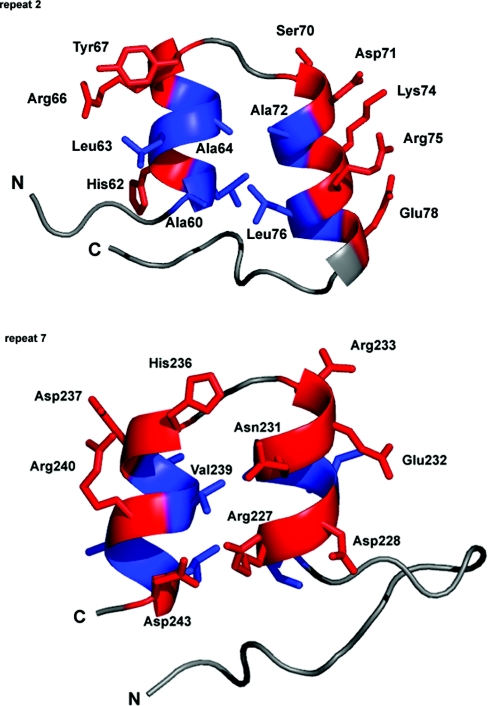
The seventh repeat, which caps the hydrophobic core at the C-terminus, is of great importance to the stability of the entire domain [28,31,32,46]. The polar and charged residues of the seventh repeat are solvent-exposed (Arg227, Asp228, Asn231, Glu232, Arg233, His236, Asp237, Arg240 and Asp243) with parts of residues Arg227 and Asn231 capping the core (Figure 4).
Figure 4. Terminal repeats differ in amino acid composition.
Repeats 2 and 7 are shown end on, looking down the long axis of the domain. The rest of the ANK domain is hidden for clarity. Residues that constitute the helices in these repeats are shown in stick-model form. Coloured red are polar and charged residues, hydrophobic residues are blue, and connecting loops are in white. Selected amino acids are labelled, the numbering referring to the position of residues in the sequence alignment in Figure 1. The hydrophobic core is exposed in repeat 2 (Ala60, Ala64, Ala72 and Leu76). In repeat 7, polar and charged residues cover the hydrophobic core (Arg227 and Asn231). The Figure was created using PyMOL.
Adjacent ANK repeats pack together to form an elongated structure that is held together predominately by hydrophobic interactions. The structures of repeat domains are dominated by local short-range interactions. Interestingly, it was assumed that the modular nature of the ANK domains would preclude the formation of long-range stabilizing interactions. However, biophysical experiments using the Notch ANK domain have shown that a modular domain can unfold co-operatively in a two-state folding pathway and that, in the case of the Notch ANK domain, even distant repeats are somehow coupled [31,32]. This is most likely to be due to the hydrophobic core that extends through the length of the domain. There appears to be a limit to the distance over which such coupling of repeats, through the hydrophobic core, can extend. It has been demonstrated that two additional repeats introduced into the Notch ANK domain slightly improve domain stability, but that three or four additional repeats represent a limit beyond which full co-operativity cannot be maintained [47], although significantly larger ANK structures are known [22].
The human Notch 1 ANK domain is slightly curved, a feature that becomes particularly apparent in structures containing a large number of repeats [48] and that appears to be the result of differing helix lengths and inter-repeat interactions between the helices. The inner helices of each repeat are shorter (by one or two residues) than the outer helices. Also, conserved small side-chain amino acids (mostly alanine and one valine) close to the short loop connecting the helices allow the inner helices to pack more closely than the outer helices, which have, with few exceptions, larger residues (leucine, methionine, phenylalanine, valine, threonine, aspartate and isoleucine) at similar positions. At the other end of the helices, towards the β-hairpin loop, residues generally have large side chains (leucine, isoleucine and, in the seventh repeat, one proline), which force the bottom of the helices apart. Each repeat is consequently rotated slightly counter-clockwise relative to the preceding repeat. The overall effect is to impart a left-handed superhelical twist to the structure.
Although Zweifel et al. [9] state that the presence of the RAM region does not enhance helix formation or ANK domain stability in Drosophila Notch, several hydrophobic residues in the second repeat of human Notch 1 ANK (Ala60, Ala64, Ala72 and Leu76) would be solvent-exposed (Figure 4). This resembles the cell-cycle regulator Swi6 ANK domain in which the N-terminal repeat has a significant hydrophobic surface that would be largely solvent exposed if a set of helices from an adjoining domain did not stack against it [29]. Similarly, exposed hydrophobic residues of the N-terminal repeat of IκBα (inhibitory κBα) are capped by part of p65 [49], whereas the IκB-family member Bcl-3 has these hydrophobic residues replaced with smaller or charged residues that form the solvent-exposed face of that ANK domain [50]. The end-to-end packing of the molecules in the human Notch 1 ANK crystal partially shields the second repeat from solvent (Figures 3B and 3C). These observations suggest that, in the cell, the core at the interface of the second repeat may be capped by another part of the NICD or another molecule, leading to a structure that is not evident for the isolated ANK domain used in the present study.
Potential binding sites
The concave surface of the ANK groove is most often the site of protein–protein interactions, with interactions usually occurring through the β-hairpins and/or the inner helices [22]. The electrostatic surface potential on either side of the Notch 1 ANK groove is negative (Figure 5). The binding site of the MAM transcriptional co-activator on the ANK domain is not known. However, given that the region that interacts with the NICD contains a large number of positively charged residues (>25% of the protein) [17], the negatively charged regions either side of the ANK groove may be an appropriate binding site for MAM.
Figure 5. Electrostatic potential of the ANK domain.
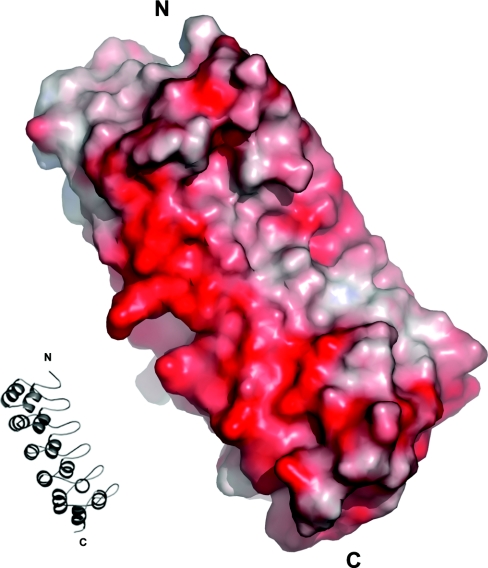
Only chain A of the asymmetric unit is shown. The orientation of chain A is that shown in the ribbon model inset. Most of the protein has negative surface potential. Potential was calculated with APBS (Adaptive Poisson–Boltzmann Solver) [45]. The Figure was created using PyMOL.
Evolutionary trace analysis [35] was used to identify possible residues that are of structural and functional importance. Several clusters of conserved residues were identified; most of them are in the antiparallel helices, the short loop that connects these helices or in the section of the loop connecting repeats that passes under the domain core, reflecting evolutionary restraints on the architecture of ANK repeat structure. Only six conserved residues are located in the hairpins (Thr56, Arg91, Asp123, Glu190, His221 and Asp223) (Figure 6A). These residues are solvent-exposed and may therefore be of functional importance.
Figure 6. Predicted binding sites and sequence conservation.
(A) Chain A is shown with repeats 2–7 individually coloured blue to red. Side chains of residues identified by the evolutionary trace analysis [35] and Crescendo [36] are shown in stick-model form with nitrogen atoms in blue, and oxygen atoms in red. In repeat 4, the side chains of residues substituted by the M1 and M2 mutations [3] are shown in stick-model form and are coloured orange (Gly124, Thr126, Leu128, Ala131 and Ala132). Binding sites predicted by Crescendo are shown as pink polygons. BS1, predicted binding site 1; BS2, predicted binding site 2. (B) Sequence conservation mapped onto the surface of the human Notch 1 ANK domain. The orientation of the molecule on the left is that shown in (A), with the β-turns parallel to the y-axis. The molecule on the right is rotated by 180 °C along the y-axis. Coloured red are residues that are absolutely conserved in the Notch homologue sequences in Figure 1. In orange are residues conserved in all but one of the homologues and in yellow are residues conserved in all but two or three homologues. Residues that are not conserved are in grey. The Figure was created using PyMOL.
In order to distinguish between structurally and functionally important residues, the multiple sequence alignment of Notch homologues was analysed using the program Crescendo as described by Chelliah et al. [36]. Two possible interaction sites were predicted, one on the concave surface of repeat 2 and one below the hairpins of repeat 6 and 7 (Figure 6A). Residues Glu58 and Arg66 are predicted to form part of the first putative binding site. These residues were also identified by the evolutionary trace analysis; however, Glu58 is not solvent-exposed. Residues Gln186 and Glu190 are part of the second putative binding site. These residues were also identified by the trace analysis and both are solvent-exposed in the structure. These residues are conserved in the Notch homologues shown in Figure 1, with the exception of Glu190, which is replaced by an aspartate residue in Drosophila Notch (Figures 1, arrows, and 6B).
The location of these predicted binding sites, on different sides of the hairpins and opposite ends of the domain, may indicate that they are responsible for binding two different molecules, or they may be part of one single binding site that covers the entire concave surface and hairpins of the domain. Several mutations that affect Notch function are located in the ANK domain. Among them are the mutations M1 and M2 in repeat 4 of the mouse Notch 1 ANK domain [3]. These are loss-of-function mutations that inhibit myogenesis [3] and result in amino acid substitutions in the domain core (Figure 6A). The secondary-structure content and unfolding transitions of a Drosophila ANK domain containing these mutations are significantly different from those of the native Drosophila ANK domain, indicating that they affect domain structure [9]. It is therefore likely that these mutations disrupt the overall fold of the ANK domain [9]. The binding-site predictions and the disruption of structure by the M1 and M2 mutations suggest that there may be an extensive interacting interface along the concave surface of the domain. Functional studies using Notch receptors in which the putative binding site residues have been substituted would shed light on their possible role in protein–protein interactions. The structures of complexes that include the Notch ANK domain and its binding partners would allow greater definition of interaction sites.
While this manuscript was under review, a paper reporting the partial structure of the mouse Notch 1 ANK domain was published [51]. Molecules in the asymmetric unit of the partial mouse Notch 1 ANK domain are oriented so as to protect the exposed hydrophobic core in a manner analogous to that observed in the crystal packing of the human Notch 1 ANK domain. The authors argue that the N- and C-terminal repeats of the ANK domain constitute functionally different subdomains, which may interact with different proteins [51]. Our predicted binding sites located at the N-terminal repeat 2 and the C-terminal repeats 6 and 7 are in broad agreement with this.
Acknowledgments
We thank the staff of station 14.1 at the Daresbury Synchrotron Radiation Source for their assistance and Vijayalakshmi Chelliah for her assistance in identifying potential binding sites. D.Y.C. is funded by a grant from the Wellcome Trust (Grant Number 064597), and M.T.E. is supported by the Cambridge Commonwealth Trust.
References
- 1.Artavanis-Tsakonas S., Rand M. D., Lake R. Notch signalling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 2.Roehl H., Bosenberg M., Blelloch R., Kimble J. Roles of the RAM and ANK domains in signalling by the C. elegans GLP-1 receptor. EMBO J. 1996;15:7002–7012. [PMC free article] [PubMed] [Google Scholar]
- 3.Kopan R., Nye J. S., Weintraub H. The intracellular domain of mouse Notch: a constitutively activated repressor of myogenesis directed at the basic helix–loop–helix region of MyoD. Development. 1994;120:2385–2396. doi: 10.1242/dev.120.9.2385. [DOI] [PubMed] [Google Scholar]
- 4.Lieber T., Kidd S., Alcamo E., Corbin V., Young M. W. Antineurogenic phenotypes induced by truncated Notch proteins indicate a role in signal transduction and may point to a novel function for Notch in nuclei. Genes Dev. 1993;7:1949–1965. doi: 10.1101/gad.7.10.1949. [DOI] [PubMed] [Google Scholar]
- 5.Rebay I., Fehon R. G., Artavanis-Tsakonas S. Specific truncations of Drosophila Notch define dominant activated and dominant negative forms of the receptor. Cell. 1993;74:319–329. doi: 10.1016/0092-8674(93)90423-n. [DOI] [PubMed] [Google Scholar]
- 6.Struhl G., Fitzgerald K., Greenwald I. Intrinsic activity of the Lin-12 and Notch intracellular domains in vivo. Cell. 1993;74:331–345. doi: 10.1016/0092-8674(93)90424-o. [DOI] [PubMed] [Google Scholar]
- 7.Hambleton S., Valeyev N. V., Muranyi A., Knott V., Werner J. M., McMichael A. J., Handford P. A., Downing A. J. Structural and functional properties of the human Notch-1 ligand binding region. Structure. 2004;12:2173–2183. doi: 10.1016/j.str.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 8.Vardar D., North C. L., Sanchez-Irizarry C., Aster J. C., Blacklow S. C. Nuclear magnetic resonance structure of a prototype Lin12-Notch repeat module from human Notch1. Biochemistry. 2003;42:7061–7067. doi: 10.1021/bi034156y. [DOI] [PubMed] [Google Scholar]
- 9.Zweifel M. E., Leahy D. J., Hughson F. M., Barrick D. Structure and stability of the ankyrin domain of the Drosophila Notch receptor. Protein Sci. 2003;12:2622–2632. doi: 10.1110/ps.03279003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mumm J. S., Kopan R. Notch signalling: from the outside in. Dev. Biol. 2000;228:151–165. doi: 10.1006/dbio.2000.9960. [DOI] [PubMed] [Google Scholar]
- 11.Kopan R. Notch: a membrane-bound transcription factor. J. Cell Sci. 2002;115:1095–1097. doi: 10.1242/jcs.115.6.1095. [DOI] [PubMed] [Google Scholar]
- 12.Fortini M. E., Artavanis-Tsakonas S. The suppressor of hairless protein participates in notch receptor signalling. Cell. 1994;79:273–282. doi: 10.1016/0092-8674(94)90196-1. [DOI] [PubMed] [Google Scholar]
- 13.Tamura K., Taniguchi Y., Minoguchi S., Sakai T., Tun T., Furukawa T., Honjo T. Physical interaction between a novel domain of the receptor Notch and transcription factor RBP-Jκ/Su(H) Curr. Biol. 1995;5:1416–1423. doi: 10.1016/s0960-9822(95)00279-x. [DOI] [PubMed] [Google Scholar]
- 14.Kato H., Taniguchi Y., Kurooka H., Minoguchi S., Sakai T., Nomura-Okazaki S., Tamura K., Honjo T. Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development. 1997;124:4133–4141. doi: 10.1242/dev.124.20.4133. [DOI] [PubMed] [Google Scholar]
- 15.Tani S., Kurooka H., Aoki T., Hashimoto N., Honjo T. The N- and C-terminal regions of RBP-J interact with the ankyrin repeats of Notch1 RAMIC to activate transcription. Nucleic Acids Res. 2001;29:1373–1380. doi: 10.1093/nar/29.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu L., Aster J. C., Blacklow S. C., Lake R., Artavanis-Tsakonas S., Griffin J. D. MAML1, a human homologue of Drosophila Mastermind, is a transcriptional co-activator for Notch receptors. Nat. Genet. 2000;26:484–489. doi: 10.1038/82644. [DOI] [PubMed] [Google Scholar]
- 17.Kitagawa M., Oyama T., Kawashima T., Yedvobnick B., Kumar A., Matsuno K., Harigaya K. A human protein with sequence similarity to Drosophila mastermind coordinates the nuclear form of notch and a CSL protein to build a transcriptional activator complex on target promoters. Mol. Cell. Biol. 2001;21:4337–4346. doi: 10.1128/MCB.21.13.4337-4346.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jefferies S., Robbins D. J., Capobianco A. J. Characterization of a high-molecular-weight complex in the nucleus of Notchic-transformed RKE cells and in a human T-cell leukaemia cell line. Mol. Cell. Biol. 2002;22:3927–3941. doi: 10.1128/MCB.22.11.3927-3941.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nam Y., Weng A. P., Aster J. C., Blacklow S. C. Structural requirements for the assembly of the CSL/intracellular Notch1/Mastermind-like 1 transcription activation complex. J. Biol. Chem. 2003;278:21232–21239. doi: 10.1074/jbc.M301567200. [DOI] [PubMed] [Google Scholar]
- 20.Kurooka H., Kuroda K., Honjo T. Role of the ankyrin repeat and C-terminal region of the mouse Notch1 intracellular region. Nucleic Acids Res. 1998;26:5448–5455. doi: 10.1093/nar/26.23.5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bork P. Hundreds of ankyrin-like repeats in functionally diverse proteins: mobile modules that cross phyla horizontally? Proteins. 1993;17:363–374. doi: 10.1002/prot.340170405. [DOI] [PubMed] [Google Scholar]
- 22.Mosavi L. K., Cammett T. J., Desrosiers D. C., Peng Z. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004;13:1435–1448. doi: 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diederich R. J., Matsuno K., Hing H., Artavanis-Tsakonas S. Cytosolic interaction between Deltex and Notch ankyrin repeats implicates Deltex in the Notch signaling pathway. Development. 1994;120:473–481. doi: 10.1242/dev.120.3.473. [DOI] [PubMed] [Google Scholar]
- 24.Matsuno K., Diederich R. J., Go M. J., Blaumueller C. M., Artavanis-Tsakonas S. Deltex acts as a positive regulator of Notch signalling through interactions with the Notch ankyrin repeats. Development. 1995;121:2633–2644. doi: 10.1242/dev.121.8.2633. [DOI] [PubMed] [Google Scholar]
- 25.Oswald F., Täuber B., Dobner T., Bourteele S., Kostezka U., Adler G., Liptay S., Schmid R. M. p300 acts as a transcription coactivator for mammalian Notch-1. Mol. Cell. Biol. 2001;21:7761–7774. doi: 10.1128/MCB.21.22.7761-7774.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wallberg A. E., Pedersen K., Lendahl U., Roeder R. G. p300 and PCAF act cooperatively to mediate transcriptional activation from chromatin templates by Notch intracellular domains in vitro. Mol. Cell. Biol. 2002;22:7812–7819. doi: 10.1128/MCB.22.22.7812-7819.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurooka H., Honjo T. Functional interaction between the mouse Notch1 intracellular region and histone acetyltransferases PCAF and GCN5. J. Biol. Chem. 2000;275:17211–17220. doi: 10.1074/jbc.M000909200. [DOI] [PubMed] [Google Scholar]
- 28.Lubman O. Y., Korolev S. V., Kopan R. Anchoring Notch genetics and biochemistry: structural analysis of the ankyrin domain sheds light on existing data. Mol. Cell. 2004;13:619–626. doi: 10.1016/s1097-2765(04)00120-0. [DOI] [PubMed] [Google Scholar]
- 29.Foord R., Taylor I. A., Sedgwick S. G., Smerdon S. J. X-ray structural analysis of the yeast cell cycle regulator Swi6 reveals variations of the ankyrin fold and has implications for Swi6 function. Nat. Struct. Biol. 1999;6:157–165. doi: 10.1038/5845. [DOI] [PubMed] [Google Scholar]
- 30.Mandiyan V., Andreev J., Schlessinger J., Hubbard S. R. Crystal structure of the ARF-GAP domain and ankyrin repeats of PYK2-associated protein β. EMBO J. 1999;18:6890–6898. doi: 10.1093/emboj/18.24.6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zweifel M. E., Barrick D. Studies of the ankyrin repeats of the Drosophila melanogaster Notch receptor. 2. Solution stability and cooperativity of unfolding. Biochemistry. 2001;40:14357–14367. doi: 10.1021/bi011436+. [DOI] [PubMed] [Google Scholar]
- 32.Bradley C. M., Barrick D. Limits of cooperativity in a structurally modular protein: responses of the Notch ankyrin domain to analogous alanine substitutions in each repeat. J. Mol. Biol. 2002;324:373–386. doi: 10.1016/s0022-2836(02)00945-2. [DOI] [PubMed] [Google Scholar]
- 33.Thompson J. D., Higgins D. G., Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cuff J. A., Clamp M. E., Siddiqui A. S., Finlay M., Barton G. J. JPred: a consensus secondary structure prediction server. Bioinformatics. 1998;14:892–893. doi: 10.1093/bioinformatics/14.10.892. [DOI] [PubMed] [Google Scholar]
- 35.Innes C. A., Shi J., Blundell T. L. Evolutionary trace analysis of TGF-β and related growth factors: implications for site-directed mutagenesis. Protein Eng. 2000;13:839–847. doi: 10.1093/protein/13.12.839. [DOI] [PubMed] [Google Scholar]
- 36.Chelliah V., Chen L., Blundell T. L., Lowell S. C. Distinguishing structural and functional restraints in evolution in order to identify interaction sites. J. Mol. Biol. 2004;342:1487–1504. doi: 10.1016/j.jmb.2004.08.022. [DOI] [PubMed] [Google Scholar]
- 37.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 38.Matthews B. W. Solvent content of protein crystals. J. Mol. Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 39.Navaza J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. Sect. A Found. Crystallogr. 1994;A50:157–163. [Google Scholar]
- 40.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 41.McRee D. E. XtalView/Xfit-A versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 42.Murshudova G. N., Vagin A. A., Dodson E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D Biol. Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 43.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. PROCHECK – a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 44.Hooft R. W. W., Vriend G., Sander C., Abola E. E. Errors in protein structures. Nature (London) 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- 45.Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zweifel M. E., Barrick D. Studies of the ankyrin repeats of the Drosophila melanogaster Notch receptor. 1. Solution conformational and hydrodynamic properties. Biochemistry. 2001;40:14344–14356. doi: 10.1021/bi011435h. [DOI] [PubMed] [Google Scholar]
- 47.Tipp K. W., Barrick D. The tolerance of a modular protein to duplication and deletion of internal repeats. J. Mol. Biol. 2004;344:169–178. doi: 10.1016/j.jmb.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 48.Michaely P., Tomchick D. R., Machius M., Anderson R. G. Crystal structure of a 12 ANK repeat stack from human ankyrinR. EMBO J. 2002;21:6387–6396. doi: 10.1093/emboj/cdf651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jacobs M. D., Harrison S. C. Structure of an IκBα/NF-κB complex. Cell. 1998;95:749–758. doi: 10.1016/s0092-8674(00)81698-0. [DOI] [PubMed] [Google Scholar]
- 50.Michel F., Soler-Lopez M., Petosa C., Cramer P., Siebenlist U., Müller C. W. Crystal structure of the ankyrin repeat domain of Bcl-3: a unique member of the IκB protein family. EMBO J. 2001;20:6180–6190. doi: 10.1093/emboj/20.22.6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lubman O. Y., Kopan R., Waksman G., Korolev S. The crystal structure of a partial mouse Notch-1 ankyrin domain: repeats 4 through 7 preserve an ankyrin fold. Protein Sci. 2005;14:1274–1281. doi: 10.1110/ps.041184105. [DOI] [PMC free article] [PubMed] [Google Scholar]