Abstract
Regulation of thrombin activity is critical for haemostasis and the prevention of thrombosis. Thrombin has several procoagulant substrates, including fibrinogen and platelet receptors, and essential cofactors for stimulating its own formation. However, thrombin is also capable of serving an anticoagulant function by activating protein C. The specificity of thrombin is primarily regulated by binding to the cofactor TM (thrombomodulin), but co-ordination of Na+ can also affect thrombin activity. The Na+-free form is often referred to as ‘slow’ because of reduced rates of cleavage of procoagulant substrates, but the slow form is still capable of rapid activation of protein C in the presence of TM. The molecular basis of the slow proteolytic activity of thrombin has remained elusive, in spite of two decades of solution studies and many published crystallographic structures. In the present paper, we report the first structure of wild-type unliganded human thrombin grown in the absence of co-ordinating Na+. The Na+-binding site is observed in a highly ordered position 6 Å (1 Å=0.1 nm) removed from that seen in the Na+-bound state. The movement of the Na+ loop results in non-catalytic hydrogen-bonding in the active site and blocking of the S1 and S2 substrate-binding pockets. Similar, if more dramatic, changes were observed in a previous structure of the constitutively slow thrombin variant E217K. The slow behaviour of thrombin in solutions devoid of Na+ can now be understood in terms of an equilibrium between an inert species, represented by the crystal structure described in the present paper, and an active form, where the addition of Na+ populates the active state.
Keywords: allostery, crystal structure, haemostasis, slow thrombin, sodium
Abbreviations: DTT, dithiothreitol; PEG, poly(ethylene glycol); PPACK, D-Phe-Pro-Arg chloromethylketone; R.M.S.D., root mean square deviation; TM, thrombomodulin
INTRODUCTION
The importance of thrombin is underscored by its position as the final serine protease that is generated in the blood coagulation cascade. Thrombin serves multiple functions in haemostasis, the generation of a blood clot (for reviews, see [1,2]), including cleavage of fibrinogen into its polymerogenic form fibrin, activation of platelets through cleavage of protease-activated receptors and stimulation of its own formation through cleavage activation of essential cofactors V and VIII. Thrombin also stabilizes fibrin clots through activation of Factor XIII, which cross-links fibrin polymers, and of TAFI (thrombin activatable fibrinolysis inhibitor), which inhibits the proteolytic breakdown of clots. However, thrombin can also act as an anticoagulant through activation of protein C, when bound to the endothelial cell-surface cofactor TM (thrombomodulin) [3]. Activated protein C shuts down thrombin formation through cleavage inactivation of Factors Va and VIIIa (for a review, see [4]). This complex network of thrombin activities is co-ordinated through competition for exosites on the surface of thrombin (for reviews, see [5,6]), and may also involve changes in specificity caused by cofactor-induced alteration of the active-site properties of thrombin. Although the structure of thrombin bound to TM did not reveal any active site conformational change [7], the structure was solved in the presence of an active-site inhibitor, and recent solution studies support a role of allostery in increasing the rate of protein C cleavage [8,9]. It has also been shown that thrombin specificity can be altered by co-ordination to the univalent cation sodium (Na+). In the absence of Na+, thrombin cleaves its prothrombotic substrates with reduced efficiency (7-fold for fibrinopeptide A), but is still capable of efficient cleavage of protein C when bound to TM [10]. Na+-free thrombin is thus considered to be an anticoagulant because of the reduced ability to cleave procoagulant substrates, while ability to activate protein C in the presence of TM is retained. The Na+-free and Na+-bound forms have become known as ‘slow’ and ‘fast’ thrombin [11] respectively, and many solution and crystallographic studies have been carried out to endeavour to discern the molecular basis for their observed catalytic differences.
Solution studies have established that the slow form has a constricted active-site cleft with several aromatic side chains in altered conformations [11–15]. Of particular importance is the proven involvement of Trp215 (template numbering is based on chymotrypsin [16]) which changes its quantum yield upon Na+ co-ordination, accounting for all of the observed 10–18% intrinsic fluorescence enhancement [17]. Na+ is octahedrally co-ordinated to the two main-chain oxygen atoms of residues Arg221a and Lys224 and four water molecules [18,19], and mutations in and adjacent to the Na+-binding loop have been made to create constitutively slow variants of thrombin for possible therapeutic use. Thus the E217K thrombin variant was discovered by scanning [20] and saturation mutagenesis [21], and was found to cleave fibrinogen 270-fold less efficiently than wild-type, while retaining near-wild-type activity towards protein C when bound to TM. Similar properties have been described for the double mutant W215A/E217A [22,23].
Recently, three structures of thrombin variants have been reported which were claimed to represent the slow form. In two reports, Na+ was excluded from the crystallization condition for thrombin variant R77aA, in the absence and presence of PPACK (D-Phe-Pro-Arg chloromethylketone) [24,25]. The authors concluded that the structures represented slow thrombin based solely on the exclusion of Na+ from the crystallization buffer; however, the structures were essentially indistinguishable from those of Na+-co-ordinated thrombin. We have published two structures of thrombin variants with structural features consistent with the observed biochemical properties of slow thrombin. The structure of S195A thrombin revealed an altered configuration of side chains for residues Trp60d, Cys168, Cys182, Trp215 and Phe227, leading to a constriction of the active-site cleft in the region of the aryl binding pocket (S2–S4 positions) [26]. This was followed by the structure of the constitutively slow variant E217K which shared the features of S195A and also revealed non-catalytic hydrogen bonding in the active site caused by a repositioning of the Na+-binding loop [27].
In the present paper, we report the 1.93 Å (1 Å=0.1 nm) structure of wild-type human thrombin solved from crystals grown in the absence of co-ordinating univalent cation. The structure is similar to that of E217K thrombin, revealing blocked S1 and S2 pockets and an altered non-catalytic hydrogen-bonding network in the active site.
EXPERIMENTAL
Escherichia coli expression of human thrombin
Human prethrombin-2 was expressed and refolded with some modifications to previously described methods [28,29]. Briefly, cDNA corresponding to the prethrombin-2 sequence was cloned from IMAGE clone ref. 6283420 and subcloned into pET23(+) vector (Novagen) pre-digested with HindIII and EcoRI. After transformation of the prethrombin-2 vector into E. coli strain BL21(DE3)pLysS, cells were cultured at 37 °C in 2×TY (tryptone/yeast extract) broth with 50 μg/ml ampicillin and 34 μg/ml chloramphenicol to a D595 of 0.6, followed by induction with 1 mM IPTG (isopropyl β-D-thiogalactoside) for 4 h. Harvested cells were stored at −80 °C. Thawed cell pellets (typically from 2 litres of cell culture) were lysed in 20 mM Tris/HCl, 1% (v/v) Triton X-100, 20 mM EDTA and 20 mM DTT (dithiothreitol), pH 7.4, and sonicated on ice in 10-s bursts for a total of 5 min. After centrifugation at 20000 g for 20 min, inclusion body pellets were sequentially washed, then centrifuged, with 20 mM Tris/HCl, 20 mM EDTA and 20 mM DTT, pH 7.4, first containing 1% (v/v) Triton X-100, then containing 1.0 M NaCl, and finally with 20 mM Tris/HCl and 20 mM EDTA, pH 7.4, alone. The washed pellet was resuspended in 20 ml of 7.0 M guanidinium chloride, 20 mM Tris/HCl, 0.5 mM EDTA and 20 mM DTT, pH 8.0, then desalted into 6 M guanidinium chloride, 20 mM Tris/HCl and 0.5 mM EDTA, pH 8.0, and glutathione was added to final concentrations of 5 mM reduced glutathione and 2 mM oxidized glutathione, before incubation at room temperature for 3 h. After adjusting the pH to 5.0, the solubilized protein was dialysed against 6.0 M guanidinium chloride and 20 mM EDTA, pH 5.0. Refolding was initiated by rapid dilution dropwise into 100 vol. of 50 mM Tris/HCl, 0.6 M arginine, 20 mM CaCl2, 10% (v/v) glycerol and 0.2% (v/v) Brij58, pH 8.5, at room temperature for 24 h. The refolded protein was concentrated to 100 ml using a 10000 Da molecular mass cut-off VivaFlow 200 module (VivaScience), then dialysed twice against 5 litres of 25 mM Tris/HCl, 2 mM EDTA, 0.1% (v/v) PEG [poly(ethylene glycol)] 6000 at 4 °C overnight. The precipitate was removed by centrifugation and filtration before purification of the correctly folded prethrombin-2 on a 5 ml heparin–Sepharose column (Amersham Biosciences) eluting with a gradient of 0–1.0 M NaCl. The elution position of the correctly folded prethrombin-2 was determined by measuring the amidolytic activity towards the chromogenic substrate S-2238 (Chromogenix) after activation of a sample of each fraction with snake venom from Echis carinatus. The peak was pooled, diluted 5-fold in 50 mM Tris/HCl, pH 7.4, and re-purified on heparin–Sepharose. Prethrombin-2 was activated by incubation for 3 h at 37 °C with snake venom from E. carinatus (10% by mass) that had been pre-treated with PMSF to inhibit serine protease activity. After 5-fold dilution with 50 mM Tris/HCl, pH 7.4, thrombin was purified on a heparin–Sepharose column as above. Correct refolding of the protein was verified by its ability to form a stoichiometric SDS-stable complex with antithrombin (results not shown).
Crystallization, data collection and refinement
Active thrombin eluted from the final heparin–Sepharose column at approx. 650 mM NaCl. The peak was immediately and exhaustively dialysed against 20 mM Tris/HCl and 650 mM LiCl, pH 7.4, then concentrated to 5.7 mg/ml using a 2 ml 10000 Da molecular-mass cut-off VivaSpin concentrator (VivaScience). Crystallization was by vapour diffusion in hanging drops consisting of 2 μl of thrombin and 2 μl of precipitant solution [24% (v/v) PEG 3350 and 250 mM magnesium acetate]. Diffraction-quality crystals grew within 2–3 days, and data were collected on the seventh day after crystal trials were established. The volume of the crystallization drops (4 μl) remained unchanged for up to 2 months, so that the conditions under which crystals were obtained were 2.85 mg/ml thrombin, 10 mM Tris/HCl, 325 mM LiCl, 125 mM magnesium acetate and 12% (v/v) PEG 3350, pH 7.0. Crystals were washed, dissolved in SDS sample buffer and run on SDS/PAGE to verify that they were composed exclusively of intact α-thrombin. Crystals were cryoprotected in 300 mM LiCl, 160 mM magnesium acetate, 22% (v/v) PEG 3350 and 10% (v/v) glycerol before flash-cooling to 100 K in a nitrogen vapour stream. Low- and high-resolution datasets were collected from a single crystal that had been annealed by blocking the cryostream three times for 3 s, followed by a fourth 10-s anneal at the Daresbury Synchrotron Radiation Source (Warrington, Cheshire, U.K.) station 9.6. The data were processed using Mosflm, Scala and Truncate [30]. The structure was solved by molecular replacement using the program MolRep [31] with the original PPACK-bound thrombin structure [16] (PDB accession code 1PPB) as the search model, and two molecules were placed in the asymmetric unit. After rigid body refinement, initial maps were generated by cycling between the automated model building utility of ARP/wARP [32] and refinement using Refmac [33]. The first rebuild in XtalView [34] was conducted on the template thrombin structure (PDB accession code 1XMN) [35] using the map produced by ARP/wARP, in order to minimize potential model bias. Further refinement was conducted using the program CNS [34] (version 1.0), and NCS restraints were not used. Data processing and refinement statistics are given in Table 1. The structure of thrombin bound to the C-terminal hirudin peptide (hirugen, PDB accession code 1HAH [36]) was chosen to represent fast thrombin because it was isomorphous to the crystal from which the Na+-binding site was first identified [19], and because it has a free active-site cleft. It thus represents an allosterically activated thrombin structure through occupation of both the Na+ site and exosite I. Figures were made using Bobscript [37], Raster3D [38] and Spock.
Table 1. Data processing, refinement and model (PDB accession code 2AFQ).
| Parameter | Value | ||
|---|---|---|---|
| Crystal | |||
| Space group | P21212 | ||
| Cell dimensions (Å) | a=153.34 | b=82.31 | c=50.93 |
| Solvent content (%) | 41.8 | ||
| Data processing statistics | |||
| Wavelength (Å) | 0.87 (SRS Daresbury, Station 9.6) | ||
| Resolution (Å) | 50.90–1.93 | 2.03–1.93 | |
| Total reflections | 403365 | 47912 | |
| Unique reflections | 49378 | 7065 | |
| Multiplicity | 8.2 | 6.8 | |
| 〈I/σ(I)〉 | 4.8 | 1.1 | |
| 〈I〉/σ(〈I〉) | 14.6 | 3.0 | |
| Completeness (%) | 99.9 | 99.4 | |
| Rmerge* | 0.10 | 0.55 | |
| Model | |||
| Atoms in asymmetric unit: | |||
| Protein | 4486 | ||
| Water | 478 | ||
| Glycerol molecules | 9 | ||
| Average B-factor (Å2) | 26.9 | ||
| Refinement statistics | 20–1.93 Å | 2.05–1.93 Å | |
| Reflections in working/free set | 47766/1513 | 7555/250 | |
| R-factor†/R-free (%) | 19.7/22.9 | 25.9/29.2 | |
| R.M.S.D. of bonds (Å)/angles (°) from ideality | 0.006/1.3 | ||
| Ramachandran plot; residues in: | |||
| Most favoured region (%) | 87.4 | ||
| Additionally allowed region (%) | 11.9 | ||
| Generously allowed region (%) | 0.2 | ||
| Disallowed region (%) | 0.4 (Cys220) | ||
* Rmerge=ΣhklΣi|Ihkl−〈Ihkl〉|/ΣhklΣi〈Ihkl〉.
† R-factor=Σhkl||Fobs|−|Fcalc||/Σhkl|Fobs|.
RESULTS AND DISCUSSION
Crystallization
Of the more than 150 structures of thrombin deposited in the PDB, none are of wild-type thrombin in the absence of an active site or exosite inhibitor. The proclivity of thrombin to undergo autolysis has necessitated the use of inhibitors or mutagenesis under the high concentrations and long incubation times required to form protein crystals. Indeed, for the wild-type human thrombin produced in our E. coli expression system, we also observed autolysis with a half-life of 48 h at 37 °C in the presence of saturating Na+ (3.6 mg/ml thrombin at pH 7.4; results not shown). In the absence of Na+, thrombin was still susceptible to autolysis, and under conditions used for crystallization (2.85 mg/ml, 325 mM LiCl, 125 mM magnesium acetate and 10 mM Tris/HCl, pH 7.0, and 21 °C) the half-life corresponded to ∼6 days (results not shown). However, crystals grew to full size within 2–3 days and were found to contain only uncleaved material by SDS/PAGE (results not shown). Although there is a reported Kd for the binding of Li+ to thrombin [39,40], older literature and recent biophysical studies have concluded that thrombin structure and activity are not affected by lithium salts [14,15,41]. In addition, we have recently solved the structure of S195A thrombin (bound to antithrombin and heparin) at 320 mM Li+ [42], and, although the Na+-binding site was formed, no density corresponding to co-ordinated univalent cation was observed (see Supplementary Figure 1S at http://www.BiochemJ.org/bj/392/bj3920021add.htm). In order to determine whether, under our current crystallization conditions, Li+ would be expected to occupy the Na+-binding site, we conducted titrations of Li+ and Na+ into solutions containing thrombin and MgCl2. We found that, although Na+ affinity and fluorescence enhancement were normal, no Li+ binding was detected up to 500 mM (see Supplementary Figure 2S at http://www.BiochemJ.org/bj/392/bj3920021add.htm). Thus the crystals we obtained were of intact wild-type human thrombin grown in the absence of co-ordinating univalent cation.
Overall structure
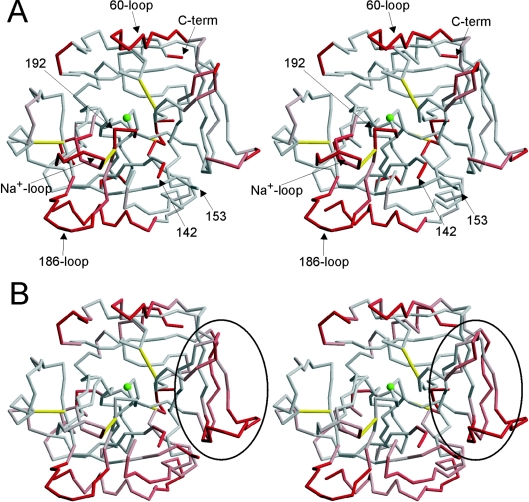
From a single flash-cooled crystal we obtained high-quality diffraction data to 1.93 Å resolution. Molecular replacement revealed two thrombin molecules in the asymmetric unit, and, as each monomer consists of a light and a heavy chain, the two monomers were denoted AB and CD to reflect the chain identifications. All of the subsequent structural analysis applies equally to the two monomers in the asymmetric unit. Interestingly, the only glycosylation site on thrombin, Asn60g, was found to mediate a crystal contact for monomer AB, indicating that the crystals could not have formed from glycosylated thrombin. The two monomers were essentially identical with respect to backbone conformation in regions containing secondary structure [R.M.S.D. (root mean square deviation) of 0.41 Å], and were also identical in these regions with thrombin structures commonly thought of as representing fast thrombin (R.M.S.D. of 0.7 Å for 1PPB, the original PPACK-bound thrombin structure, and for 1HAH, active-site free thrombin inhibited by the C-terminal hirudin peptide [36]). However, significant differences in the flexible surface loops resulted in Cα R.M.S.D.s of approx. 3 Å when comparing monomer AB with CD, 1PPB or 1HAH. When the flexible light chain was removed from the calculation, this value reduced to 0.77 Å for CD and approx. 2 Å for 1PPB and 1HAH. The Cα trace of AB coloured according to R.M.S.D. with 1HAH is given in Figure 1(A). The regions which commonly differ between thrombin structures are the N-terminal light chain, the C-terminus and the two loops which frame the active site, the 60-loop and the γ-loop (also known as the 147-loop or the autolysis loop). Indeed, the γ-loop is entirely disordered in both AB and CD, with no density observed for residues 143–152, although the presence of the intact loop was verified by SDS/PAGE (results not shown). The lack of density for the γ-loop in this structure is consistent with its increased proteolytic susceptibility in the absence of co-ordinated Na+ [15]. Importantly, several other regions are found in conformations which deviate from those found in fast thrombin. The Na+-binding loop (Cys220–Tyr225) has shifted 6 Å (Asp222), and the adjacent 186-loop has moved 5 Å (Lys186d). Movement of the Na+-binding loop has also affected the conformation of the active-site loop (Gly193–Ser195) because of their covalent connection through the disulphide bond between Cys220 and Cys191. This shift has been observed previously in the structure of the recombinant slow thrombin variant E217K [26].
Figure 1. Structural comparison of Na+-free wild-type thrombin with fast thrombin.
(A) The stereo representation of the Cα trace of monomer AB coloured according to R.M.S.D. with thrombin in its normal fast conformation (1HAH) reveals an overall conserved fold, but with significant conformational changes in certain regions. The colour ramp is from 1 to 3 Å R.M.S.D., from grey to red. Thrombin is shown in the standard orientation, with disulphide bonds in yellow, significant regions labelled, and the active-site Ser195 represented as a green ball. (B) Stereo representation of the Cα trace of monomer AB (in the same orientation as above, with the green ball representing Ser195, and yellow rods indicating disulphide bonds), coloured according to B-factor, from grey to red for B-factors from 20 to 40 Å2. Exosite I is indicated by the oval.
The overall flexibility of thrombin has been shown to increase in the absence of co-ordinated Na+ [15], and, consistent with this finding, we observed several regions with high B-factors (Figure 1B). Interestingly, these regions correspond to those which have shifted conformation with respect to fast thrombin (Figure 1A). An inability to model the γ-loop represents a significant increase in mobility, while more modest increases in modelled regions are coloured according to their B-factor. The two loops adjacent to the Na+-binding loop (the 186-loop and the γ-loop) are highly disordered, with density lacking for the entire γ-loop, and the 186-loop difficult to model in the poor density. Both loops form stabilizing contacts with the Na+-binding loop in the fast form, but the repositioning of the Na+ loop observed in the present study has broken the contacts. Interestingly, the Na+-binding and active-site loops are not highly mobile, in spite of a significant shift in their conformations. This suggests a concerted flipping between two stable conformations for these loops, with two distinct sets of stabilizing contacts. The high-quality electron density for this region and the observed network of hydrogen bonds support this hypothesis (see Figure 2). Another region for which shift in position does not correlate with high B-factor is exosite I. TM binds at exosite I with high affinity and can convert slow thrombin into an effective activator of protein C. The allosteric change induced by TM binding to slow thrombin is equivalent to the change induced by hirugen (which also binds in exosite I), by Na+ or by active-site inhibitors [13]. The mobility of exosite I observed in AB could explain the allosteric effect of exosite I-binding ligands on slow thrombin.
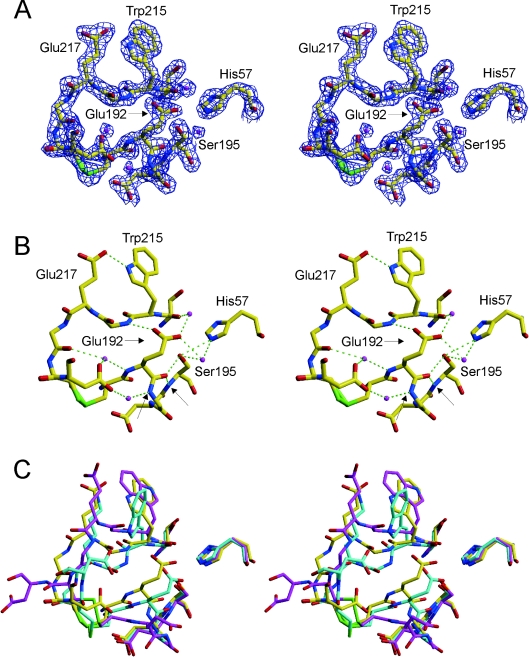
Figure 2. The active site of Na+-free wild-type thrombin is non-catalytic.
(A) The stereo representation of electron density (blue) surrounding residues 57, 191–195 and 214–221 contoured at 2σ indicates the high quality of the structure in the active-site region. Carbon atoms are shown in yellow, oxygen in red, nitrogen in blue, sulphur in green, and waters are magenta. (B) The hydrogen-bonding interactions in the active site are extensive and non-catalytic (green broken lines). Of particular importance for the activity of thrombin is the movement of Glu192, which results in a main-chain hydrogen bond to the side-chain Hγ and main-chain N–H of Ser195, and the flipping of the oxyanion hole partner Gly193. The atoms which normally constitute the oxyanion hole (Gly193 and Ser195 nitrogens) are indicated by arrows. (C) A superposition of the same active-site residues for Na+-free wild-type thrombin (yellow), recombinant slow thrombin E217K variant (cyan) and normal fast thrombin (magenta) illustrates the radical nature of the active site perturbations caused by the absence of Na+. The active-site geometry is similarly perturbed for the E217K variant, but the perpendicular orientation of the side chain of Trp215 further blocks the active site. The orientation of the Trp215 side chain in Na+-free thrombin is parallel to that of fast thrombin, but has flipped 180° to form a hydrogen bond with Glu217 (see B).
The active site
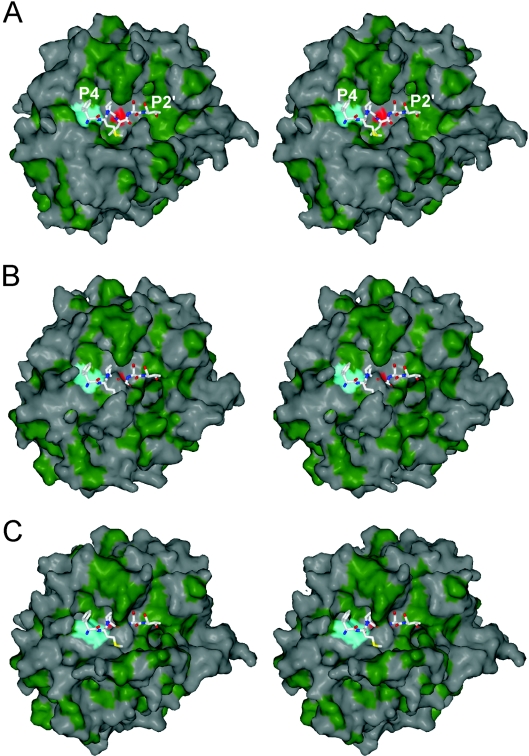
Thrombin in the absence of Na+ co-ordination is ‘slow’ due to an increased Km and decreased kcat towards peptide and protein substrates. These features are explained nicely by the active-site conformation observed in this structure. Active-site residues 57, 191–195 and 214–221 are shown in Figure 2(A), with 2Fo–Fc electron density contoured at twice the R.M.S.D. of the map (2σ) to illustrate the high quality of the structure in this region. The low B-factors and high-quality density are due to a network of hydrogen-bonding interactions, shown in Figure 2(B), which result in a significant blocking of the active-site cleft and a rearrangement of the active-site geometry. Glu192 has shifted approx. 3 Å to make a water-mediated hydrogen bond between its side chain oxygens and the side chains of Ser195 and His57 and the main-chain oxygen of Ser214, and a direct hydrogen bond with main-chain nitrogen of Gly216. In addition, the backbone conformation of Glu192 now places the main-chain oxygen in position to hydrogen-bond with both the side chain and main chain of Ser195, thus rendering Ser195 non-catalytic. The oxyanion hole is also significantly reordered in this structure, with the main-chain nitrogen of Ser195 hydrogen-bonded to the main-chain oxygen of Glu192, and, additionally, the movement of Glu192 has reoriented Gly193 so that it hydrogen-bonds to Asp221 (through a water molecule). This new active-site hydrogen-bonding configuration explains the significantly reduced kcat of thrombin in the absence of co-ordinated Na+. The increased Km is similarly explained. Figure 3 shows surface representations of thrombin in the standard orientation (active-site-facing, so that substrates bind from left to right from N- to C-termini). A comparison of fast thrombin (1HAH; Figure 3A) with that of thrombin monomer AB (Figure 3B) reveals a constriction of the S1 and S2 pockets which would cause extensive steric overlap with the P1 and P2 residues of a potential substrate. Thus the structure of Na+-free thrombin presented here has an altered active-site architecture leading to a non-catalytic hydrogen-bonding network and the steric blocking of the substrate-binding pocket.
Figure 3. Surface representations of thrombin reveal the blocking of the substrate-binding pocket.
(A) A stereo view of the surface of fast thrombin (PDB accession code 1HAH; coloured green for hydrophobicity), in the standard orientation, shows an open active-site cleft which is competent in binding a substrate peptide (shown here as rods is the P4–P2′ region of the reactive centre loop of HCII from PDB accession code 1JMO). The active site is indicated by colouring the Oγ atom of Ser195 red, and the aryl-binding pocket is indicated in a similar fashion by colouring the entire Trp215 residue cyan. (B) In contrast, the active-site cleft of Na+-free wild-type thrombin is in a more closed conformation with the S1 and S2 pockets significantly blocked. The restriction is evident from the overlap of the substrate and the burying of the catalytic serine (red). (C) The same representation of recombinant slow thrombin (E217K) shows an even greater occlusion of the active site, and the further blocking of the aryl-binding pocket (S2–S4) by the reorienting of the Trp215 side chain (cyan).
We recently solved a structure of the constitutively slow thrombin variant E217K, shown for comparison in Figure 2(C), in which we observed a similar occlusion of the active site cleft due to the movement of the Na+-binding loop and residue Glu192. Although the oxyanion hole was disrupted by the same interactions, the active-site Ser195 was observed in direct contact with the side chain of Glu192. The effect was a somewhat more dramatic blocking of the S1 pocket (Figure 3C). In addition, Trp215 was observed in a perpendicular orientation to its normally observed conformation, resulting in a partial obstruction of the aryl-binding pocket (S2–S4) (Figure 3C). We observed a similar orientation of Trp215 in an earlier structure of S195A thrombin [25]. In the structure presented here, we do not see an obstruction of the aryl-binding pocket, as Trp215 is in a parallel configuration. However, as seen in Figure 2(C), the side chain of Trp215 is flipped 180° with respect to its conformation in fast thrombin. The reason for this new conformation is a hydrogen bond with Glu217, illustrated in Figure 2(B). This interaction cannot be formed by the E217K variant, causing Trp215 to adopt a conformation which further occludes the active-site cleft and helps explain the reduced activity of the variant relative to Na+-free wild-type thrombin.
Conclusions
The structure of wild-type human thrombin in the Na+-free state presented here is consistent with the properties ascribed to the slow form of thrombin: (i) it has a more closed active-site cleft with the blocking of the S1 and S2 pockets, explaining the increased Km; (ii) the active site is found in a non-catalytic hydrogen-bonding network, explaining the reduced kcat; (iii) Trp215 is in a conformation different from that of the fast form, explaining the change in fluorescence quantum yield; (iv) the surface loops are more flexible, consistent with recent biochemical data [15]; and (v) the Na+-binding loop is in a different conformation to that found in the Na+-bound state. In addition, the structure of Na+-free wild-type thrombin is similar to that of the recombinant slow thrombin variant E217K. These structures suggest that the poor catalytic efficiency of thrombin in the absence of Na+ reflects an equilibrium between an inert state, like that described here, and an active state, where the effect of Na+ addition is to pull the equilibrium towards the right and promote population of the active state. Stopped flow data support the equilibrium model by demonstrating that one-third of thrombin in the absence of Na+ was incapable of binding to either Na+ or an active-site inhibitor [13]. High concentrations of Na+ or inhibitor resulted in the same limiting rate, which was interpreted to be the zero-order rate of conversion of the inert state to a binding-competent state. The conformation of the active state has been known for decades; that of the inert state has now been revealed.
Online data
Acknowledgments
Funding for J.A.H. was provided by the Medical Research Council (UK) and the British Heart Foundation, and T.E.A. was supported by a grant from the Isaac Newton Trust (Cambridge, U.K.). We thank R.J. Read for help with running ARP/wARP.
References
- 1.Stubbs M. T., Bode W. A player of many parts: the spotlight falls on thrombin's structure. Thromb. Res. 1993;69:1–58. doi: 10.1016/0049-3848(93)90002-6. [DOI] [PubMed] [Google Scholar]
- 2.Bode W. The structure of thrombin, a chameleon-like proteinase. J. Thromb. Haemostasis. 2005 doi: 10.1111/j.1538-7836.2005.01356.x. 10.1111/j.1538-7836.2005.01356.x. [DOI] [PubMed]
- 3.Esmon C. T. The regulation of natural anticoagulant pathways. Science. 1987;235:1348–1352. doi: 10.1126/science.3029867. [DOI] [PubMed] [Google Scholar]
- 4.Esmon C. T. The protein C pathway. Chest. 2003;124:26S–32S. doi: 10.1378/chest.124.3_suppl.26s. [DOI] [PubMed] [Google Scholar]
- 5.Huntington J. A. Molecular recognition mechanisms of thrombin. J. Thromb. Haemostasis. 2005;3:1861–1872. doi: 10.1111/j.1538-7836.2005.01363.x. [DOI] [PubMed] [Google Scholar]
- 6.Lane D. A., Philippou H., Huntington J. A. Directing thrombin. Blood. 2005;106:2605–2612. doi: 10.1182/blood-2005-04-1710. [DOI] [PubMed] [Google Scholar]
- 7.Fuentes-Prior P., Iwanaga Y., Huber R., Pagila R., Rumennik G., Seto M., Morser J., Light D. R., Bode W. Structural basis for the anticoagulant activity of the thrombin–thrombomodulin complex. Nature (London) 2000;404:518–525. doi: 10.1038/35006683. [DOI] [PubMed] [Google Scholar]
- 8.Rezaie A. R., Yang L. Thrombomodulin allosterically modulates the activity of the anticoagulant thrombin. Proc. Natl. Acad. Sci. U.S.A. 2003;100:12051–12056. doi: 10.1073/pnas.2135346100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu G., Chhum S., Krishnaswamy S. The affinity of protein C for the thrombin–thrombomodulin complex is determined in a primary way by active site-dependent interactions. J. Biol. Chem. 2005;280:15471–15478. doi: 10.1074/jbc.M500881200. [DOI] [PubMed] [Google Scholar]
- 10.Dang O. D., Vindigni A., Di Cera E. An allosteric switch controls the procoagulant and anticoagulant activities of thrombin. Proc. Natl. Acad. Sci. U.S.A. 1995;92:5977–5981. doi: 10.1073/pnas.92.13.5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wells C. M., Di Cera E. Thrombin is a Na+-activated enzyme. Biochemistry. 1992;31:11721–11730. doi: 10.1021/bi00162a008. [DOI] [PubMed] [Google Scholar]
- 12.Orthner C. L., Kosow D. P. Evidence that human α-thrombin is a monovalent cation-activated enzyme. Arch. Biochem. Biophys. 1980;202:63–75. doi: 10.1016/0003-9861(80)90406-3. [DOI] [PubMed] [Google Scholar]
- 13.Lai M. T., Di Cera E., Shafer J. A. Kinetic pathway for the slow to fast transition of thrombin: evidence of linked ligand binding at structurally distinct domains. J. Biol. Chem. 1997;272:30275–30282. doi: 10.1074/jbc.272.48.30275. [DOI] [PubMed] [Google Scholar]
- 14.Villanueva G. B., Perret V. Effects of sodium and lithium salts on the conformation of human α-thrombin. Thromb. Res. 1983;29:489–498. doi: 10.1016/0049-3848(83)90344-4. [DOI] [PubMed] [Google Scholar]
- 15.De Filippis V., De Dea E., Lucatello F., Frasson R. Effect of Na+ binding on the conformation, stability and molecular recognition properties of thrombin. Biochem. J. 2005;390:485–492. doi: 10.1042/BJ20050252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bode W., Mayr I., Baumann U., Huber R., Stone S. R., Hofsteenge J. The refined 1.9 Å crystal structure of human α-thrombin: interaction with D-Phe-Pro-Arg chloromethylketone and significance of the Tyr-Pro-Pro-Trp insertion segment. EMBO J. 1989;8:3467–3475. doi: 10.1002/j.1460-2075.1989.tb08511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arosio D., Ayala Y. M., Di Cera E. Mutation of W215 compromises thrombin cleavage of fibrinogen, but not of PAR-1 or protein C. Biochemistry. 2000;39:8095–8101. doi: 10.1021/bi0006215. [DOI] [PubMed] [Google Scholar]
- 18.Di Cera E., Guinto E. R., Vindigni A., Dang Q. D., Ayala Y. M., Wuyi M., Tulinsky A. The Na+ binding site of thrombin. J. Biol. Chem. 1995;270:22089–22092. doi: 10.1074/jbc.270.38.22089. [DOI] [PubMed] [Google Scholar]
- 19.Zhang E., Tulinsky A. The molecular environment of the Na+ binding site of thrombin. Biophys. Chem. 1997;63:185–200. doi: 10.1016/s0301-4622(96)02227-2. [DOI] [PubMed] [Google Scholar]
- 20.Gibbs C. S., Coutre S. E., Tsiang M., Li W. X., Jain A. K., Dunn K. E., Law V. S., Mao C. T., Matsumura S. Y., Mejza S. J. Conversion of thrombin into an anticoagulant by protein engineering. Nature (London) 1995;378:413–416. doi: 10.1038/378413a0. [DOI] [PubMed] [Google Scholar]
- 21.Tsiang M., Paborsky L. R., Li W. X., Jain A. K., Mao C. T., Dunn K. E., Lee D. W., Matsumura S. Y., Matteucci M. D., Coutre S. E., et al. Protein engineering thrombin for optimal specificity and potency of anticoagulant activity in vivo. Biochemistry. 1996;35:16449–16457. doi: 10.1021/bi9616108. [DOI] [PubMed] [Google Scholar]
- 22.Cantwell A. M., Di Cera E. Rational design of a potent anticoagulant thrombin. J. Biol. Chem. 2000;275:39827–39830. doi: 10.1074/jbc.C000751200. [DOI] [PubMed] [Google Scholar]
- 23.Gruber A., Cantwell A. M., Di Cera E., Hanson S. R. The thrombin mutant W215A/E217A shows safe and potent anticoagulant and antithrombotic effects in vivo. J. Biol. Chem. 2002;277:27581–27584. doi: 10.1074/jbc.C200237200. [DOI] [PubMed] [Google Scholar]
- 24.Pineda A. O., Carrell C. J., Bush L. A., Prasad S., Caccia S., Chen Z. W., Mathews F. S., Di Cera E. Molecular dissection of Na+ binding to thrombin. J. Biol. Chem. 2004;279:31842–31853. doi: 10.1074/jbc.M401756200. [DOI] [PubMed] [Google Scholar]
- 25.Pineda A. O., Savvides S., Waksman G., Di Cera E. Crystal structure of the anticoagulant slow form of thrombin. J. Biol. Chem. 2002;277:40177–40180. doi: 10.1074/jbc.C200465200. [DOI] [PubMed] [Google Scholar]
- 26.Huntington J. A., Esmon C. T. The molecular basis of thrombin allostery revealed by a 1.8 Å structure of the “slow” form. Structure. 2003;11:469–479. doi: 10.1016/s0969-2126(03)00049-2. [DOI] [PubMed] [Google Scholar]
- 27.Carter W. J., Myles T., Gibbs C. S., Leung L. L., Huntington J. A. Crystal structure of anticoagulant thrombin variant E217K provides insights into thrombin allostery. J. Biol. Chem. 2004;279:26387–26394. doi: 10.1074/jbc.M402364200. [DOI] [PubMed] [Google Scholar]
- 28.Soejima K., Mimura N., Yonemura H., Nakatake H., Imamura T., Nozaki C. An efficient refolding method for the preparation of recombinant human prethrombin-2 and characterization of the recombinant-derived α-thrombin. J. Biochem. (Tokyo) 2001;130:269–277. doi: 10.1093/oxfordjournals.jbchem.a002982. [DOI] [PubMed] [Google Scholar]
- 29.DiBella E. E., Maurer M. C., Scheraga H. A. Expression and folding of recombinant bovine prethrombin-2 and its activation to thrombin. J. Biol. Chem. 1995;270:163–169. doi: 10.1074/jbc.270.1.163. [DOI] [PubMed] [Google Scholar]
- 30.Leslie A. W. G. Warrington: SERC, Daresbury Laboratory; 1992. Joint CCP4 and ESF-EACMB Newsletter on Protein Crystallography No. 26. [Google Scholar]
- 31.Vagin A., Teplyakov A. An approach to multi-copy search in molecular replacement. Acta Crystallogr. Sect. D Biol. Crystallogr. 2000;56:1622–1624. doi: 10.1107/s0907444900013780. [DOI] [PubMed] [Google Scholar]
- 32.Perrakis A., Morris R., Lamzin V. S. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 33.Murshudov G. N., Vagin A. A., Dodson E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 34.Brunger A. T., Adams P. D., Clore G. M., Delano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 35.Carter W. J., Cama E., Huntington J. A. Crystal structure of thrombin bound to heparin. J. Biol. Chem. 2004;280:2745–2749. doi: 10.1074/jbc.M411606200. [DOI] [PubMed] [Google Scholar]
- 36.Skrzypczak-Jankun E., Carperos V. E., Ravichandran K. G., Tulinsky A., Westbrook M., Maraganore J. M. Structure of the hirugen and hirulog 1 complexes of α-thrombin. J. Mol. Biol. 1991;221:1379–1393. [PubMed] [Google Scholar]
- 37.Esnouf R. M. An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graphics Modell. 1997;15:132. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
- 38.Merritt E. A., Murphy M. E. P. Raster3D Version 2.0: a program for photorealistic molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 39.Guinto E. R., Di Cera E. Critical role of W60d in thrombin allostery. Biophys. Chem. 1997;64:103–109. doi: 10.1016/s0301-4622(96)02211-9. [DOI] [PubMed] [Google Scholar]
- 40.Prasad S., Cantwell A. M., Bush L. A., Shih P., Xu H., Di Cera E. Residue Asp-189 controls both substrate binding and the monovalent cation specificity of thrombin. J. Biol. Chem. 2004;279:10103–10108. doi: 10.1074/jbc.M312614200. [DOI] [PubMed] [Google Scholar]
- 41.Landis B. H., Koehler K. A., Fenton J. W. Human thrombins: group IA and IIA salt-dependent properties of α-thrombin. J. Biol. Chem. 1981;256:4604–4610. [PubMed] [Google Scholar]
- 42.Li W., Johnson D. J., Esmon C. T., Huntington J. A. Structure of the antithrombin–thrombin–heparin ternary complex reveals the antithrombotic mechanism of heparin. Nat. Struct. Mol. Biol. 2004;11:857–862. doi: 10.1038/nsmb811. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.