Abstract
The main function of CAs (carbonic anhydrases) is to participate in the regulation of acid–base balance. Although 12 active isoenzymes of this family had already been described, analyses of genomic databases suggested that there still exists another isoenzyme, CA XV. Sequence analyses were performed to identify those species that are likely to have an active form of this enzyme. Eight species had genomic sequences encoding CA XV, in which all the amino acid residues critical for CA activity are present. However, based on the sequence data, it was apparent that CA XV has become a non-processed pseudogene in humans and chimpanzees. RT-PCR (reverse transcriptase PCR) confirmed that humans do not express CA XV. In contrast, RT-PCR and in situ hybridization performed in mice showed positive expression in the kidney, brain and testis. A prediction of the mouse CA XV structure was performed. Phylogenetic analysis showed that mouse CA XV is related to CA IV. Therefore both of these enzymes were expressed in COS-7 cells and studied in parallel experiments. The results showed that CA XV shares several properties with CA IV, i.e. it is a glycosylated glycosylphosphatidylinositol-anchored membrane protein, and it binds CA inhibitor. The catalytic activity of CA XV is low, and the correct formation of disulphide bridges is important for the activity. Both specific and non-specific chaperones increase the production of active enzyme. The results suggest that CA XV is the first member of the α-CA gene family that is expressed in several species, but not in humans and chimpanzees.
Keywords: bioinformatics, carbonic anhydrase XV, glycosylphosphatidylinositol (GPI) anchor
Abbreviations: CA, carbonic anhydrase; CA-RP, carbonic anhydrase-related protein; DTT, dithiothreitol; EndoH, endoglycosidase H; EST, expressed sequence tag; GPI, glycosylphosphatidylinositol; NP40, Nonidet P40; p-AMBS, p-aminomethylbenzenesulphonamide; PBA, 4-phenylbutyric acid; PI-PLC, phosphoinositide-specific phospholipase C; poly(A)+, polyadenylated; RT, reverse transcriptase
INTRODUCTION
CAs (carbonic anhydrases) are zinc-containing metalloenzymes that catalyse the reversible hydration of carbon dioxide according to the following reaction: CO2+H2O↔HCO3−+H+ [1]. This reaction forms the basis for the regulation of acid–base balance in organisms. In addition to this main function, CAs participate in a number of other physiological processes, such as CO2 and HCO3− transport, bone resorption, production of body fluids, gluconeogenesis, ureagenesis and lipogenesis [2]. During evolution, at least 12 active CA isoenzymes have emerged in both rodents and humans. The isoenzymes have differences in their tissue distribution, kinetic properties and subcellular localizations: CAs I, II, III, VII and XIII [3,4] are cytoplasmic, IV, IX, XII and XIV [5–8] are membrane-bound, VI is secreted [9] and VA (as well as VB) are located in mitochondria [10]. Three CA-RPs (CA-related proteins), VIII, X, XI, are closely similar to CAs, but they lack one or more of the critical histidine residues in their active site. Since these histidines are required to bind the zinc ion, which is essential for CO2 hydration activity, the CA-RPs lack enzymatic activity [11].
The aim of the present study was to characterize the novel CA XV, the cDNA sequence of which was submitted to the National Center for Biotechnology Information by Hewett-Emmett and Shimmin in 2000 (GenBank® accession no. AF231122). Genome-wide sequence analyses were performed to identify the species that might express an active CA XV enzyme in their proteome. The expression of this novel isoenzyme was studied in human and mouse tissues by RT (reverse transcriptase)-PCR, and the three-dimensional structure of the murine enzyme was predicted by computer modelling. Recombinant mouse CA XV was produced in COS-7 cells and Escherichia coli bacteria. Biochemical characterization of CA XV revealed that it is enzymatically active and is bound to the plasma membrane through a GPI (glycosylphosphatidylinositol) anchorage, as found for CA IV. The production of catalytically active recombinant enzyme in transfected COS-7 cells was markedly increased by treatment with both specific and non-specific chaperones, as recently reported for human CA IV [12].
EXPERIMENTAL
Bioinformatics analyses
Sequence analyses were performed in order to find out which species may produce an active CA XV. The procedure was started with the search and retrieval of known CA15 sequences [Mus musculus (house mouse)]. Subsequently, BLAT [13] searches were performed in all selected genomes using sequences found in the previous step as query sequences. The genomic sequences were obtained, and these were translated in three frames. The translations were aligned with known sequences, and the exon locations were visually identified. The gene models were confirmed and fine-tuned using GeneWise [14]. Finally, the final best transcripts and protein sequences were assembled manually.
The following sequences were obtained from Ensembl Genome Browser (http://www.ensembl.org): Mus musculus (ENSMUSP00000012152) and Rattus norvegicus (brown rat) (ENSRNOP00000000312). The UCSC Genome Browser (http://genome.ucsc.edu) showed two mRNAs for the CA15 of Gallus gallus (chicken): of these two, the accession number BX929589 was selected and translated into protein, because it was in closest accordance with other species. For Danio rerio (zebrafish), there was an EST (expressed sequence tag) sequence (CO960501) representing CA15 that was mainly of high quality. Because the latter part of the EST sequence was of lower quality, it was constructed manually from the genome. The sequence for Tetraodon nigroviridis (green spotted pufferfish) was also obtained from UCSC Genome Browser, Genscan Gene Prediction (GSCT00001777001). The CA15 of Fugu rubripes (fugu puffer-fish) was constructed manually by using the information of Ensembl Genome Browser (SINFRUP00000165581 and SINFRUP00000175429). The UCSC Genome Browser also showed some EST sequences for this gene. The gene for the CA15 of Canis familiaris (dog) could not be found in the biological databases, and it was constructed manually from UCSC Genome Browser from region chr26: 32168776–32171692. The CA XV amino acid sequence of Xenopus tropicalis (pipid frog) was constructed manually with the help of two EST sequences (BX734706 and AL890846). The sequence alignment including all the species was constructed with T-Coffee version 2.11 [15].
For humans (Homo sapiens) or chimpanzees (Pan troglodytes), there were no mRNAs or EST sequences representing CA15 in the Ensembl Genome Browser. However, the human genome was shown to have three copies of CA15 located to chromosome 22q11.21: positions 17393598–17396941, 18860411–18863739 and 20034808–20038136. For convenience, they will be referred to as human CA15 candidate genes 1, 2, and 3 respectively. In chimpanzee (Pan troglodytes), two gene candidates were observed in chromosome 23, at positions 17310504–17312161 and 19990225–19993434. These are referred as gene candidates 1 and 3 respectively. The second chimpanzee gene candidate that should be syntenic with the human gene candidate 2 was in a sequence gap of the genome. All of these gene candidates were observed to be pseudogenic, because they contained several frame-shifts and point mutations. Detailed information on these pseudogenes can be found in Supplementary Figures 1, 2 and 3 (see http://www.BiochemJ.org/bj/392/bj3920083add.htm).
The prediction of N-glycosylation sites for mouse CA XV was performed by using NetNGlyc 1.0 Server with default parameters (http://www.cbs.dtu.dk/services/NetNGlyc/).
The sequence alignment of CAs for phylogenetic analysis was performed with ClustalW [17]. Phylogenetic analysis was carried out with PAUP* 4.0 [18]. The majority rule consensus tree was obtained by three bootstrap runs with different random seeds. The dataset was bootstrapped 1000 times for each run.
The structural prediction for mouse CA XV was based on human CA IV at 2.8 Å (1 Å≡0.1 nm) resolution (PDB entry 1ZNC) [19]. In structural modelling, amino acids 25–304 (Figure 1) from the mouse CA XV were included. The model was constructed with the program InsightII (Acelrys Inc., San Diego, CA, U.S.A.). Amino acid substitutions were built using a side chain rotamer library. The initial model was refined with Discover™ in a stepwise manner by energy minimization using the Amber™ force field. The newly built loops were refined with 500 steps of minimization with a fixed and a free backbone respectively. Subsequently, all side chains with a constrained backbone were minimized for 500 steps, followed by a further 1000 steps of minimization for the whole model.
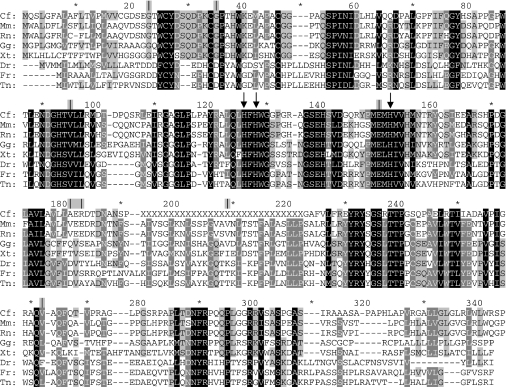
Figure 1. The results of sequence analyses.
The alignment of CA XV in eight species, showing its conservation throughout evolution. The three histidine residues co-ordinating the zinc atom are pointed out by arrows. Exon boundaries are indicated by vertical lines above the alignment; asterisks denote every tenth residue not labelled in the Figure. The only exon boundary having interspecies differences is located at amino acid residues 181–183. Abbreviations: Cf, Canis familiaris; Mm, Mus musculus; Rn, Rattus norvegicus; Gg, Gallus gallus; Xt, Xenopus tropicalis; Dr, Danio rerio; Fr, Fugu rubripes; Tn, Tetraodon nigroviridis. X, sequencing gap in the genome.
RT-PCR experiments, sequencing of the PCR products and in situ hybridization
The RT-PCR method was used to reveal those murine and human tissues that express CA15 mRNA. The expression studies were carried out using the commercial cDNA kits purchased from BD Biosciences (Palo Alto, CA, U.S.A.). The mouse MTC™ panel I contained first-strand cDNA preparations produced from total poly(A)+ (polyadenylated) RNAs isolated from 12 different murine tissues. In addition, mRNA was isolated from six mouse tissues absent in the panel (stomach, duodenum, jejunum, ileum, colon and blood) by using TRIzol® reagent (Invitrogen, Carlsbad, CA, U.S.A.). Reverse transcription was performed with Mo-MuLV reverse transcriptase (Finnzymes, Espoo, Finland) using random primers (500 μg/ml). The procedures were conducted according to the principles of the Declaration of Helsinki and approved by the institutional animal care committee (University of Tampere, Finland). The human MTC™ Panels I and II were used to study the expression of CA15 mRNA in 15 human tissues.
To study mRNA in mouse, sequence-specific primers for murine CA15 were designed using the information published in GenBank (accession number NM_030558). The forward primer (MF1) was 5′-TACCTGGTGCTACGACTC-3′ (nt 148–165) and the reverse primer (MR1) was 5′-TATCGGTAGTACCGCAAG-3′ (nt 739–756), and thus the resulting amplification product size was 609 bp.
Sequence analyses revealed that the human genome contains three copies of CA15 genes which have most likely become pseudogenes. In order to obtain experimental data concerning whether any of these candidate genes are expressed, primers were designed for each of them, and additionally, one primer pair was designed to recognize all of them. More detailed information on the primers used for human studies, as well as exact PCR conditions for all reactions, can be found in the Supplementary methods section (see http://www.BiochemJ.org/bj/392/bj3920083add.htm).
The results of the PCR reactions were analysed using a 1.5% agarose gel containing 0.1 μg/ml ethidium bromide with a DNA standard (100 bp DNA Ladder; New England Biolabs, Beverly, MA, U.S.A.). The 609 and 713 bp PCR products were sequenced in order to confirm their identity and to reveal possible unspecific binding of primers. The detailed protocol of the sequencing can be found in the Supplementary Methods section (http://www.BiochemJ.org/bj/392/bj3920083add.htm).
In situ hybridization was performed for mouse tissues as described by Heikinheimo et al. [20].
Expression of CA XV in COS-7 cells and CA assay
The full-length mouse CA15 cDNA in pME 18S-FL3 (I.M.A.G.E Consortium, clone ID 1908347, MRC Geneservice, Cambridge, U.K.) and, as a control, mouse CA4 cDNA in pCXN [21] was used for transient transfection of COS-7 cells using the DEAE–dextran procedure [22]. After 12 h of transfection, cells were treated with 100 mM chloroquine for 1 h [23]. After 72 h of transfection, COS-7 cells were harvested in cold PBS and the cell pellets were homogenized in 25 mM Tris/SO4 buffer, pH 7.5, containing protease inhibitors by sonication. The membrane-associated enzymes were separated after centrifugation at 100000 g for 30 min. The protein concentration was determined by the micro-Lowry protein assay [24].
The CA activity from cell lysates or membrane suspension was determined in duplicate by the procedure of Maren, as described previously [25]. The CA activity was expressed as units/mg of cell protein or membrane protein.
Binding of CA XV to the CA-inhibitor affinity resin
p-AMBS (p-aminomethylbenzenesulphonamide) affigel (Sigma–Aldrich, St Louis, MO, U.S.A.) affinity resin was equilibrated with 10 mM Hepes/NaOH, pH 7.5. Cell membrane extracts in 10 mM Hepes/NaOH, pH 7.5/1% (v/v) NP40 (Nonidet P40) buffer were mixed with the affinity resin at 4 °C for 30 min or on ice with intermittent mixing. Unbound proteins were removed and the resin–enzyme complex was washed with 25 mM Tris/SO4, pH 7.5, containing protease inhibitor buffer. The bound enzyme was eluted with SDS/PAGE sample buffer and analysed by Western blot. SDS/PAGE was carried out according to Laemmli [26] under reducing/non-reducing conditions. The polypeptides were characterized by Western blotting, as described previously [7]. For the detection of CA XV, the antibody for CA IV was used, because mouse CA XV shares a distinct homology with mouse CA IV. Affinity-purified recombinant mouse CA IV enzyme was used to raise antibodies in rabbit, as described previously [8,27]. The goat anti-rabbit IgG–peroxidase conjugate was purchased from Sigma–Aldrich. The intensity of the bands was quantified and expressed as the percentage enzyme bound of the total enzyme present in the membrane extract.
Deglycosylation of mouse CA XV and PI-PLC (phosphoinositide-specific phospholipase C) treatment
COS-7 cell membranes equivalent to 50 μg of protein were treated with EndoH (endoglycosidase H) from Boehringer Mannheim (Mannheim, Germany) as described in [28]. COS-7 cell membranes were treated with PI-PLC from ICN Biomedical Research Products (Costa Mesa, CA, U.S.A.) for 2 h at room temperature [29]. For a control, the membranes were treated with buffer alone. Both deglycosylation as well as GPI anchoring were analysed by SDS/PAGE followed by Western blotting.
In vitro refolding of CA XV by oxidized glutathione and in vivo refolding by chemical chaperones
CA XV protein was refolded using 10 mM oxidized glutathione (GSSG; Sigma-Aldrich) as described previously [30]. COS-7 cell membrane extract in 25 mM Tris/SO4, pH 7.5, containing 1% NP40 and protease inhibitors was incubated with 10 mM GSSG at 4 °C for 24 h before the enzyme assay. COS-7 cell membrane extracts overexpressing mouse CA IV were used as a positive control.
The transfected COS-7 cells, just after chloroquine treatment, were incubated with different concentrations of PBA (4-phenylbutyric acid; Sigma–Aldrich) and dorzolamide (Trusopt, Walgreen's Pharmacy, Deerfield, IL, U.S.A.) for 72 h. The cell lysates were used to isolate the cell membranes after centrifugation at 30000 g. The cell membranes were washed with 20 mM sodium acetate buffer, pH 5.5, to remove bound CA inhibitor. The membrane pellets were recovered after washing and suspended in 20 mM Tris/SO4 buffer, pH 7.5, for the CA activity measurement. The enzyme activity was measured in duplicate, and average enzyme activity was used to calculate the fold increase in the CA activity.
Expression and purification of recombinant CA XV produced in E. coli
The fragment of CA15 was amplified by PCR from a full-length CA15 into a pCXN vector. The NdeI and BamHI sites were introduced. The amplified product encoding amino acids 22–292 of the full-length native sequence was cloned into TA vector (Invitrogen) and sequenced. The DNA fragment, isolated by restriction-enzyme digestion, was cloned into the bacterial expression vector pET11a (Novagen) at the NdeI/BamHI sites to construct CA XV P293X pET11a. E. coli host cells [BL21(DE3)pLysS or Origami (DE3)], from Novagen, were transformed with the vector.
E. coli host cells BL21(DE3)pLysS or Origami (DE3) containing CA XV P293X pET11a were grown as described for human CA II [31]. For Origami (DE3) cells, the antibiotics used were kanamycin, tetracycline and ampicillin. The Origami host strains are K-12 derivatives that have mutations in both the thioredoxin reductase (trxB) and glutathione reductase (gor) genes, which enhance disulphide-bond formation in the cytoplasm. The trx and gor mutations are selectable on kanamycin and tetracycline respectively. For BL21(DE3)pLysS cells, the culture medium contained ampicillin and chloramphenicol. The overnight cultures were diluted 100–200-fold in Luria–Bertani medium containing appropriate antibiotics, and grown at 37 °C with shaking to an attenuance at 600 nm (D600) of 0.5. The expression of mouse CA XV was induced by 0.5 mM IPTG (isopropyl β-D-thiogalactoside) and 0.6 mM zinc sulphate, and the culture was allowed to continue for a further 3 h. The bacterial cells were recovered after centrifugation.
The bacterial cell pellet was lysed in 10 mM Hepes/NaOH, pH 7.5/0.5% (v/v) Triton X-100 containing 1 mM each of PMSF, o-phenanthroline, iodoacetamide and EDTA, as well as 6 μM ZnSO4, using a Brinkman Polytron at 4 °C. The mouse CA XV in the cell lysate was refolded by incubating with 10 mM oxidized glutathione at 4 °C for 72 h, as described previously for CA IV [30]. The clear supernatant recovered after centrifugation at 30000 g for 30 min was mixed with one-tenth the volume of 1 M Tris/SO4 buffer, pH 9.0. The cell supernatant containing CA XV was applied to a p-AMBS Affigel column equilibrated with 10 mM Hepes/NaOH buffer, pH 7.5. The unbound proteins were removed by equilibration buffer, followed by 50 mM NaCl in equilibration buffer and 100 mM Tris/SO4 buffer, pH 7.5. The affinity-resin-bound proteins were eluted with 100 mM sodium acetate, pH 5.5, containing 0.5 M sodium perchlorate. The fractions containing the enzymes were concentrated on Amicon tubes and dialysed against 10 mM Tris/SO4 buffer, pH 7.5. The enzyme was purified further on a Sephacryl S-300 sizing column. The fractions were analysed by SDS/PAGE. The polypeptides were visualized by silver staining or by Western blot analysis.
RESULTS
Bioinformatics analyses
The sequence analyses were applied to 10 species in order to reveal those that could possess an active CA15 gene in their genome. The results of the sequence analyses showed that eight animal species are likely to have an active CA15. The alignment of the predicted CA XV sequence of these species is shown in Figure 1. The amino acid sequence for CA XV, as well as its exon structure, seems to be well maintained throughout evolution, because species from different taxons showed conserved sequences. Surprisingly, humans were shown to have three copies of CA15, and chimpanzees at least two copies of the gene. Chimpanzees may also possess one more copy, since there was a sequencing gap in the region where the third gene might reside. However, all of these human and chimpanzee CA15 genes seem to have become pseudogenes for various reasons: an AluY repeat splits exon 8, and there are several frame-shifts resulting in the loss of many CA hallmark residues conserved in all active isoenzymes. There is also an insertion in the middle of the active site, and one intron has lost the essential GT dinucleotide in the beginning. In addition, each copy shows unique point mutations and frame-shifts. Figure 2 shows those disrupting features that were common to all of these five gene copies of CA15. Each of these defects is alone likely to be able to make this CA isoenzyme non-functional. More detailed information on these errors can be found in Supplementary Figure 1 (http://www.BiochemJ.org/bj/392/bj3920083add.htm), where the sequences for all of these gene copies have been presented. Supplementary Figure 2 shows an alignment where a reconstruction of human CA XV has been included. Supplementary Figure 3 presents this gene in the human genome, and shows that the automatic gene prediction algorithms in genome databases had been unsuccessful in finding the correct exons for these pseudogenes. In databases, no mRNAs or EST sequences have been reported for any of these genes. In all species, except humans and chimpanzees, the three histidine residues critical for CA activity have been conserved. Thus it can be summarized that humans and chimpanzees seem to have lost their CA XV during evolution.
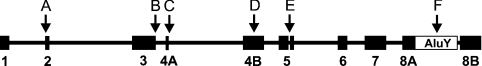
Figure 2. Organization of human and chimpanzee CA XV pseudogenes.
Numbered black boxes show reconstructed exons. Exon and intron lengths are presented in the correct scale. Arrows highlight those defects that are common to all five copies. Arrows A and B: frame-shifts. Arrow C, the beginning of the intron after exon 4A has GA instead of the conserved GT dinucleotide. Arrow D, a 9 bp insertion in exon 4B, disrupting the active centre. Arrow E, a 4-bp insertion in exon 5, leading to a frame-shifted sequence in a region which is highly conserved in all CAs. Arrow F, insertion of an AluY-repeat sequence which splits exon 8, duplicating 17 bp of the exon sequence (duplicated part seen as a gap after AluY).
The draft assembly of the genome of rhesus monkey (Macaca mulatta) was inspected at the UCSC genome browser. The quality of the genomic sequence was low and therefore not all exons were found. At the active site, the second histidine was mutated to asparagine, identical with human and chimpanzee pseudogenes, and in addition, there were several in-frame stop codons and one frame-shift in the sequences. There was no AluY sequence insertion corresponding to human and chimpanzee sequences. However, from the pieces of CA15 sequence that we could retrieve, we concluded that CA15 has probably become a pseudogene also in the rhesus monkey.
Because humans did not show active CA XV, mouse CA XV was characterized in order to reveal the properties of this new enzyme. The mouse CA XV protein sequence contains three potential glycosylation sites (Asn-not Pro-Ser/Thr) at asparagine residues 189, 201 and 210 (numbering shown in Figure 1). The NetNGlyc 1.0 server predicted all three sites to be capable of being glycosylated (potential>threshold 0.5). The potentials were 0.5977, 0.6636 and 0.8056 respectively. This result is also supported by endoglycosidase digestion data in the present paper.
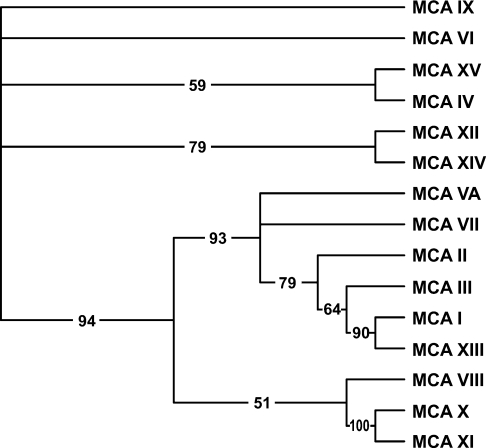
A phylogenetic analysis was performed to estimate evolutionary relationship of CA XV to the other known CA isoforms. The results in Figure 3 show that CA XV is most closely related to the extracellular, GPI-anchored CA IV.
Figure 3. The phylogenetic tree of 15 mouse CAs and CA-RPs.
The numbers at the branches show confidence levels in the bootstrap analysis. The tree implies that mouse CA XV is most closely related to CA IV.
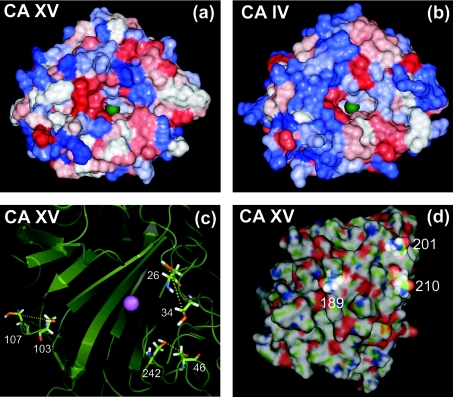
A prediction for the structure of mouse CA XV was carried out using computer modelling. The results are illustrated in Figure 4. Panels (a) and (b) show the surfaces of mouse CA XV and human CA IV coloured according to their hydrophobicity profiles. The green sphere represents the zinc atom in the active site of the enzyme. The pictures illustrate that the active site is conserved in mouse CA XV. In panel (c), all the cysteine residues are shown. In the molecular model the distances between cysteine pairs 26/34, 46/242 and 103/107 (numbering according to Figure 1) are compatible with disulphide-bridge formation, even though the disulphides were not forced in our model. The C-terminal part of the molecule, which contains a cysteine residue at position 326, is cleaved off in the process of GPI anchoring and therefore is not available to form a disulphide bond with the cysteines present in the mature molecule. The locations of the asparagine residues that were predicted to be glycosylated are pointed out in panel (d). All of these residues are predicted to be located on the surface of the molecule, and thus they have the potential to have oligosaccharide chains attached.
Figure 4. The prediction of the structure for CA XV.
Panels (a) and (b): comparison of the surfaces of CA XV and CA IV, coloured according to hydropathy (blue represents hydrophilic, and red hydrophobic). The green sphere represents the zinc atom crucial for CA activity. Panel (c): secondary structures and cysteine pairs. The zinc atom is shown by a purple sphere. Panel (d): highlighted asparagines predicted to represent glycosylation sites; residue numbers are the same as shown as in Figure 1.
CA15 mRNA expression in murine tissues
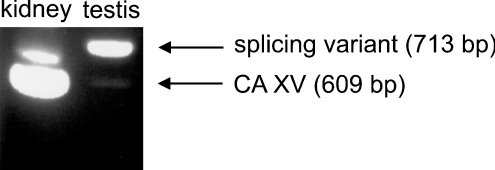
In mouse tissues the most abundant CA15 mRNA expression was found in the kidney. Sequencing confirmed that the positive band detected by PCR corresponded to the correct amplification product. The PCR results in the kidney and testes are shown in Figure 5. The rest of the results are summarized in Table 1. The kidney showed the strongest PCR reaction; weak bands were observed in the brain, testes, 7-day-old embryo and 17-day-old embryo. Testes contained an additional band, which was 104 bp longer than the expected CA15 product. This PCR product was also sequenced, and it appeared to represent a CA15 splicing variant which contains a longer second exon, i.e. 104 nt of the following intron are included in this exon, which causes a frame-shift and thus results in a stop codon near the beginning of the third exon. Therefore this splicing variant would not produce a functional CA.
Figure 5. Results of RT-PCR.
The results of the kidney and testis RT-PCR are shown in the Figure. The kidney has a strong band for CA XV (609 bp), whereas in the testis the band is much weaker. In testis, however, there is a stronger band for a splicing variant (713 bp). The rest of the results are summarized in Table 1.
Table 1. Expression of CA15 mRNA in mouse tissues.
Scores in RT-PCR (denoted by the band intensity) were as follows: +, strong band; +/− weak band; − no band observed.
| Tissue | Band intensity |
|---|---|
| Heart | − |
| Skeletal muscle | − |
| Brain | +/− |
| Kidney | + |
| Liver | − |
| Lung | − |
| Spleen | − |
| Testis | +/− |
| Stomach | − |
| Duodenum | − |
| Jejunum | − |
| Ileum | − |
| Colon | − |
| Blood | − |
| 7-Day embryo | +/− |
| 11-Day embryo | − |
| 15-Day embryo | − |
| 17-Day embryo | +/− |
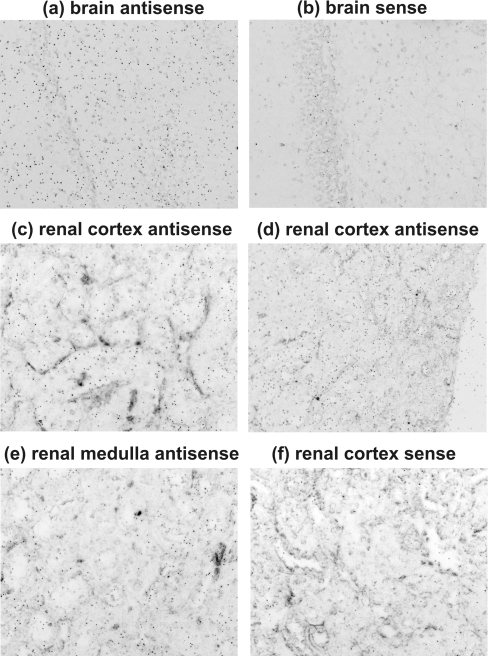
In addition to RT-PCR, in situ hybridization was also performed to reveal the murine tissues that express CA15 mRNA. In accordance with the RT-PCR results, the kidney and brain showed a positive signal (Figure 6). The signal density varied within the renal parenchyma such that high expression was observed in the renal cortex and lower expression in the medulla.
Figure 6. In situ hybridization revealing CA15 mRNA in mouse tissues.
The signal was present in brain (a), whereas the negative control had a much lower signal density (b). High expression was observed in the renal cortex (c, d) and lower expression in the medulla (e). The sense control shows the background level of the signal (f). Original magnifications of the panels: a, b, c, e and f, ×400; d, ×200.
The following human tissues were screened for the expression of three CA15 candidate genes: kidney, heart, lung, brain, pancreas, spleen, thymus, small intestine, colon, skeletal muscle, prostate, testis, ovary, placenta and peripheral blood leukocytes. All the human tissues remained negative in RT-PCR experiments using a primer pair designed for detection of all the candidate genes (results not shown). In addition, gene-specific primers for the putative CA15 genes were used to re-examine the kidney, brain and heart samples, and also these results remained negative (results not shown). Taken together, these findings and the sequence data indicate that all three CA15 candidate genes are pseudogenes in the human genome.
Expression of CA XV in COS-7 cells
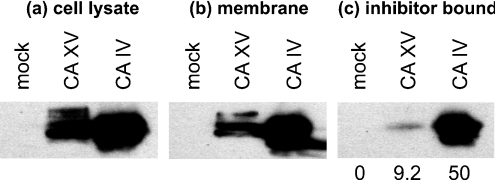
CA XV was expressed in COS-7 cells transfected with mouse CA15 cDNA (Figure 7a). The CA XV polypeptides migrated on SDS/PAGE with an apparent molecular mass of 34–36 kDa. For a positive control, mouse CA IV, a glycosylated GPI-anchored protein, was also expressed in COS-7 cells. The results in Figure 7(b) show that the majority of the CA XV protein was associated with the membrane fraction, suggesting a membrane localization for the enzyme. The affinity of the CA XV enzyme towards p-AMBS was studied using p-AMBS Affigel resin. The results in Figure 7(c) show that 9% of the total membrane-associated mouse CA XV and 50% of the mouse CA IV were retained on the affinity resin. This difference could be due to the presence of an extra pair of thiol groups in CA XV, and/or weak retention of CA XV over p-AMBS affinity resin. Part of this hypothesis has been tested in the latter part of the present study.
Figure 7. Expression of functional mouse CA XV in the membrane of transfected COS-7 cells.
(a) Total cell lysates of non-transfected COS-7 cell (mock) and transfected COS-7 cells (with CA15 or CA4 cDNAs) were analysed by Western blotting using anti-mouse CA IV antibodies. (b) The cell lysates were centrifuged at 100000 g for 30 min. The membranes were analysed by Western blotting. (c) The membrane extracts in 10 mM Hepes/NaOH, pH 7.5, containing 1% NP40 and protease inhibitors were incubated with CA inhibitor-affinity resin, and bound enzyme was analysed by Western blotting. The numbers at the bottom of the gel show percentages of inhibitor-bound CA XV or CA IV.
Mouse CA XV is a glycosylated and GPI-anchored protein
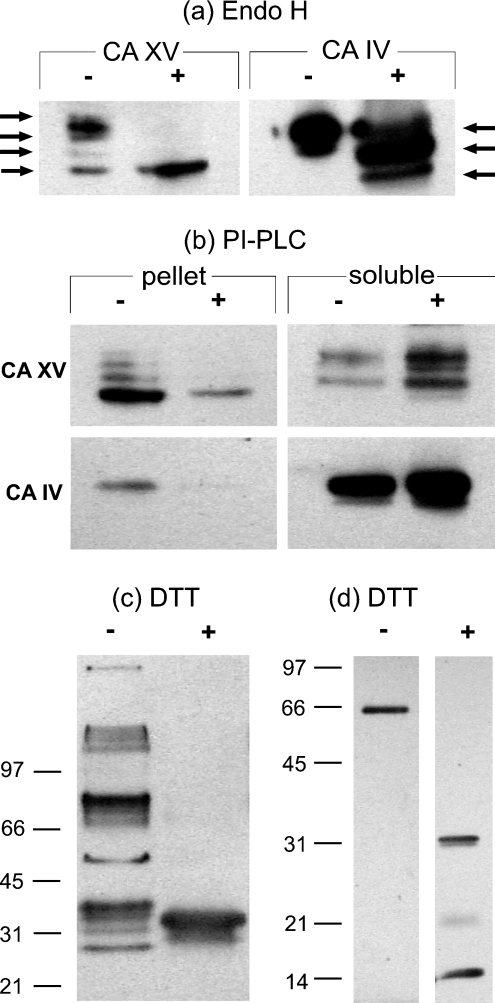
Figure 8(a) shows that CA XV without EndoH treatment contains one major polypeptide of 36 kDa and three minor polypeptides of 34, 32 and 29 kDa respectively. Upon EndoH treatment, the higher-apparent-molecular-mass forms of CA XV were reduced to 29 kDa. These results suggested that expression of CA XV results in glycosylated (36, 34 and 32 kDa) and non-glycosylated (29 kDa) forms, and also that the fully glycosylated mouse CA XV contains at least three N-linked oligosaccharides. This result is in accordance with the bioinformatics predictions. Mouse CA IV contains two N-linked oligosaccharides, as was evident from one intermediate polypeptide between 34 and 29 kDa. The results in Figure 8(b) show that, after treatment with PI-PLC, the membrane pellet contained little residual CA XV compared with non-treated membranes, indicating that the extracellular domain of CA XV can be sheared from the COS-7 cell membrane by PI-PLC. The mouse CA IV was similarly removed from the cell membrane by PI-PLC treatment. From these results, we conclude that CA XV, like CA IV, is a GPI-anchored protein.
Figure 8. Biochemical properties of mouse CA XV.
(a) EndoH treatment resulted in shifts of high-molecular-mass polypeptide of CA XV and CA IV. The CA XV showed two intermediate polypeptides, whereas CA IV showed only one intermediate polypeptide, as indicated by arrows. (b) COS-7 cell membranes were treated without (−) and with (+) PI-PLC enzyme. The enzymes remaining in the membrane pellet and soluble fraction after PI-PLC treatment were analysed by Western blotting. The decrease in the membrane-associated CA XV was compensated by an increase in the soluble fraction after PI-PLC treatment, suggesting that CA XV is a GPI-anchored protein. (c) COS-7 cell lysates expressing mouse CA XV were analysed by Western blotting under non-reducing (−) and reducing (+) conditions. Under non-reducing conditions, the mouse CA XV formed high-molecular-mass aggregates, which upon reducing with DTT resulted in a single immunoreactive polypeptide. (d) Affinity pure recombinant mouse CA XV from E. coli was treated with non-reducing (−) and reducing (+) sample buffer and analysed by SDS/PAGE, followed by Western blotting.
Mouse CA XV forms disulphide-bond-linked, high-molecular-mass quaternary structures
CA XV forms several high-molecular-mass aggregates in COS-7 cells under non-reducing conditions (Figure 8c). However, in the presence of a reducing compound, DTT (dithiothreitol), CA XV migrated as a diffuse 29–34 kDa protein band. These results suggested that CA XV in COS-7 cells could form disulphide-bond-linked aggregates. When affinity-purified mouse CA XV from E. coli was analysed on SDS/PAGE in the absence and presence of DTT, CA XV migrated as 66 kDa disulphide-linked dimer, which turned into a 31 kDa monomer upon reduction with DTT. Proteolytically cleaved fragments of 14–16 and 21 kDa were also seen (Figure 8d).
Functional activity and enhancement by chemical chaperones
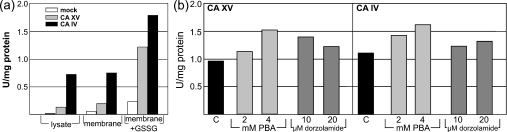
The results in Figure 9(a) show that the cell lysates expressing mouse CA XV contained a detectable amount of CA activity, which was enriched in membrane pellets. Mouse CA IV, a positive control, showed significantly higher activity than CA XV in the membrane suspension and cell lysates. Since CA XV contains an additional pair of thiol residues, a slower or less perfect folding or a stronger tendency to form aggregates may reduce the activity of the enzyme. This can especially occur in a cell culture overexpressing the protein. When the membrane lysates were treated with 10 mM GSSG to facilitate rearrangement of disulphide bonds, mouse CA XV showed significantly higher enzymatic activity, due to deaggregation and/or refolding. Similarly, mouse CA IV activity was also enhanced, but not as much as CA XV. The results in Figure 9(b) show that the enzymatic activity of CA XV was also significantly higher when the transfected COS-7 cells were incubated with 2 or 4 mM PBA and 10 or 20 μM dorzolamide. Mouse CA IV activity was also increased by PBA and dorzolamide treatment. However, the enhancement in CA IV activity by dorzolamide was lower than that seen for CA XV. This difference could be due to stronger binding of dorzolamide with CA IV, which prevented it from being completely removed by washing before the CA assay.
Figure 9. Refolding of CA XV by oxidized glutathione and chemical chaperones.
(a) The cell lysates of COS-7 cells transfected with CA15 or CA4 cDNAs and untransfected cells (mock) were analysed for CA activity. The membrane suspensions from each cell line were also used for CA activity measurement. The results suggested that the cell membranes contained detectable CA XV and IV activities. Upon treatment with 10 mM GSSG, both CA XV and CA IV were refolded further into more active enzymes. (b) The COS-7 cells, after transfection with CA15 or CA4 cDNAs, were treated with 2 and 4 mM PBA or 10 and 20 μM dorzolamide for 72 h. After removing the PBA or dorzolamide, the cell membranes were used for CA activity measurement. Both non-specific (PBA) and specific (dorzolamide) chemical chaperones were able to refold the mouse CA XV and IV into more active enzymes. Abbreviation: U, units.
Recombinant mouse CA XV from E. coli is functionally active
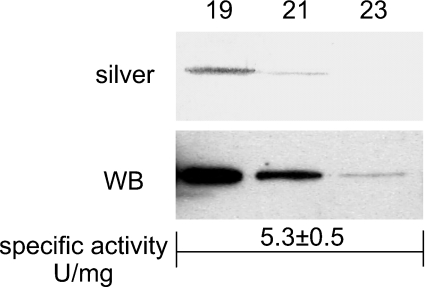
Since CA XV from the COS-7 cell extract showed CA activity that was increased by treatment with oxidized glutathione (Figure 10), we expressed recombinant mouse CA XV in E. coli. The CA XV expressed in E. coli was refolded with oxidized glutathione and purified to homogeneity using a combination of CA-inhibitor affinity resin and sizing column chromatography over Sephacyl S-300. The results in Figure 10 show that homogeneous mouse CA XV from E. coli is functionally active, showing specific activity of 5.3±0.5 units/mg of enzyme. By way of comparison, the high-activity enzymes such as human CA II and CA IV show activities between 2000–3000 units/mg [32]. The activity of CA XV is in the same range as that of CA III (1–5 units/mg).
Figure 10. Recombinant affinity-purified mouse CA XV from E. coli.
The fractions from the Sephacryl S-300 sizing column were analysed by SDS/PAGE. The polypeptides were visualized by silver staining (silver) or Western blot analysis (WB). The numbers show the fractions used for SDS/PAGE. The homogeneous pooled enzyme showed a specific activity of 5.3±0.5 units/mg.
DISCUSSION
New tools of bioinformatics have allowed genome-wide analyses in which novel genes can be efficiently identified and aligned with other homologous counterparts. The present study was initiated when we found a new CA-related gene, named CA15, in the databases (submitted by Hewett-Emmett and Shimmin in 2000; see GenBank® accession no. AF231122). Sequence comparisons indicated that the CA15 gene can be found in genomes of several species. Interestingly, three copies of CA15 genes were identified in the human chromosome band 22q11.21. In the chimpanzee genome, we found two copies, and it is possible that one copy of the gene is missing, due to incomplete genomic data. We concluded that, in both species, all the CA15 genes represent pseudogenes, because of frame-shifts, insertions, point mutations and the lack of mRNAs and EST sequences. In contrast, all the other genomes exhibited only single CA15 genes, which apparently encode catalytic CA XV enzymes with conserved active-site residues. This prediction was verified in mice by studies reported here. The full-length murine cDNA produced enzymatically active CA XV in COS-7 cells. Thus CA XV is the only active CA isoenzyme thus far known that is expressed in several vertebrate species, but has been lost in humans and chimpanzees. This is a novel and unique observation in terms of evolution. In a recent paper, only 27 genes that are active in rodents have been reported to become pseudogenes in humans and chimpanzees; CA15 was not among the reported genes [33].
CAs are ancient enzymes [34]. The efficient regulation of acid–base balance mediated by CAs appears to be extremely important in biological processes, since numerous isoenzymes of this family catalyse the same fundamental reaction in different organs, tissues and cell compartments. Prior to this example, 12 active isoenzymes had been characterized in humans and mice, and all are highly conserved in evolution. CA XV is the first CA isoenzyme that is expressed in many other vertebrates, but not in chimpanzees and humans. Conservation of all key residues and a well-conserved intron–exon structure suggest that CA XV is functionally important in these non-primate vertebrates. Perhaps functional redundancy may explain why it was lost in chimpanzees and humans.
Phylogenetic studies suggested that CA XV and CA IV are closely related isoenzymes, which share distinct sequence similarity. The relatedness of CA XV and CA IV was supported by several experimental results. Until now, CA IV was a unique member of the CA family because of its GPI anchorage to cell membranes. CA XV was shown to be similarly anchored to cell membranes by a GPI anchor. Two disulphide linkages were shown to be critical to stabilize the conformation of the extracellular CA IV protein [19]. The experiments using oxidized glutathione reported here showed that the correct formation of disulphide bridges is also important for maturation of CA XV. Finally, like murine CA IV, CA XV is an N-glycosylated protein.
The expression of CA IV has been studied most intensively in humans and rats. In both species, CA IV is expressed in a variety of tissues: in kidney, CA IV is present mainly on the apical brush border membrane of the proximal tubular cells and on the cells of the thick ascending limbs of Henle, where its physiological role is to facilitate bicarbonate reabsorption [27,35]. In lung, CA IV is localized on the luminal surface of pulmonary endothelial cells, where it catalyses the dehydration of bicarbonate in the serum to yield CO2 [27,36]. CA IV is localized in the capillary endothelium of skeletal and heart muscle, and in the latter it can be also found in special sarcolemmal structures and sarcoplasmic reticulum [37,38]. In distal small and large intestine, CA IV participates in ion and fluid transport [5]. CA IV participates in acidification of epididymal fluid, and is also expressed in the capillary endothelial cells of brain [39]. In addition, CA IV has been reported to be expressed in human pancreas, salivary glands [40], gall-bladder epithelium [41], choriocapillaris of the eye [42] and erythrocytes [43]. CA IV has been shown to form physical complexes with chloride/bicarbonate exchange proteins, and therefore it facilitates the rate of bicarbonate transportation [44]. CA IV is also crucial to the function of NBC1 (the Na+–HCO3− co-transporter) [45]. Recently, an apoptosis-inducing mutation has been identified in the signal sequence of CA4 gene that causes an RP17 form of retinitis pigmentosa [12,46].
The similarity in properties of CA IV and CA XV raises the question of whether CA IV makes CA XV redundant and dispensable in chimpanzees and humans. For instance, the reabsorption of bicarbonate in the kidney is a major process that may require the assistance of CA XV in mice. Mice have a relatively low activity of CA IV. The high activity of human CA IV may have made CA XV redundant in humans, and explains why this gene has become a pseudogene in the course of evolution. In further studies, it would be interesting to study the expression of these isoenzymes in several species in order to gain a better understanding of their roles in normal physiology. It would be attractive to develop single knockout mice for CA IV and CA XV, as well as double knockout mice in which both of these enzyme activities are missing, in order to deepen our knowledge of the physiological importance of these two GPI-anchored enzymes in physiology.
Online data
Acknowledgments
This work was supported by grants from the Finnish Cultural Foundation, Sigrid Juselius Foundation, Academy of Finland and National Institutes of Health (GM34182 and DK40163). The use of computing and software resources provided by CSC, the Finnish IT Center for Science, partly funded by the Finnish Ministry of Education, is gratefully acknowledged. We also thank Dr Markku Heikinheimo for advising with in situ hybridization.
References
- 1.Lindskog S., Silverman D. N. The catalytic mechanism of mammalian carbonic anhydrases. In: Chegwidden W. R., Carter N. D., Edwards Y. H., editors. The Carbonic Anhydrases: New Horizons. Basel: Birkhäuser Verlag; 2000. pp. 175–195. [DOI] [PubMed] [Google Scholar]
- 2.Sly W. S., Hu P. Y. Human carbonic anhydrases and carbonic anhydrase deficiencies. Annu. Rev. Biochem. 1995;64:375–401. doi: 10.1146/annurev.bi.64.070195.002111. [DOI] [PubMed] [Google Scholar]
- 3.Hewett-Emmett D. Evolution and distribution of the carbonic anhydrase gene families. In: Chegwidden W. R., Carter N. D., Edwards Y. H., editors. The Carbonic Anhydrases: New Horizons. Basel: Birhkhäuser Verlag; 2000. pp. 29–76. [DOI] [PubMed] [Google Scholar]
- 4.Lehtonen J., Shen B., Vihinen M., Casini A., Scozzafava A., Supuran C. T., Parkkila A. K., Saarnio J., Kivela A. J., Waheed A., et al. Characterization of CA XIII, a novel member of the carbonic anhydrase isoenzyme family. J. Biol. Chem. 2004;279:2719–2727. doi: 10.1074/jbc.M308984200. [DOI] [PubMed] [Google Scholar]
- 5.Fleming R. E., Parkkila S., Parkkila A. K., Rajaniemi H., Waheed A., Sly W. S. Carbonic anhydrase IV expression in rat and human gastrointestinal tract regional, cellular, and subcellular localization. J. Clin. Invest. 1995;96:2907–2913. doi: 10.1172/JCI118362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pastorekova S., Parkkila S., Parkkila A. K., Opavsky R., Zelnik V., Saarnio J., Pastorek J. Carbonic anhydrase IX, MN/CA IX: analysis of stomach complementary DNA sequence and expression in human and rat alimentary tracts. Gastroenterology. 1997;112:398–408. doi: 10.1053/gast.1997.v112.pm9024293. [DOI] [PubMed] [Google Scholar]
- 7.Kyllonen M. S., Parkkila S., Rajaniemi H., Waheed A., Grubb J. H., Shah G. N., Sly W. S., Kaunisto K. Localization of carbonic anhydrase XII to the basolateral membrane of H+-secreting cells of mouse and rat kidney. J. Histochem. Cytochem. 2003;51:1217–1224. doi: 10.1177/002215540305100912. [DOI] [PubMed] [Google Scholar]
- 8.Parkkila S., Parkkila A. K., Rajaniemi H., Shah G. N., Grubb J. H., Waheed A., Sly W. S. Expression of membrane-associated carbonic anhydrase XIV on neurons and axons in mouse and human brain. Proc. Natl. Acad. Sci. U.S.A. 2001;98:1918–1923. doi: 10.1073/pnas.98.4.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leinonen J., Parkkila S., Kaunisto K., Koivunen P., Rajaniemi H. Secretion of carbonic anhydrase isoenzyme VI (CA VI) from human and rat lingual serous von Ebner's glands. J. Histochem. Cytochem. 2001;49:657–662. doi: 10.1177/002215540104900513. [DOI] [PubMed] [Google Scholar]
- 10.Shah G. N., Hewett-Emmett D., Grubb J. H., Migas M. C., Fleming R. E., Waheed A., Sly W. S. Mitochondrial carbonic anhydrase CA VB: differences in tissue distribution and pattern of evolution from those of CA VA suggest distinct physiological roles. Proc. Natl. Acad. Sci. U.S.A. 2000;97:1677–1682. doi: 10.1073/pnas.97.4.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishimori I. Acatalytic CAs: Carbonic anhydrase-related proteins. In: Supuran C. T., Scozzafava A., Conway J., editors. Carbonic Anhydrase: Its Inhibitors and Activators. Boca Raton: CRC Press; 2004. pp. 25–43. [Google Scholar]
- 12.Bonapace G., Waheed A., Shah G. N., Sly W. S. Chemical chaperones protect from effects of apoptosis-inducing mutation in carbonic anhydrase IV identified in retinitis pigmentosa 17. Proc. Natl. Acad. Sci. U.S.A. 2004;101:12300–12305. doi: 10.1073/pnas.0404764101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kent W. J. BLAT — the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Birney E., Clamp M., Durbin R. GeneWise and Genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Notredame C., Higgins D. G., Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 16. Reference deleted.
- 17.Thompson J. D., Higgins D. G., Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swofford D. L. Version 4. Sunderland, MA: Sinauer Associates; 2002. PAUP*, Phylogenetic Analysis Using Parsimony (* and Other Methods) [Google Scholar]
- 19.Stams T., Nair S. K., Okuyama T., Waheed A., Sly W. S., Christianson D. W. Crystal structure of the secretory form of membrane-associated human carbonic anhydrase IV at 2.8-Å resolution. Proc. Natl. Acad. Sci. U.S.A. 1996;93:13589–13594. doi: 10.1073/pnas.93.24.13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heikinheimo M., Scandrett J. M., Wilson D. B. Localization of transcription factor GATA-4 to regions of the mouse embryo involved in cardiac development. Dev. Biol. 1994;164:361–373. doi: 10.1006/dbio.1994.1206. [DOI] [PubMed] [Google Scholar]
- 21.Whittington D. A., Grubb J. H., Waheed A., Shah G. N., Sly W. S., Christianson D. W. Expression, assay, and structure of the extracellular domain of murine carbonic anhydrase XIV: implications for selective inhibition of membrane-associated isoenzymes. J. Biol. Chem. 2004;279:7223–7228. doi: 10.1074/jbc.M310809200. [DOI] [PubMed] [Google Scholar]
- 22.Lopata M. A., Cleveland D. W., Sollner-Webb B. High level transient expression of a chloramphenicol acetyl transferase gene by DEAE-dextran mediated DNA transfection coupled with a dimethyl sulfoxide or glycerol shock treatment. Nucleic Acids Res. 1984;12:5707–5717. doi: 10.1093/nar/12.14.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luthman H., Magnusson G. High efficiency polyoma DNA transfection of chloroquine treated cells. Nucleic Acids Res. 1983;11:1295–1308. doi: 10.1093/nar/11.5.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 25.Sundaram V., Rumbolo P., Grubb J., Strisciuglio P., Sly W. S. Carbonic anhydrase II deficiency: diagnosis and carrier detection using differential enzyme inhibition and inactivation. Am. J. Hum. Genet. 1986;38:125–136. [PMC free article] [PubMed] [Google Scholar]
- 26.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Zhu X. L., Sly W. S. Carbonic anhydrase IV from human lung. Purification, characterization, and comparison with membrane carbonic anhydrase from human kidney. J. Biol. Chem. 1990;265:8795–8801. [PubMed] [Google Scholar]
- 28.Waheed A., Parkkila S., Zhou X. Y., Tomatsu S., Tsuchihashi Z., Feder J. N., Schatzman R. C., Britton R. S., Bacon B. R., Sly W. S. Hereditary hemochromatosis: effects of C282Y and H63D mutations on association with beta2-microglobulin, intracellular processing, and cell surface expression of the HFE protein in COS-7 cells. Proc. Natl. Acad. Sci. U.S.A. 1997;94:12384–12389. doi: 10.1073/pnas.94.23.12384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waheed A., Zhu X. L., Sly W. S. Membrane-associated carbonic anhydrase from rat lung. Purification, characterization, tissue distribution, and comparison with carbonic anhydrase IVs of other mammals. J. Biol. Chem. 1992;267:3308–3311. [PubMed] [Google Scholar]
- 30.Waheed A., Pham T., Won M., Okuyama T., Sly W. S. Human carbonic anhydrase IV: in vitro activation and purification of disulfide-bonded enzyme following expression in Escherichia coli. Protein Expr. Purif. 1997;9:279–287. doi: 10.1006/prep.1996.0691. [DOI] [PubMed] [Google Scholar]
- 31.Hu P. Y., Waheed A., Sly W. S. Partial rescue of human carbonic anhydrase II frameshift mutation by ribosomal frameshift. Proc. Natl. Acad. Sci. U.S.A. 1995;92:2136–2140. doi: 10.1073/pnas.92.6.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karhumaa P., Parkkila S., Waheed A., Parkkila A. K., Kaunisto K., Tucker P. W., Huang C. J., Sly W. S., Rajaniemi H. Nuclear NonO/p54(nrb) protein is a nonclassical carbonic anhydrase. J. Biol. Chem. 2000;275:16044–16049. doi: 10.1074/jbc.275.21.16044. [DOI] [PubMed] [Google Scholar]
- 33.International Human Genome Sequencing Consortiuim. Finishing the euchromatic sequence of the human genome. Nature (London) 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 34.Tripp B. C., Smith K., Ferry J. G. Carbonic anhydrase: new insights for an ancient enzyme. J. Biol. Chem. 2001;276:48615–48618. doi: 10.1074/jbc.R100045200. [DOI] [PubMed] [Google Scholar]
- 35.Brown D., Zhu X. L., Sly W. S. Localization of membrane-associated carbonic anhydrase type IV in kidney epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 1990;87:7457–7461. doi: 10.1073/pnas.87.19.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fleming R. E., Crouch E. C., Ruzicka C. A., Sly W. S. Pulmonary carbonic anhydrase IV: developmental regulation and cell-specific expression in the capillary endothelium. Am. J. Physiol. 1993;265:L627–L635. doi: 10.1152/ajplung.1993.265.6.L627. [DOI] [PubMed] [Google Scholar]
- 37.Sender S., Gros G., Waheed A., Hageman G. S., Sly W. S. Immunohistochemical localization of carbonic anhydrase IV in capillaries of rat and human skeletal muscle. J. Histochem. Cytochem. 1994;42:1229–1236. doi: 10.1177/42.9.8064130. [DOI] [PubMed] [Google Scholar]
- 38.Sender S., Decker B., Fenske C. D., Sly W. S., Carter N. D., Gros G. Localization of carbonic anhydrase IV in rat and human heart muscle. J. Histochem. Cytochem. 1998;46:855–861. doi: 10.1177/002215549804600709. [DOI] [PubMed] [Google Scholar]
- 39.Ghandour M. S., Langley O. K., Zhu X. L., Waheed A., Sly W. S. Carbonic anhydrase IV on brain capillary endothelial cells: a marker associated with the blood-brain barrier. Proc. Natl. Acad. Sci. U.S.A. 1992;89:6823–6827. doi: 10.1073/pnas.89.15.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fujikawa-Adachi K., Nishimori I., Sakamoto S., Morita M., Onishi S., Yonezawa S., Hollingsworth M. A. Identification of carbonic anhydrase IV and VI mRNA expression in human pancreas and salivary glands. Pancreas. 1999;18:329–335. doi: 10.1097/00006676-199905000-00001. [DOI] [PubMed] [Google Scholar]
- 41.Parkkila S., Parkkila A. K., Juvonen T., Waheed A., Sly W. S., Saarnio J., Kaunisto K., Kellokumpu S., Rajaniemi H. Membrane-bound carbonic anhydrase IV is expressed in the luminal plasma membrane of the human gallbladder epithelium. Hepatology. 1996;24:1104–1108. doi: 10.1002/hep.510240521. [DOI] [PubMed] [Google Scholar]
- 42.Hageman G. S., Zhu X. L., Waheed A., Sly W. S. Localization of carbonic anhydrase IV in a specific capillary bed of the human eye. Proc. Natl. Acad. Sci. U.S.A. 1991;88:2716–2720. doi: 10.1073/pnas.88.7.2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wistrand P. J., Carter N. D., Conroy C. W., Mahieu I. Carbonic anhydrase IV activity is localized on the exterior surface of human erythrocytes. Acta Physiol. Scand. 1999;165:211–218. doi: 10.1046/j.1365-201x.1999.00478.x. [DOI] [PubMed] [Google Scholar]
- 44.Sterling D., Alvarez B. V., Casey J. R. The extracellular component of a transport metabolon. Extracellular loop 4 of the human AE1 Cl−/HCO3− exchanger binds carbonic anhydrase IV. J. Biol. Chem. 2002;277:25239–25246. doi: 10.1074/jbc.M202562200. [DOI] [PubMed] [Google Scholar]
- 45.Alvarez B. V., Loiselle F. B., Supuran C. T., Schwartz G. J., Casey J. R. Direct extracellular interaction between carbonic anhydrase IV and the human NBC1 sodium/bicarbonate co-transporter. Biochemistry. 2003;42:12321–12329. doi: 10.1021/bi0353124. [DOI] [PubMed] [Google Scholar]
- 46.Rebello G., Ramesar R., Vorster A., Roberts L., Ehrenreich L., Oppon E., Gama D., Bardien S., Greenberg J., Bonapace G., et al. Apoptosis-inducing signal sequence mutation in carbonic anhydrase IV identified in patients with the RP17 form of retinitis pigmentosa. Proc. Natl. Acad. Sci. U.S.A. 2004;101:6617–6622. doi: 10.1073/pnas.0401529101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.