Abstract
RTP801 is a newly discovered stress-response gene that is induced by hypoxia and other cell stress signals. Arsenic is a heavy metal that is linked to carcinogenesis in humans. Here, we investigated the mechanism by which arsenic induces RTP801 transcription. In HaCaT human keratinocytes, arsenite was able to induce a rapid rise in the RTP801 mRNA level. Correspondingly, arsenite treatment was capable of stimulating a 2.5 kb human RTP801 promoter. Such a stimulatory effect was inhibited by co-expression of superoxide dismutase or glutathione peroxidase, and was abrogated by N-acetylcysteine, implying that ROS (reactive oxygen species) were involved in transcriptional regulation of the RTP801 gene. A series of deletion studies with the promoter revealed a critical arsenic-responsive region between −1057 and −981 bp of the promoter. Point mutations of the putative Elk-1 site and the C/EBP (CCAAT/enhancer-binding protein) site within this region were able to reduce the stimulatory effect of arsenite, indicating that Elk-1 and C/EBP are involved in transcriptional regulation of the RTP801 gene by arsenite. Furthermore, a gel mobility-shift assay demonstrated that arsenite was able to mount the rapid formation of a protein complex that bound the arsenic-responsive region as well as the C/EBP-containing sequence. The arsenite stimulation on RTP801 transcription was partly mediated by the ERK (extracellular-signal-regulated kinase) pathway, since the effect of RTP801 was inhibited by a selective ERK inhibitor. In addition, overexpression of Elk-1 and C/EBPβ was able to elevate the promoter activity. Therefore these studies indicate that RTP801 is a transcriptional target of arsenic in human keratinocytes, and that arsenic and ROS production are linked to Elk-1 and C/EBP in the transcriptional control.
Keywords: arsenite, CCAAT/enhancer-binding protein (C/EBP), reactive oxygen species (ROS), RTP801 gene
Abbreviations: C/EBP, CCAAT/enhancer-binding protein; EMSA, electrophoretic mobility-shift assay; ERK, extracellular-signal-regulated kinase; G3PDH, glyceraldehyde-3-phosphate dehydrogenase; GPX1, glutathione peroxidase 1; HIF-1, hypoxia-inducible factor-1; HNF-4, hepatic nuclear factor-4; HRP, horseradish peroxidase; JNK, c-Jun N-terminal kinase; MAPK, mitogen-activated protein kinase; NAC, N-acetylcysteine; NF-κB, nuclear factor κB; ROS, reactive oxygen species; RT-PCR, reverse transcription-PCR; SOD1, superoxide dismutase 1
INTRODUCTION
Metals are trace elements that are essential for life. However, certain metals, including arsenic, and their derivatives confer toxic effects on the body, which are manifested in carcinogenesis, neurotoxicity and immunotoxicity [1]. Two mechanisms are implicated in the toxicity of metals. One of them is through interruption of the normal physiology of cells by displacing the physiological metals from their natural binding sites. The second mechanism involves the ability of metals to serve as a catalytic centre for redox reactions that generate ROS (reactive oxygen species), including superoxide anion (O2•−), hydroxyl radical (•OH) and hydrogen peroxide (H2O2). ROS are able to damage a variety of cellular macromolecules, as well as altering particular signalling pathways in the cell, leading to cancer and other toxic effects [2]. High concentrations of ROS impose oxidative damage on cellular components such as DNA, proteins and lipids. ROS-mediated DNA damage is probably the most biologically significant event that is implicated in cancer formation. For example, arsenic has been shown to function as a strong dose-dependent mutagen and to induce mostly large deletion mutations via ROS [3].
Arsenic has long been considered as a toxic metal [1]. Four major types of arsenic have been found in mammals, including arsenate [As(V)], arsenite [As(III)], monomethylarsonic acid (MMA) and dimethylarsenic acid (DMA). As(III) is the reduced form of As(V). The form of arsenic that is more carcinogenic to humans remains controversial, an issue which is largely complicated by the fact that different forms of arsenic can be interconverted into each other in vivo. Studies over the past decade have revealed that the reduction of arsenic is associated with in vivo and in vitro ROS production, which plays an important role in mediating arsenic-induced carcinogenesis [3]. The carcinogenic effect of arsenic is linked to activation of cell-proliferative signalling pathways. For example, arsenic is able to activate the MAPK (mitogen-activated protein kinase) cascade at concentrations ranging from 0.1 to 500 μM [4]. Arsenic differentially activates ERKs (extracellular-signal-regulated kinases), p38 and JNKs (c-Jun N-terminal kinases) to mediate various effects on carcinogenesis, depending on the time, dose, chemical form and cell type [4]. Activation of ERKs by arsenic can induce cell proliferation and transformation, whereas p38 and JNK activation are associated with cell growth arrest and apoptosis. In addition to the mitogenic pathway, arsenic is also able to activate NF-κB (nuclear factor κB), which plays a critical role in the inflammatory response as well as cancer formation [5]. It has been shown that low concentrations of As(III) (1–10 μM) are able to activate NF-κB, and that arsenic-mediated generation of ROS is involved in NF-κB activation [6]. It has also been suggested that the SEK1 (stress-activated protein kinase 1/ERK1)–JNK pathway may partly contribute to the activation of NF-κB by arsenic [6].
Studies over the past 3 years have indicated that RTP801 (also referred to as REDD1 or dig2) is a novel stress-response gene [7–9]. RTP801 was originally isolated as an HIF-1 (hypoxiainducible factor-1)-responsive gene [7], and is likely to play a role in hypoxia-mediated cell survival downstream of phosphoinositide 3-kinase [10]. RTP801 is strongly induced by hypoxia both in vitro, such as in mammalian cells, and in vivo, such as in an animal model of ischaemic stroke. In addition to hypoxia, RTP801 has been found to be induced by several signals that mediate cell stress, apoptosis and DNA damage. For example, RTP801 is induced by dexamethasone, thapsigargin, tunicamycin and heat shock in murine T-cell lymphoma cells [9]. It appears that RTP801 is a transcriptional target of p63 and p53, and it is implicated in ROS regulation by these two tumour suppressor proteins [8]. In addition, recent studies have suggested that RTP801 is implicated in the regulation of mTOR (mammalian target of rapamycin) that is also dependent on TSC1/TSC2 (tuberous sclerosis complex 1/2) [11–13]. Even though the biological function of RTP801 remains to be defined, increasing evidence from recent studies has suggested that RTP801 is a novel cell stress-responsive protein and plays a role in the regulation of cellular ROS and apoptosis.
Because arsenic is a cell stress signal that mediates ROS generation and carcinogenesis, we investigated the ability of arsenic to induce RTP801 transcription. We found that arsenic is able to employ previously identified cellular pathways to activate the transcription of RTP801.
MATERIALS AND METHODS
Materials, cell culture and transfection
HaCaT cells, a spontaneously immortalized human epidermal keratinocyte cell line [14], were grown in DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% heat-inactivated (56 °C, 30 min) fetal bovine serum, 1% non-essential amino acids (Gibco), 100 μg/ml streptomycin and 100 units/ml penicillin. Transient cell transfection was performed using SuperFect reagent (Qiagen). The three inhibitors of the MAPK cascade, including PD98059, SB203580 and JNK Inhibitor II, were purchased from Calbiochem. The trivalent arsenic(III) oxide and 30% H2O2 used in the experiments were from Sigma. The anti-Flag antibody was purchased from Sigma. The antibodies against Elk-1, C/EBPβ (CCAAT/enhancer-binding protein β), ERK and phosphorylated ERK were from Santa Cruz Biotechnology.
RNA isolation and RT-PCR (reverse transcription-PCR)
Total RNA was prepared from HaCaT cells with an RNA isolation kit (Ambion). The RNA was reverse-transcribed with oligo(dT) primer using the SuperScript First-Strand Synthesis System (Invitrogen) to generate first-strand cDNA. The products were diluted and used in PCR to detect the expression of RTP801 and G3PDH (glyceraldehyde-3-phosphate dehydrogenase). The sequences of the PCR primers are as follows: 5′-TGGGCAAAGAACTACTGCGCCTGG-3′ and 5′-TCAACACTCCTCAATGAGCAGCTG-3′ for human RTP801, 5′-GTCTTCACCACCATGGAGAAGG-3′ and 5′-TCGCTGTTGAAGTCAGAGGAGA-3′ for G3PDH. The PCR products were separated on a 2% agarose gel containing 0.1 μg/ml ethidium bromide. The band intensity was quantified using densitometric analysis by the NIH Image program.
Plasmid construction
The 2.5 kb human RTP801 promoter was cloned by PCR with the human genomic DNA as template, using two primers [5′-CAGCCCTTCCCATTTGAGACAAGTTGTT-3′ (for the 5′ end) and 5′-ATGAACTCAGAGTGCCGGAGCGTAGA-3′ (for the 3′ end)]. The resultant PCR fragment was first cloned into pCR4Blunt-TOPO (Invitrogen). The PCR insert was confirmed by DNA sequencing and then subcloned into the KpnI and SacI sites of pGL3-basic luciferase vector (Promega). All deletion constructs of the promoter were generated using PCR. The 5′ primers for these constructs were: 5′-TGCAGTTCCAGCTTCTCGGGA-3′ (for ΔA); 5′-TGTCAAATGAGAGTAGGGTGCA-3′ (for ΔB); 5′-GTCTGGATCTTTTACACAAAGCG-3′ (for ΔC); 5′-CCCATATCCCG AGCCTTTGCAA-3′ (for ΔD); 5′-CCTTGATCCTCCCCTGCCGCGA (for ΔE); 5′-CCTCTTCCATCAGGGTCCTCCT-3′ (for ΔF); 5′-CCTCTGAGCACTGCTGCCAGGC-3′ (for ΔG); 5′-CCGCCTGAATGATGAAACACGG-3′ (for ΔH); and 5′-CCTGGCTTTTCCAGAGATCTC G-3′(for ΔI). The 3′ primer used for these deletion constructs was the same as the one for the 2.5 kb promoter. The PCR products were cloned into pGL3-basic vector and confirmed by DNA sequencing. Point mutations of the RTP801 promoter were introduced into the putative transcription factor-binding sites using PCR. In short, two PCR fragments were first generated with a point mutation introduced by the PCR primers. These two fragments were annealed and used as a template for PCR with two outside primers. The Elk-1 sequence ATTCCTGTG was replaced by ATTggTGTG, the C/EBP sequence GATGAAACAC by GATGccACAC and the HNF-4 (hepatic nuclear factor-4) sequence ATGGCCATTGCA by ATGGatATTGCA (the mutated nucleotides are shown as lowercase letters). These mutant PCR products were cloned into pGL3-basic vector and confirmed by DNA sequencing. The FLAG-tagged RTP801 protein was generated by PCR. The human RTP801 cDNA coding region was obtained by PCR with primers 5′-TATCGATCCTAGCCTTTGGGACCGCTTC-3′ and 5′-CTTCTAGAAGTTCAACACTCCTCAATGAGCAG-3′ using human liver cDNA as template. The PCR product was cloned into a pRC/CMV (Invitrogen) that contains a FLAG tag (generated by ligation with a double-stranded oligonucleotide that contains a FLAG sequence) at the ClaI and XbaI restriction sites. A fusion protein of RTP801 with green fluorescence protein (eGFP-C1, from ClonTech) was generated by ligation of a PCR fragment containing the whole coding region of RTP801 with eGFP-C1 digested with EcoRI and XbaI. The subcloning was later confirmed by DNA sequencing. The full-length human Elk-1 and C/EBPβ cDNAs were cloned by RT-PCR with the RNA samples isolated from HaCaT cells. Primers of 5′-AGCGATGGACCCATCTGTGACG-3′ and 5′-TGGAGAGAGCATGGATGGAGTGACC-3′ were used for cloning Elk-1, and primers of 5′-CGTTCATGCAACGCCTGGTGGCCTG-3′ and 5′-CTAGCAGTGGCCGGAGGAGGCGA-3′ were used for cloning of C/EBPβ. Both cDNA clones were cloned into pcDNA3 for mammalian cell expression and confirmed by DNA sequencing. To generate the minimal promoter construct, which contains −1057 to −981 bp of the As(III)-responsive region of the human RTP801 promoter, the corresponding region of the promoter was cloned by PCR with primers of 5′-GCCGGTACCTCTTCCATCAGGGTCCTCCT-3′ and 5′-CTCGAGCTCGAGATCTCTGGAAAAGCCAGG-3′. The PCR fragment was cloned into the KpnI and SacI sites of the pGL3-basic vector that contains an E1B TATA box previously described by us [15].
Promoter assay
HaCaT cells were seeded in 24-well plates at a density of 2×105 cells/ml and transfected by SuperFect (Qiagen). A Renilla luciferase vector phRL-SV40 (Promega) was co-transfected to serve as an internal control for transfection efficiency. The cells were harvested at 24–48 h after transfection by lysis with 100 μl of PBS with 0.1% (v/v) Triton X-100 and 1 mM PMSF. Aliquots (10 μl) of the lysate was used in a dual-luciferase assay (Promega). The samples were counted for 10 s in a FB12 luminometer (Zylux). Data are represented in the present paper as relative light units/s. The Student's t test was used to compare the values of luciferase data from different groups.
EMSA (electrophoretic mobility-shift assay)
EMSA was performed using nuclear extracts from HaCaT cells. The nuclear extracts were prepared as described previously [16]. Three probes were used in EMSA: (1) a PCR-generated DNA fragment spanning −1038 to −982 bp of the RTP801 promoter; (2) a C/EBP-containing oligonucleotide probe of sequence AATGATGAAACACGGGAT; and (3) an Elk-1-containing oligonucleotide probe of sequence CCAGATTCCTGTGGCCC. All probes were labelled with [γ-32P]dATP by T4 polynucleotide kinase. The labelled probes (approx. 5×104 c.p.m.) were incubated with 2 μg of nuclear extract in a buffer containing a final concentration of 4% (v/v) glycerol, 10 mM Tris/HCl, pH 7.5, 1 mM MgCl2, 0.5 mM EDTA, 0.5 mM dithiothreitol, 50 mM NaCl and 0.1 μg/μl poly(dI-dC). For supershift EMSA, antibodies against Elk-1 and C/EBPβ (0.2 μg/reaction, from Santa Cruz Biotechnology) were added to the binding reaction. The reaction was incubated at 4 °C for 30 min after addition of the probe, then separated by electrophoresis with a 5% non-denaturing polyacrylamide gel [in 0.5×TBE (Tris/borate/EDTA) buffer] and detected by autoradiography with the dried gel.
Primer extension assay
The primer extension assay was performed using the total RNA isolated from HaCaT cells with a method previously described by us [17]. The oligonucleotide primer 5′-ATGAACTCAGAGTGCCGGAGCGTAGA-3′ was used in this assay.
Western blotting analysis
HaCaT cells cultured on six-well plates were lysed with 2×SDS/PAGE loading buffer. The DNA was sheared using a 22-gauge needle and equal volumes were loaded on to SDS/10%-PAGE gels. The protein was transferred on to PVDF and the membrane was blocked with 5% non-fat dry milk in a buffer containing 10 mM Tris/HCl, pH 7.6, 0.9% NaCl and 0.1% (v/v) Tween 20. The membrane was then probed with an antibody specific for Tyr-205-phosphorylated ERK (sc-7383, 1:200 dilution; Santa Cruz Biotechnology), followed by incubation with a goat–anti-mouse HRP (horseradish peroxidase) antibody (1:2500 dilution; Bio-Rad) and ECL (enhanced chemiluminescence; PerkinElmer). The blots were stripped using Western blot stripping buffer from Pierce, and re-probed for total ERK (sc-94, 1:200 dilution; Santa Cruz Biotechnology) followed by incubation with goat–anti-rabbit HRP (1:5000; Bio-Rad) and ECL.
Immunofluorescence staining of the cells
HaCaT cells were cultured at 2×105 cells/ml on glass coverslips in six-well plates before transient transfection. Cell were cultured for the indicated times before fixation, which was performed with 3.7% formaldehyde and 0.1% Triton X-100. The cells were incubated with anti-FLAG antibody (1 μg/μl; Sigma), washed, and then incubated with Alexa-546-conjugated anti-mouse antibody (Molecular Probes). Nuclear staining was performed with Hoechst 33342 (Sigma). The coverslips were mounted and the stained cells were analysed under a fluorescence microscope (DM5000B; Leica).
RESULTS
As(III) augments human RTP801 mRNA levels in HaCaT cells
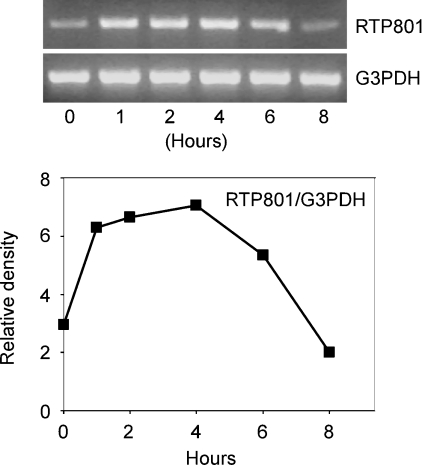
Recent studies have indicated that RTP801 is a novel stress-response gene that is induced by hypoxia and other cell stress signals [7–9]. As arsenic is able to elicit a stress response in the cells [1,4], we first determined whether or not the trivalent arsenic, arsenite [As(III)], is able to induce RTP801 expression at the mRNA level. This was performed in HaCaT cells, which are from a spontaneously immortalized human epidermal keratinocyte cell line [14]. In addition, arsenic has been shown to be able to initiate ROS production and induce apoptosis in these cells, a phenomenon that is related to the carcinogenic effect of arsenic on skin [18]. In our experiment, HaCaT cells were treated with As(III) for various periods of time and the total RNA was isolated from the cells and used in RT-PCR. Primers specific for the human RTP801 cDNA were used in the PCR. The same samples were also subjected to RT-PCR with primers for the housekeeping gene, G3PDH, to monitor the relative RNA levels used in the experiment. As shown in Figure 1, As(III) was not able to cause discernible changes in the G3PDH level. However, As(III) was capable of inducing a rapid up-regulation of the RTP801 mRNA level. This stimulatory effect reached a maximal level at 4 h after As(III) treatment. These data provided the first evidence that As(III) is able to regulate RTP801 at the mRNA level in HaCaT keratinocytes.
Figure 1. Effect of As(III) on mRNA level of RTP801.
HaCaT cells were treated with 10 μM As(III) for various periods of time. The total RNAs were isolated and used for RT-PCR with specific primers for RTP801 and G3PDH. The PCR products were run on a 2% agarose gel and stained with ethidium bromide (top panel). The relative density of the PCR product of RTP801 was normalized to G3PDH level and is shown in the lower panel.
Determination of the transcription initiation site and isolation of a human RTP801 promoter
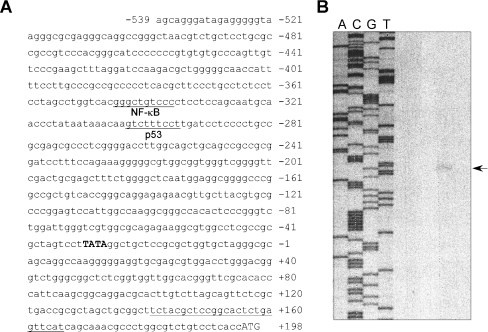
To identify the putative transcription initiation site, we performed a primer extension assay with total RNA isolated from HaCaT cells. As shown in Figure 2, a single extension band was detected at 166 bp upstream of the oligonucleotide primer, indicating that transcription starts 195 bp upstream of the translation initiation site, i.e. the first codon encoding ATG. With this information, we isolated a 2.5 kb human genomic RTP801 fragment immediately in front of the coding region. This promoter region corresponds to −2548 to +166 bp relative to the transcription initiation site identified by the primer extension assay. Sequence analysis indicated that there is a putative TATA box at position −21 to −18 bp (Figure 2A).
Figure 2. The human RTP801 promoter sequence and identification of the transcription initiation site.
(A) A fragment of the human RTP801 promoter sequence spanning the region −539 to +198 bp is shown. The translation initiation site (ATG) is shown in uppercase letters. The putative TATA box is shown in bold uppercase letters. The putative binding sites for transcription factors were determined through the internet TESS program (provided by the Computational Biology and Informatics Laboratory at the University of Pennsylvania Schools of Medicine Engineering and Applied Science). The putative NF-κB- and p53-binding sites are indicated. The position of the primer used for the primer extension assay is marked by an underline at the end of the sequence. (B) Results of the primer extension assay. Total RNA isolated from HaCaT cells was used in this assay. The position of the transcription initiation sites was determined by a DNA sequencing reaction (shown on left) and marked by an arrow on the right.
Regulation of the RTP801 promoter by As(III)
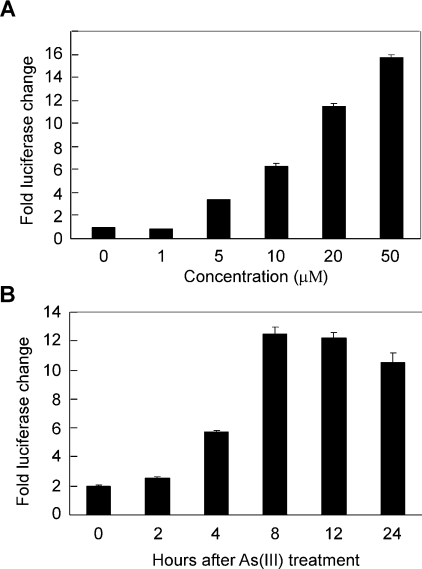
To determine whether or not the isolated RTP801 promoter is regulated by As(III), we analysed the transcriptional activity of the promoter. The 2.5 kb RTP801 promoter (−2548 to +166 bp) was fused with a firefly luciferase reporter gene. This construct was transiently transfected into HaCaT cells and treated with various concentrations of As(III) for various periods of time. As shown in Figure 3(A), As(III) was able to stimulate RTP801 promoter activity in a dose-dependent manner. At a concentration of 10 μM, As(III) was able to activate the promoter to approx. 7-fold relative to the untreated samples. At 50 μM, As(III) was capable of inducing the transcription to approx. 16-fold, even though most cells were dead at this high concentration (results not shown). In addition, we analysed the effect of various treatment times with As(III). We found that As(III) treatment for 8 h was able to activate the promoter to the maximal level (Figure 3B), equivalent to those treated for 12 and 24 h. Taken together, these data indicated that As(III) was indeed able to strongly activate the RTP801 promoter, and that such stimulation took place rapidly upon treatment. These data, together with our findings that As(III) rapidly induced the RTP801 mRNA level by RT-PCR assay (Figure 1), provided convincing evidence that RTP801 is a transcriptional target regulated by arsenic in HaCaT keratinocytes.
Figure 3. Stimulation of the RTP801 promoter by arsenite.
(A) Effect of different As(III) concentrations. HaCaT cells were transiently transfected with RTP801 promoter construct (−2548/+166 bp). An SV40 (simian virus 40)-driven Renilla luciferase was used to delineate the transfection efficiency. At 24 h after transfection, cells were treated with As(III) for 8 h in various concentrations, as indicated. The whole cell lysate was used in a dual-luciferase assay. The fold change of luciferase activity normalized by Renilla luciferase activity is shown as the means±S.D. (B) Effect of various treatment times. The same promoter construct was used as above, but the cells were treated with 10 μM As(III) for different periods of time. The fold changes in luciferase activity are shown as the means±S.D. At least three independent experiments were performed for the assays described above, and only one representative result is shown here.
Activation of the RTP801 promoter by As(III) is mediated by ROS production
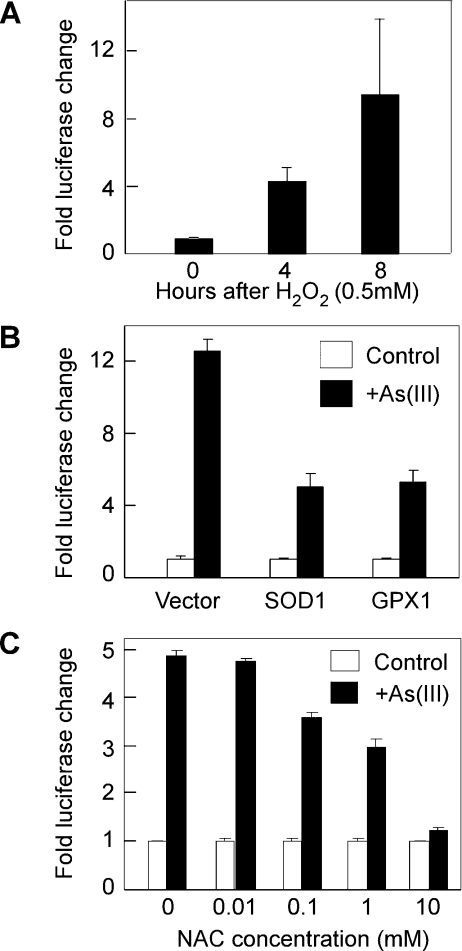
The biological toxicity of arsenic is mainly caused by production of ROS catalysed by the redox reactions of the metal [1,4]. Since As(III) is able to activate RTP801 transcription, we next examined if the effect of arsenic is mediated by ROS. We first analysed the effect of one of the ROS, hydrogen peroxide (H2O2), on RTP801 transcription. The 2.5 kb RTP801 promoter construct was transiently transfected into HaCaT cells, and the cells were treated with H2O2 for various periods of time. As shown in Figure 4(A), H2O2 was able to rapidly activate the promoter activity, suggesting that H2O2 itself is sufficient to activate RTP801 transcription. We next analysed the activity of two ROS-reducing enzymes, SOD1 (superoxide dismutase 1) and GPX1 (glutathione peroxidase 1), in modulating the As(III)-mediated stimulation of RTP801 transcription. SOD1 catalyses the reaction to reduce the cellular amount of superoxide anion (O2•−) [19]. GPX1 reduces H2O2 by oxidizing glutathione to glutathione disulphide [2]. Therefore both SOD1 and GPX1 are able to protect the cells against oxidative damage by reducing the intracellular amount of ROS. When either SOD1 or GPX1 was co-transfected with the 2.5 kb RTP801 promoter into HaCaT cells, the As(III)-induced RTP801 promoter activity was inhibited by approx. 50% (Figure 4B). These data indicate that ROS production is at least partly implicated in the stimulation of RTP801 transcription by arsenic. To further confirm that ROS generation is required for the effect of arsenic on the RTP801 promoter, we employed an ROS scavenger, NAC (N-acetylcysteine), in the experiment. NAC is able to block ROS production and suppress the cytotoxic effects of ROS. As shown in Figure 4(C), treatment of HaCaT cells with NAC was able to abrogate the stimulation of RTP801 transcription by As(III) in a dose-dependent manner. Taken together, these experiments provided profound evidence that the arsenic-mediated activation of RTP801 transcription is mediated by ROS production.
Figure 4. Stimulation of the RTP801 promoter by As(III) is mediated by ROS.
(A) Activation of the RTP801 promoter by H2O2. HaCaT cells were transiently transfected with RTP801 promoter construct (−2548/+166 bp) and Renilla luciferase vector. The cells were treated with 0.5 mM H2O2 for different periods of time, followed by a dual luciferase assay. The fold changes in luciferase activity are shown as the means±S.D. (B) Effect of SOD1 and GPX1 expression on As(III) stimulation of the RTP801 promoter activity. HaCaT cells were transiently transfected with RTP801 promoter construct together with SOD1 or GPX1 expression plasmids (0.2 μg each), as indicated. At 24 h after transfection, the cells were treated with 10 μM As(III) for 8 h before the luciferase assay. The fold changes in luciferase activity are shown as the means±S.D. (C) Effect of NAC on RTP801 promoter activation by As(III). HaCaT cells were transiently transfected with RTP801 promoter construct as described above. At 24 h after transfection, the cells were treated with 10 μM As(III) for 8 h in the presence or absence of NAC at various concentrations. The fold changes of luciferase activity are shown as the means±S.D. At least three independent experiments were performed for the assays described above, and only one representative result is shown here.
Identification of an arsenic-responsive region at the RTP801 promoter
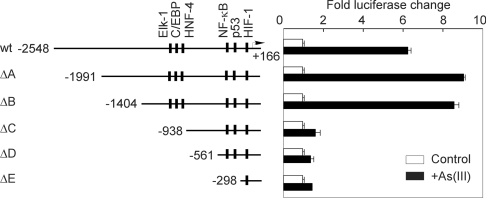
The human RTP801 promoter contains multiple putative transcription factor-binding sites that are likely to be involved in the transcriptional regulation of the gene. For example, the promoter contains a putative NF-κB site around −350 bp and a p53 site around −300 bp (Figure 2A). It has been shown that arsenic is able to activate NF-κB to mediate its cellular effect [6]. In addition, a recent study also indicated that RTP801 is a transcriptional target of p63 and p53 [8]. In order to delineate the region(s) of the RTP801 promoter that is/are implicated in arsenic-mediated activation, we analysed the effect of As(III) treatment on a series of RTP801 promoter deletion mutants which had been fused to the luciferase reporter. As shown in Figure 5, the two deletion constructs, −1991/+166 bp (ΔA) and −1404/+166 bp (ΔB), displayed arsenic stimulation to a level similar to or even slightly higher than that of the 2.5 kb promoter (−2548/+166 bp). However, three other deletion mutants, −938/+166 bp (ΔC), −561/+166 bp (ΔD) and −298/+166 bp (ΔE), demonstrated greatly reduced As(III)-induced stimulation. These data suggest that the promoter sequences between −1404 and −938 bp probably contain a major arsenic-responsive region (Figure 5). In addition, the putative NF-κB (around −350 bp) and p53 (around −300 bp) sites do not appear to be directly involved in the arsenic-mediated stimulation of RTP801 transcription in these cells (Figure 5).
Figure 5. Characterization of the human RTP801 promoter.
The left panel showed a graphic representation of the length of different promoter constructs and the relative location of putative binding sites for p53, NF-κB and HIF-1. HaCaT cells were transiently transfected with various deletion constructs of the RTP801 promoter as indicated, together with Renilla luciferase vector. At 24 h after transfection, the cells were treated with 10 μM As(III) for 8 h and the fold changes in luciferase activity are shown as the means±S.D. At least three independent experiments were performed for this assay, and only one representative result is shown here.
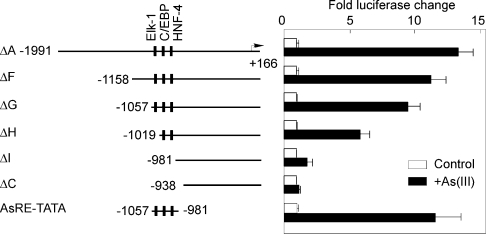
To characterize further the arsenic-responsive region of the RTP801 promoter, we analysed a few more deletion mutants of the promoter, including −1158/+166 bp (ΔF), −1057/+166 bp (ΔG), −1019/+166 bp (ΔH) and −981/+166 bp (ΔI). These constructs were transfected into HaCaT cells, followed by treatment with As(III). We found that both the −1158/+166 bp (ΔF) and −1057/+166 (ΔG) constructs showed only a slight reduction in As(III)-mediated transcriptional stimulation (Figure 6). However, the construct −1019/+166 bp (ΔH) markedly lost the ability to mediate arsenic activation of the promoter (Figure 6). Furthermore, the −981/+166 bp construct (ΔI) only possessed a residual ability to mediate As(III) stimulation (Figure 6). These data therefore delineated further the arsenic-responsive region to −1057 to −981 bp of the RTP801 promoter. In addition, we analysed the responsiveness of the identified arsenic-responsive region to arsenite by using a minimal promoter construct that contains a −1057 to −981 bp fragment of the promoter fused with a TATA box that was previously described by us [15]. As shown in Figure 6, this region (AsRE, standing for arsenic-responsive element) is able to mediate arsenite stimulation at a level even higher than the ΔF construct. These data indicated that this region is not only required but also sufficient to mediate the stimulatory effect of arsenite on the human RTP801 promoter.
Figure 6. Identification of a critical arsenic-responsive region.
HaCaT cells were transiently transfected with various deletion constructs of the RTP801 promoter, as indicated. The cells were treated with 10 μM As(III) for 8 h. The activity of a minimal RTP801 promoter was also analysed. The promoter region −1057 to −981 bp was fused to a TATA box (AsRE-TATA) and used in the luciferase assay. Data are shown as the fold changes in luciferase activities (means±S.D). At least three independent experiments were performed for the assay, and only one representative result is shown here.
Identification of an Elk-1 site and a C/EBP site that mediate arsenic activation of the RTP801 promoter
As our experiments revealed a critical arsenic-responsive region between −1057 and −981 bp, we carefully analysed the putative transcription factor-binding sites in this area. There are three major putative transcription-factor binding sites that are well conserved in both human and mouse genomic sequences: an Elk-1 site (ATTCCTGTG at −1031/−1023 bp), a C/EBP site (GATGAAACAC at −1009/−999 bp) and an HNF-4 site (ATGGCCATTGCA at −996/−985 bp). Elk-1 is a regulator of the c-Fos proto-oncogene and belongs to the ETS-domain family of transcriptional factors [20] and functions as a substrate of MAPK [21]. C/EBP was originally identified as a heat-stable transcription factor in rat liver nuclei that was able to interact with the CCAAT motif present in several cellular gene promoters and certain viral enhancers [22] and plays a pivotal role in many cellular responses [23]. HNF-4 is a potent transcriptional activator that controls the expression of a wide variety of genes important for metabolism [24].
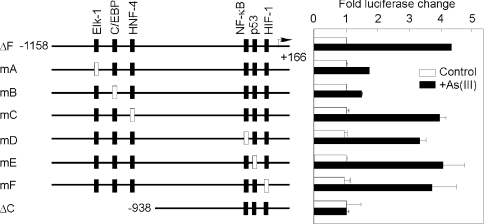
To analyse the potential contribution of these sites to the arsenic-mediated transcription of the RTP801 gene, we introduced point mutations into these sites. The Elk-1 sequence ATTCCTGTG was replaced by ATTggTGTG, the C/EBP sequence GATGAAACAC by GATGccACAC and the HNF-4 sequence ATGGCCATTGCA by ATGGatATTGCA (the mutated nucleotides are shown in lowercase letters). These mutations were incorporated into the −1158/+166 bp region (ΔF) of the RTP801 promoter and linked to a luciferase reporter. Transient transfection of these mutants was carried out in HaCaT cells, followed by As(III) treatment. As shown in Figure 7, mutation of the Elk-1 site was able to partially abrogate the arsenic-mediated stimulation of RTP801 promoter. Meanwhile, mutation of the C/EBP site profoundly reduced the As(III)-mediated transcriptional activation. However, mutation of the putative HNF-4 site revealed no obvious effect on As(III)-initiated stimulation of the promoter. Taken together, these data provided significant evidence that the Elk-1 and C/EBP sites at the RTP801 promoter are implicated in the transcriptional activation by arsenic.
Figure 7. Effect of different point mutations of RTP801 promoter.
The left panel shows a graphic representation of the relative location of putative transcription factor-binding sites for Elk-1, C/EBP, HNF-4, NF-κB, p53 and HIF-1. The filled-in bars represent the wild-type sites and the open bars are for the mutated sites. HaCaT cells were transiently transfected with various mutation constructs of the RTP801 promoter as indicated, together with Renilla luciferase vector. The ΔC and ΔF constructs were the same as shown in Figure 6. The cells were treated with 10 μM As(III) for 8 h at 24 h after transfection. The fold changes in luciferase activity normalized by Renilla luciferase activity are shown as means±S.D. At least three independent experiments were performed for the assay, and only one representative result is shown here.
In addition, we also analysed the contribution of other putative transcription factor-binding sites to the transactivation of the promoter upon arsenite stimulation, including NF-κB, p53 and HIF-1-responsive element. A conserved NF-κB site at −346/−337 bp was mutated from gggctgtccc to aAActgtccc. A p53 site was found at −310/−301 bp, and this sequence was previously identified to be responsible for p53 activation of the RTP801 promoter [8]. The p53-responsive sequence aaacaagtct was replaced by aaaTaaAtct. Furthermore, we mutated a putative HIF-1-responsive site at −127/−122 bp from acgtga to acAAgc, and such a site was shown to be responsive to hypoxia in a gel mobility-shift assay [7]. These mutations were integrated into the −1158/+166 bp region (ΔF) of the RTP801 promoter and used in luciferase assays in HaCaT cells. As shown in Figure 7, mutations of these sites had no effect on As(III)-mediated stimulation of the RTP801 promoter. Therefore these data suggest that the NF-κB-, p53- and HIF-1-responsive sites studied here are not directly involved in the promoter response to As(III) treatment in HaCaT cells.
Characterization of C/EBP and Elk-1 binding by EMSA
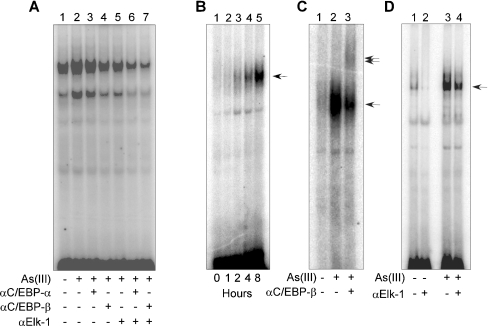
We next employed a gel-shift assay to analyse further the association of the putative transcription factors with the critical arsenic-responsive region of the RTP801 promoter. As a first step, we analysed whether or not As(III) treatment is able to induce protein binding to this critical region. HaCaT cells were treated with As(III) for various periods of time and the nuclear extracts were isolated for EMSA. A 57 bp DNA fragment spanning −1038 to −982 bp that contains both the C/EBP and Elk-1 sites was used as a probe in the experiment. We found that arsenite treatment in these cells was able to induce binding of nuclear proteins to this region at 4 and 8 h after treatment (results not shown). The C/EBP family is composed of at least six members [23]. Among these members, C/EBPβ has been reported to play an important role in keratinocyte differentiation involving growth arrest and keratin expression [25]. Therefore we analysed whether C/EBPβ is present in the protein complex that binds the C/EBP site-containing probe in HaCaT keratinocytes. As shown in Figure 8(A), addition of an anti-C/EBPβ antibody was able to reduce the binding of the protein complex to the 57 bp probe, indicating that C/EBPβ is indeed present in the complex (Figure 8A, lane 4). We also tested an anti-C/EBPα antibody and found that it did not affect the protein binding by itself (Figure 8A, lane 3). An anti-Elk-1 antibody was able to partially reduce the protein binding (Figure 8A, lane 5), indicating that Elk-1 is also present in the protein complex that binds the RTP801 promoter. Interestingly, the anti-C/EBPα antibody or the anti-C/EBPβ antibody, when combined with the anti-Elk-1 antibody, was able to significantly reduce the protein binding to the 57 bp probe (Figure 8A, lanes 6 and 7), suggesting a synergistic effect between C/EBP and Elk-1 in binding the arsenic-responsive region of the RTP801 promoter.
Figure 8. EMSA using nuclear extracts.
(A) HaCaT cells were treated with 10 μM As(III) for 4 h, and the nuclear extracts were isolated from the cells and used in an EMSA with a 57 bp 32P-labelled probe that covers the critical As(III)-responsive region of RTP801 promoter (from −1038 to −982 bp). Different anti-C/EBP or anti-Elk-1 antibodies were used in EMSA, as indicated. (B) The nuclear extracts isolated as described above were used in EMSA with an oligonucleotide probe (AATGATGAAACACGGGAT) that contains a C/EBP-binding site in the middle. The single arrow indicates the binding complex induced by As(III). (C) Supershift assay with an anti-C/EBPβ antibody. The nuclear extracts isolated from HaCaT cells treated with 10 μM As(III) for 4 h were used in an EMSA with the oligonucleotide probe containing the C/EBP-binding site. The single arrow indicates the band induced by As(III), and the double arrows mark the band shifted by the anti-C/EBPβ antibody (lane 3). (D) Effect of an anti-Elk-1 antibody in EMSA. The nuclear extracts were isolated from HaCaT cells treated with 10 μM As(III) for 4 h. An Elk-1-containing oligonucleotide probe (CCAGATTCCTGTGGCCC) was used in EMSA. The single arrow indicates the band induced by As(III). Note the reduction in the intensity of the band in the presence of the anti-Elk-1 antibody (lane 4).
To analyse further the binding of the transcription factors to the RTP801 promoter, we utilized an oligonucleotide probe (AATGATGAAACACGGGAT) that contains the consensus C/EBP site at the RTP801 promoter. Similarly, As(III) treatment was able to initiate a time-dependent binding of a protein complex to this sequence, with a maximal binding at 4 and 8 h after arsenite treatment (Figure 8B). Addition of an anti-C/EBPβ antibody was able to induce a ‘supershift’ in EMSA, indicating further that C/EBPβ is present in the complex (Figure 8C, lane 3). We later analysed the presence of Elk-1 protein in EMSA with an oligonucleotide probe (CCAGATTCCTGTGGCCC) that contains the consensus Elk-1 sequence at the RTP801 promoter. Arsenite treatment was able to induce protein complex formation with this probe (Figure 8C, lane 3), and an anti-Elk-1 antibody was able to partially reduce such binding (lane 4). These data provided additional evidence that Elk-1 is present in the protein complex that binds the RTP801 promoter upon arsenic treatment.
Arsenic-stimulated RTP801 transcription is mediated by MAPK
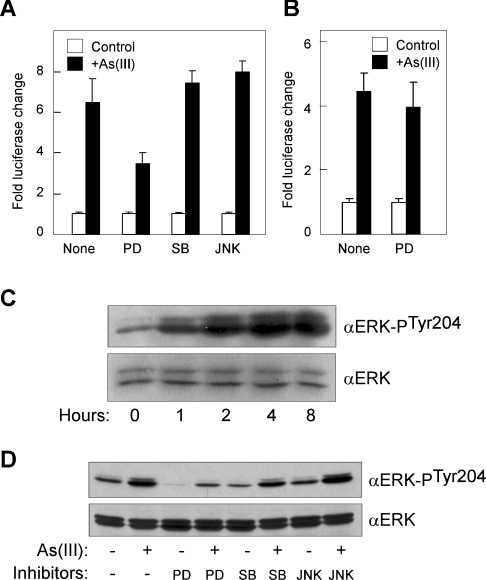
It has been reported that arsenic differentially activates ERKs, p38 and JNKs to mediate various effects on carcinogenesis [1,4]. Activation of ERKs by arsenic is able to induce cell proliferation and transformation, whereas p38 and JNK activation are associated with cell growth arrest and apoptosis. To elucidate a role for these kinases in the arsenic-mediated activation of the RTP801 promoter, we examined the effects of various MAPK inhibitors. PD98059 is a selective inhibitor of ERK and MEK (MAPK/ERK kinase). SB203580 is able to selectively inhibit p38 MAPK, and JNK inhibitor II potently inhibits JNK. In the experiment, HaCaT cells were transfected with a −1158/+166 bp RTP801 promoter construct and treated with these inhibitors. As shown in Figure 9(A), SB203580 and JNK inhibitor II at 10 μM had no obvious effect on As(III)-mediated activation of the promoter. However, PD98059 at 10 μM was able to partially abrogate the arsenic effect, indicating that ERK is probably involved in arsenic stimulation of the RTP801 promoter. Such an inhibitory effect of PD98059 could be observed at a concentration as low as 1 μM (results not shown). To confirm that ERK is activated by arsenite in HaCaT cells, we analysed the phosphorylation status of ERK in these cells by Western blotting. As shown in Figure 9(C), arsenite treatment was able to induce a time-dependent elevation of ERK phosphorylation. In addition, we found that treatment of HaCaT cells with PD98059 was able to reduce the arsenite-mediated ERK phosphorylation (Figure 9D). However, the p38 inhibitor (SB203580) and JNK inhibitor (JNK inhibitor II) had a minimal effect on the phosphorylation of ERK upon arsenite exposure. Furthermore, we analysed the effect of the MAPK inhibitor PD98059 on the RTP801 promoter with a point mutation of the putative Elk-1 site at the arsenite-responsive region, as described in Figure 7. As shown in Figure 9(B), such a mutant promoter construct was no longer inhibited by PD98059, indicating further that the conserved Elk-1 site in this region is implicated in MAPK-mediated As(III) stimulation of the promoter. Taken together, these data suggest that activation of ERK by arsenic is implicated in the signalling pathway that stimulates RTP801 transcription. This is consistent with our findings that Elk-1, a transcription factor downstream of MAPK, is involved in arsenic stimulation of the RTP801 promoter.
Figure 9. Effect of MAPK and JNK inhibitors.
(A) HaCaT cells were transiently transfected with RTP801 promoter construct (−2548/+166bp) and Renilla luciferase vector. At 24 h after transfection, cells were treated with 10 μM As(III) for 8 h in the presence of 10 μM inhibitors, as indicated. PD, PD98059; SB, SB203580; JNK, JNK inhibitor II. The whole cell lysate was used in a dual luciferase assay. The fold changes in luciferase activity normalized by Renilla luciferase activity are shown as means±S.D. (B) The mA promoter construct used in Figure 7 with a mutation of the conserved Elk-1 site was used in the luciferase assay, together with PD98059 and As(III) treatment, as above. (C) Activation of ERK by arsenite. HaCaT cells were treated with 10 μM As(III) for various periods of time, as indicated. The cell lysate was used in Western blotting analysis with an anti-ERK antibody and an antibody specific for phosphorylation of ERK at Tyr-204. (D) Inhibition of ERK activation by PD98059. HaCaT cells were treated for 4 h with or without 10 μM As(III) and/or 10 μM inhibitors, as indicated. The cell lysate was used in Western blotting analysis as above.
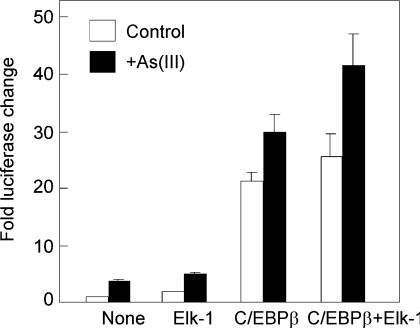
Effect of overexpression of Elk-1 and C/EBPβ on the RTP801 promoter
To provide further evidence that Elk-1 and C/EBP sites are involved in the regulation of RTP801 transcription upon arsenic treatment, we next analysed the effect of overexpression of Elk-1 or C/EBPβ on the human RTP801 promoter. Both human Elk-1 and C/EBPβ were cloned and co-transfected with the human RTP801 promoter in HaCaT cells. As shown in Figure 10, overexpression of Elk-1 has a small effect in raising the basal promoter activity (approx. 2-fold). Overexpression of C/EBPβ was able to dramatically elevate the promoter activity by more than 20-fold. Interestingly, co-expression of Elk-1 with C/EBPβ appeared to have a synergistic effect on the promoter activity. These data therefore provided additional evidence to implicate Elk-1 and C/EBPβ in the regulation of the human RTP801 promoter.
Figure 10. Effect of overexpression of Elk-1 and C/EBPβ on RTP801 transcription.
The ΔF promoter construct was co-transfected with Elk-1 or C/EBPβ (0.25 μg of each), as indicated. The cells were treated with 10 μM As(III) for 8 h at 24 h after the transfection. The fold changes in luciferase activity are shown as the means±S.D. At least two independent experiments were performed for this assay, and only one representative result is shown here.
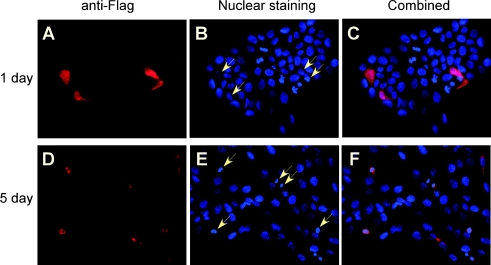
Overexpression of RTP801 induces apoptosis in HaCaT cells
RTP801 was originally isolated as a target gene of HIF-1 [7]. It has been demonstrated that the biological activity of RTP801 overexpression is dependent on the cellular context, and may promote or inhibit cell death [7]. To determine the biological effect of RTP801 on HaCaT keratinocytes, we carried out an immunofluorescence assay to analyse the cellular effect of RTP801 overexpression. A FLAG-tagged RTP801 expression plasmid was transfected into HaCaT cells, and protein expression was analysed by immunofluorescence with an anti-FLAG antibody. Nuclear staining was also performed with these cells. At 24 h after transfection, RTP801 was evenly expressed in both cytoplasm and nucleus (Figures 11A–11C). Interestingly, at 5 days after transfection, most RTP801-expressing cells (stained red) revealed condensed pycnotic nuclei (Figures 11D–11F), characteristic of apoptotic cells. Taken together, these data clearly indicate that overexpression of RTP801 was able to rapidly induce apoptosis in HaCaT keratinocytes.
Figure 11. Induction of apoptosis by overexpression of RTP801.
(A–C) HaCaT cells were transfected with FLAG-tagged human RTP801 expression plasmid (2.0 μg). Fixation and fluorescence staining of the cells were performed at various times after the transfection. An anti-FLAG antibody and a secondary Alexa-546-conjugated anti-mouse antibody were used to determine the expression of RTP801 (shown by the red colour). Nuclear staining was performed with Hoechst 33342 (shown by the blue colour). The arrows indicate the cells expressing RTP801. Note the change in nuclear shapes in RTP801-expressing cells at 5 days after transfection (D–F).
DISCUSSION
In the present study, we investigated the activity of arsenic in regulating a newly discovered gene, RTP801, that is induced by hypoxia and other cell stress signals. In HaCaT human keratinocytes, we found that arsenic was able to induce a rapid rise of RTP801 mRNA level. Similarly, arsenic treatment was capable of stimulating the human RTP801 promoter in HaCaT cells, and such an effect was mediated by ROS production in these cells. Through studies with a series of deletion constructs of the promoter, we identified a critical arsenic-responsive region between −1057 and −981 bp of the promoter. When this critical arsenic-responsive region was linked to a TATA box, it was able to mediate arsenite stimulation, indicating that such a region is not only required, but also sufficient, for the transcriptional regulation of the promoter by arsenite. Further point mutation studies with the putative Elk-1 site and C/EBP site within this region revealed that Elk-1 and C/EBP are involved in transcriptional regulation of the RTP801 gene by arsenic. This was confirmed further by a gel mobility-shift assay. The involvement of the MAPK pathway in the effect of arsenic was supported by our findings that a selective ERK inhibitor was able to partially antagonize the arsenic effect on RTP801 transcription. To elucidate further the effect of Elk-1 and C/EBP on RTP801 transcription, we overexpressed these two genes in HaCaT cells and found that they were able to elevate the promoter activity. Therefore these studies provided the first evidence that RTP801 is a transcriptional target of arsenic in keratinocytes, and that Elk-1 and C/EBP are involved in transcriptional regulation.
One of the interesting findings from this study is that C/EBP is involved in arsenic-mediated gene regulation. Our experiments revealed a critical region in the RTP801 promoter that mediates transcriptional regulation by arsenic. Further deletion and mutation analyses of this region indicate that a consensus C/EBP site is implicated in the regulation. Furthermore, treatment of HaCaT cells with arsenic was able to rapidly induce a protein complex that binds the C/EBP consensus sequence, and such binding was partially ‘supershifted’ by an anti-C/EBPβ antibody. It is noteworthy that the arsenic-induced protein binding at the promoter reached a maximal level (Figure 8) later than the maximal induction of RTP801 mRNA by arsenic (Figure 1), indicating that the transcription of the RTP801 gene might be initiated at an earlier stage, before maximal C/EBP and Elk-1 complexes are formed at the promoter. C/EBPs are a family of transcription factors containing a highly conserved basic leucine-zipper domain at the C-terminus that participates in dimerization and DNA binding [23]. At least six C/EBP members have been characterized so far, including C/EBPα, C/EBPβ, C/EBPγ, C/EBPδ, C/EBPϵ and C/EBPζ. These factors play pivotal roles in the control of cell proliferation, cell differentiation, inflammation, metabolism and many other cellular responses. C/EBP has not been previously implicated in ROS-mediated gene regulation. Regulation of C/EBP could occur at the transcriptional, translational and post-translational levels [26]. It will be of great importance in the future to elucidate at which level C/EBP is involved in arsenic- and ROS-mediated transcriptional control.
RTP801 has been shown to play a role in the regulation of ROS involved in the p53-dependent DNA damage response. Ellisen et al. [8] recently reported that RTP801 is a transcriptional target of p63, a family member of p53. During mouse development, the expression pattern of RTP801 closely correlates with that of p63. ROS production is largely abrogated in p63-deficient mouse embryonic fibroblasts [8]. Meanwhile, expression of RTP801 is able to greatly induce H2O2-mediated ROS production in these cells [8]. It appears that RTP801 is required for the production of ROS in a p63-dependent manner, and it may play a role in epithelial differentiation. However, the regulation of ROS by RTP801 is probably dependent on the cellular context. In human breast cancer (MCF-7) cells and neuronal PC12 cells, overexpression of RTP801 is not able to change the basal level of ROS [7]. On the contrary, RTP801 can abrogate ROS production induced by H2O2 or hypoxia, consistent with the ability of RTP801 to protect these cells from apoptosis after treatment with these agents [7]. In the present study, we found that the transcription of RTP801 itself is regulated by ROS following arsenite treatment in HaCaT keratinocytes. The arsenic-mediated RTP801 transactivation is inhibited by overexpression of enzymes that reduce cellular ROS concentration, or by incubation with the ROS scavenger NAC. We propose that RTP801 is implicated in the complex regulation of ROS production in the cells. On the one hand, RTP801 is directly involved in the production of ROS, with an effect that varies from cell type to cell type. On the other hand, the transcription of RTP801 is under the control of the ROS level inside the cell, which probably had a role in feedback regulation to further modulate the production and function of intracellular ROS. As the protein primary structure of RTP801 does not share any similarity to the enzymes that regulate the production and conversion of ROS, it is unlikely that RTP801 is directly involved in the generation of ROS in the cells. It remains to be determined whether RTP801 serves as a modulator that functionally collaborates with or antagonizes enzymes that are directly involved in the regulation of ROS.
At present, the biological function of RTP801 is largely unknown. One of the primary functions suggested by recent studies is its cell-type-dependent role in apoptosis. In some types of cells, RTP801 has a protective effect. Overexpression of RTP801 is able to protect MCF7 and PC12 cells from hypoxia- and H2O2-triggered apoptosis [7]. In addition, overexpression of RTP801 in WEH17.2 murine T-lymphoma cells was able to protect the cells from dexamethasone-induced apoptosis [9]. However, RTP801 may also function in promoting cell death in other types of cells. It has been shown that RTP801 is able to increase the sensitivity of non-dividing PC12 cells to the insults of hypoxia and oxidative stress [7]. Concomitantly, liposomal delivery of RTP801 to mouse lungs leads to massive cell death [7]. In our studies, overexpression of RTP801 is able to induce nuclear condensation as well as apoptosis in HaCaT keratinocytes. Therefore the apoptotic effect of RTP801 seems to be dependent on the cellular context. It is likely that RTP801 interacts with the apoptotic machinery differently in a cell-dependent manner. Bearing this in mind, detailed biochemical analysis of RTP801 function in the apoptosis pathway in various cell types will undoubtedly aid in clarifying this issue in the future.
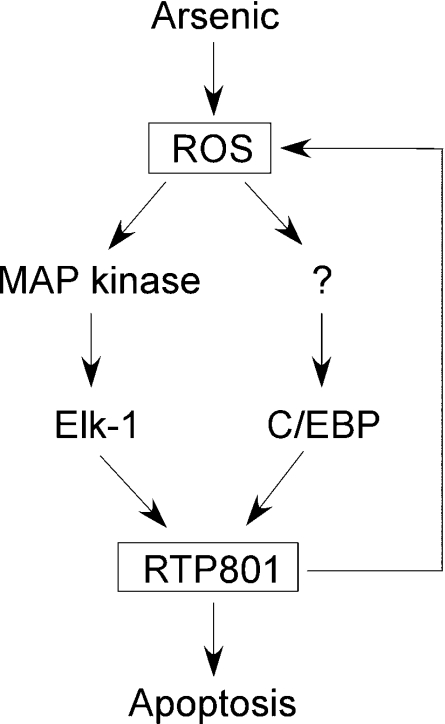
Taken together, our studies have shown that arsenic is able to regulate RTP801 transcription through ROS production and the transcription factors Elk-1 and C/EBP, as illustrated in Scheme 1. In this model, arsenic increases cellular ROS, which in turn activate Elk-1 via MAPK. Meanwhile, C/EBP is activated via an unknown mechanism. Both transcription factors then act on the promoter of RTP801 and induce transcription of the gene. The change of intracellular level of RTP801 will have an effect on the apoptotic potential of the cells, either enhancing or inhibiting apoptosis in a cell-type-dependent manner. In addition, RTP801 may participate in the regulatory circuit that controls the production of intracellular ROS, which will modulate further the transcription of RTP801 as well as its effect on apoptosis in a cell-context-dependent manner.
Scheme 1. A hypothetical circuit that links arsenic and RTP801 transcription to ROS production and apoptosis.
Acknowledgments
We thank Dr Fei Chen for a critical reading of, and constructive comments on, this manuscript. We thank Dr Jeffrey B. Travers for providing the HaCaT cells. This work was supported by a research grant from the American Cancer Society (PRG-00-273-01-MGO), a grant from the National Institute of Diabetes and Digestive & Kidney Diseases (R01-DK55991) and a Scientist Development Award from the American Heart Association to Y.C.
References
- 1.Qian Y., Castranova V., Shi X. New perspectives in arsenic-induced cell signal transduction. J. Inorg. Biochem. 2003;96:271–278. doi: 10.1016/s0162-0134(03)00235-6. [DOI] [PubMed] [Google Scholar]
- 2.Droge W. Free radicals in the physiological control of cell function. Physiol. Rev. 2002;82:47–95. doi: 10.1152/physrev.00018.2001. [DOI] [PubMed] [Google Scholar]
- 3.Hei T. K., Liu S. X., Waldren C. Mutagenicity of arsenic in mammalian cells: role of reactive oxygen species. Proc. Natl. Acad. Sci. U.S.A. 1998;95:8103–8107. doi: 10.1073/pnas.95.14.8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bode A. M., Dong Z. The paradox of arsenic: molecular mechanisms of cell transformation and chemotherapeutic effects. Crit. Rev. Oncol. Hematol. 2002;42:5–24. doi: 10.1016/s1040-8428(01)00215-3. [DOI] [PubMed] [Google Scholar]
- 5.Baldwin A. S. Control of oncogenesis and cancer therapy resistance by the transcription factor NF-kappaB. J. Clin. Invest. 2001;107:241–246. doi: 10.1172/JCI11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen F., Shi X. Signaling from toxic metals to NF-kappaB and beyond: not just a matter of reactive oxygen species. Environ. Health Perspect. 2002;110:807–811. doi: 10.1289/ehp.02110s5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shoshani T., Faerman A., Mett I., Zelin E., Tenne T., Gorodin S., Moshel Y., Elbaz S., Budanov A., Chajut A., et al. Identification of a novel hypoxia-inducible factor 1-responsive gene, RTP801, involved in apoptosis. Mol. Cell. Biol. 2002;22:2283–2293. doi: 10.1128/MCB.22.7.2283-2293.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellisen L. W., Ramsayer K. D., Johannessen C. M., Yang A., Beppu H., Minda K., Oliner J. D., McKeon F., Haber D. A. REDD1, a developmentally regulated transcriptional target of p63 and p53, links p63 to regulation of reactive oxygen species. Mol. Cell. 2002;10:995–1005. doi: 10.1016/s1097-2765(02)00706-2. [DOI] [PubMed] [Google Scholar]
- 9.Wang Z., Malone M. H., Thomenius M. J., Zhong F., Xu F., Distelhorst C. W. Dexamethasone-induced gene 2 (dig2) is a novel pro-survival stress gene induced rapidly by diverse apoptotic signals. J. Biol. Chem. 2003;278:27053–27058. doi: 10.1074/jbc.M303723200. [DOI] [PubMed] [Google Scholar]
- 10.Schwarzer R., Tondera D., Arnold W., Giese K., Klippel A., Kaufmann J. REDD1 integrates hypoxia-mediated survival signaling downstream of phosphatidylinositol 3-kinase. Oncogene. 2005;24:1138–1149. doi: 10.1038/sj.onc.1208236. [DOI] [PubMed] [Google Scholar]
- 11.Brafman A., Mett I., Shafir M., Gottlieb H., Damari G., Gozlan-Kelner S., Vishnevskia-Dai V., Skaliter R., Einat P., Faerman A., et al. Inhibition of oxygen-induced retinopathy in RTP801-deficient mice. Invest. Ophthalmol. Vis. Sci. 2004;45:3796–3805. doi: 10.1167/iovs.04-0052. [DOI] [PubMed] [Google Scholar]
- 12.Brugarolas J., Lei K., Hurley R. L., Manning B. D., Reiling J. H., Hafen E., Witters L. A., Ellisen L. W., Kaelin W. G., Jr Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18:2893–2904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corradetti M. N., Inoki K., Guan K. L. The stress-inducted proteins RTP801 and RTP801L are negative regulators of the mammalian target of rapamycin pathway. J. Biol. Chem. 2005;280:9769–9772. doi: 10.1074/jbc.C400557200. [DOI] [PubMed] [Google Scholar]
- 14.Boukamp P., Petrussevska R. T., Breitkreutz D., Hornung J., Markham A., Fusenig N. E. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol. 1988;106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagarajan R. P., Chen F., Li W., Vig E., Harrington M. A., Nakshatri H., Chen Y. Repression of transforming-growth-factor-beta-mediated transcription by nuclear factor kappaB. Biochem. J. 2000;348:591–596. [PMC free article] [PubMed] [Google Scholar]
- 16.Andrews N. C., Faller D. V. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagarajan R. P., Zhang J., Li W., Chen Y. Regulation of smad7 promoter by direct association with smad3 and smad4. J. Biol. Chem. 1999;274:33412–33418. doi: 10.1074/jbc.274.47.33412. [DOI] [PubMed] [Google Scholar]
- 18.Shi H., Hudson L. G., Ding W., Wang S., Cooper K. L., Liu S., Chen Y., Shi X., Liu K. J. Arsenite causes DNA damage in keratinocytes via generation of hydroxyl radicals. Chem. Res. Toxicol. 2004;17:871–878. doi: 10.1021/tx049939e. [DOI] [PubMed] [Google Scholar]
- 19.Deby C., Goutier R. New perspectives on the biochemistry of superoxide anion and the efficiency of superoxide dismutases. Biochem. Pharmacol. 1990;39:399–405. doi: 10.1016/0006-2952(90)90043-k. [DOI] [PubMed] [Google Scholar]
- 20.Rao V. N., Huebner K., Isobe M., ar-Rushdi A., Croce C. M., Reddy E. S. elk, tissue-specific ets-related genes on chromosomes X and 14 near translocation breakpoints. Science. 1989;244:66–70. doi: 10.1126/science.2539641. [DOI] [PubMed] [Google Scholar]
- 21.Rao V. N., Reddy E. S. elk-1 proteins interact with MAP kinases. Oncogene. 1994;9:1855–1860. [PubMed] [Google Scholar]
- 22.Graves B. J., Johnson P. F., McKnight S. L. Homologous recognition of a promoter domain common to the MSV LTR and the HSV tk gene. Cell. 1986;44:565–576. doi: 10.1016/0092-8674(86)90266-7. [DOI] [PubMed] [Google Scholar]
- 23.McKnight S. L. McBindal l – a better name for CCAAT/enhancer binding proteins? Cell. 2001;107:259–261. doi: 10.1016/s0092-8674(01)00543-8. [DOI] [PubMed] [Google Scholar]
- 24.Sladek F. M., Zhong W. M., Lai E., Darnell J. E., Jr Liver-enriched transcription factor HNF-4 is a novel member of the steroid hormone receptor superfamily. Genes Dev. 1990;4:2353–2365. doi: 10.1101/gad.4.12b.2353. [DOI] [PubMed] [Google Scholar]
- 25.Zhu S., Oh H. S., Shim M., Sterneck E., Johnson P. F., Smart R. C. C/EBPbeta modulates the early events of keratinocyte differentiation involving growth arrest and keratin 1 and keratin 10 expression. Mol. Cell. Biol. 1999;19:7181–7190. doi: 10.1128/mcb.19.10.7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramji D. P., Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem. J. 2002;365:561–575. doi: 10.1042/BJ20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]