Abstract
Phys (phytochromes) are a superfamily of photochromic photoreceptors that employ a bilin-type chromophore to sense red and far-red light. Although originally thought to be restricted to plants, accumulating genetic and genomic analyses now indicate that they are also prevalent among micro-organisms. By a combination of phylogenetic and biochemical studies, we have expanded the Phy superfamily and organized its members into distinct functional clades which include the phys (plant Phys), BphPs (bacteriophytochromes), Cphs (cyanobacterial Phys), Fphs (fungal Phys) and a collection of Phy-like sequences. All contain a signature GAF (cGMP phosphodiesterase/adenylate cyclase/FhlA) domain, which houses the bilin lyase activity. A PHY domain (uppercase letters are used to denote the PHY domain specifically), which helps stabilize the Pfr form (far-red-light-absorbing form of Phy), is downstream of the GAF region in all but the Phy-like sequences. The phy, Cph, BphP and Fph families also include a PLD [N-terminal PAS (Per/Arnt/Sim)-like domain] upstream of the GAF domain. Site-directed mutagenesis of conserved residues within the GAF and PLD motifs supports their importance in chromophore binding and/or spectral activity. In agreement with Lamparter, Carrascal, Michael, Martinez, Rottwinkel and Abian [(2004) Biochemistry 43, 3659–3669], a conserved cysteine within the PLD of several BphPs was found to be necessary for binding the chromophore via the C-3 vinyl side chain on the bilin A ring. Phy-type sequences were also discovered in the actinobacterium Kineococcus radiotolerans and collections of microorganisms obtained from marine and extremely acidic environments, thus expanding further the range of these photoreceptors. Based on their organization and distribution, the evolution of the Phy superfamily into distinct photoreceptor types is proposed.
Keywords: bilin, histidine kinase, light perception, phytochrome, phylogeny
Abbreviations: Phy, phytochrome; PHY, phytochrome domain; BphP, bacteriophytochrome; Cph, cyanobacterial Phy; Fph, fungal Phy; phy, plant Phy; AthphyA, etc., Arabidopsis thaliana phy isoform A, etc; BV, biliverdin; EnvSeq, environmental sequence related to the Phy-type protein superfamily; FR, far-red light; Pfr, FR-absorbing form of Phy; R, red light; Pr, R-absorbing form of Phy; Et, ethyl; GAF, cGMP phosphodiesterase/adenylate cyclase/FhlA; HK, histidine kinase; HO, haem oxygenase; PAS domain, Per/Arnt/Sim domain; PCB, phycocyanobilin; PΦB, phytochromobilin; PLD, PAS-like domain; PP2C, protein phosphatase-2C; RR, response regulator; TC, two-component receptor
INTRODUCTION
Light is an important environmental signal for almost all cellular organisms, providing both the energy necessary for growth and metabolism and the sensory information helpful for adaptation. Detection of light is achieved by a suite of photoreceptors that work either singly or in various combinations [1]. One pervasive set of regulatory photoreceptors is the Phys (phytochromes), which are collectively defined by the use of a bilin (or linear tetrapyrrole) chromophore for detecting R (red light) and/or FR (far-red light) [2–4]. This bilin is generated by oxidative cleavage of haem by an HO (haem oxygenase) to generate BV (biliverdin) IXα, and either used directly or further modified depending on the organism [5,6]. Once bound to Phy apoprotein, the bilin can phototransform between two stable conformers, an R-absorbing form (Pr) and an FR-absorbing form (Pfr). By having one form behave as ‘active’ and the other as ‘inactive’, Phys can act as R/FR-regulated reversible switches. This photochromicity also provides a crude form of colour vision through measurement of the Pr/Pfr ratio, which varies depending on the relative amounts of R and FR [3].
Phy-type pigments were first discovered in higher plants over 50 years ago, based on the ability of R and FR to control many aspects of plant growth and development [2,3]. Arabidopsis thaliana, for example, contains five Phy isoforms (AthphyA–E) that work individually and in concert to regulate photomorphogenesis. phys (plant Phys) contain an N-terminal domain that binds a single bilin, PΦB (phytochromobilin), through a distinctive GAF (cGMP phosphodiesterase/adenylate cyclase/FhlA) domain, and a C-terminal domain that is required for dimerization. The chromophore is linked autocatalytically to the GAF domain via a thioether linkage between the A-ring ethylidene side chain of PΦB and a positionally conserved Cys (Cys-323 in Ar. thaliana phyA). Immediately distal to the GAF domain is a PHY domain (uppercase letters are used to denote the Phy domain specifically) motif that together with the GAF domain is needed for the unusual photochemical properties of this photoreceptor type [7–10]. The C-terminal half contains two PAS (Per/Arnt/Sim) domains distantly related to GAFs, which are important for signal transmission [2]. Although the mechanism of action for plant phys remains unclear, it has been proposed that they act as light-regulated HKs (histidine kinases) based on limited sequence similarity of their extreme C-termini to the HK domain present in bacterial TC (two-component receptor)-HKs [11,12]. However, the facts that the HK-related sequence of plant phys is missing several important residues that typify a TC-HK, including the His residue that serves as the acceptor for the initial phosphorylation step [4], and that the HK region is partially dispensable in vivo [10,13,14], suggest that either plant phys are not HKs or work by an alternative mechanism. The possibility that plant phys are Ser/Thr kinases has been put forward [11].
More recently, genetic and genomic analyses have dramatically expanded the distribution of Phys to other domains of life, with the discovery of similar photoreceptors in proteobacteria, cyanobacteria, fungi and possibly slime moulds [4,15]. Kehoe and Grossman [16] first realized this potential with the identification of RcaE, a locus required for complementary chromatic adaptation in the cyanobacterium Fremyella diplosiphon. The N-terminal region of FdRcaE is similar in sequence to the bilin-binding GAF domain of plant phys. Downstream of the GAF domain is a canonical HK domain, implying that FdRcaE is a bona fide histidine phosphotransferase. Subsequent BLAST searches identified proteins related to FdRcaE in several other cyanobacterial genomes. For example, Synechocystis PCC6803 encodes five such sequences [8,16–21]. SyCph1 (where ‘Cph’ represents cyanobacterial Phy), which has both the GAF/PHY and the HK domains, was shown to bind PΦB and PCB (phycocyanobilin) in vitro to generate a photochromic photoreceptor with a light-regulated HK activity [17]. SyCph1 could donate this phosphate to an RR (response regulator) encoded downstream in the SyCph operon, which would demonstrate that it participates directly in a TC-HK phospho-relay.
We and others also found related Phys in searches of various proteobacterial and fungal genomes, thus expanding further the distribution of Phys to non-photosynthetic organisms [5,22–25]. In most cases, both the GAF and PHY motifs were evident in the N-terminal region followed by a HK domain. However, these sequences were distinguishable from plant phys and some relatives from cyanobacteria by the absence of the positionally conserved Cys in the GAF domain, which is used by plant phys to bind bilins. Holoprotein assembly studies subsequently implicated both a GAF-domain His immediately distal to the Cys used by plant phys [22] and a conserved Cys within an N-terminal PLD (PAS-like domain) as alternative attachment site(s) [26,27]. Surprisingly, the proteobacterial forms appear to prefer BV IXα instead of PΦB or PCB, which would simplify chromophore biosynthesis [5]. In several cases, the HO needed to convert haem into BV IXα is within or near the Phy operon, thus linking genetically chromophore and apoprotein production [5,28].
Clearly, the discovery of numerous Phy-type proteins in the microbial world now allows us to exploit the evolutionary distance to help define common structural features critical to this photoreceptor superfamily. In the present paper, we used the exponentially expanding wealth of genomic data to help define phylogenetically the breadth and diversity of microbial Phy-type photoreceptors and to predict important distinguishing features. Using the signature GAF, PHY and PLD sequences from these photoreceptors as queries [8,26], we identified a large collection of Phy-type proteins in numerous bacterial, fungal, and as yet to be classified species. By combining both sequence alignments and motif searches, we tentatively divided this superfamily into several distinct clades that better reflect their photobiological properties and distribution within the bacterial and fungal kingdoms. The alignments also identified numerous conserved amino acids, some of which were confirmed by site-directed mutagenesis to be important for bilin binding and/or photochemistry. Collectively, these analyses show that the Phy superfamily includes a diverse set of photoreceptors present in many ecological niches and provide structural information for predicting the behaviour of new Phy-related sequences.
EXPERIMENTAL
Identification of Phy-related sequences
The SMART database (http://smart.embl-heidelberg.de) was used to locate the GAF domain (also referred to as P3 [7]) in several representative Phy proteins from micro-organisms and the five plant phy polypeptides from Ar. thaliana. These sequences were used as queries in exhaustive NCBI-BLASTP searches of all available cyanobacterial, fungal and bacterial genomic databases (completed 1 June 2004) and 40 microbial sequences with an E-value of below 0.02 were identified. BLAST searches were repeated using the SMART and PFAM-predicted GAF domains from this collection as queries, which recovered 16 additional loci with sequences scores beneath the 0.02 cut off. This process was a repeated a third time with these additional sequences and recovered two more loci for a total of 58 representatives. The SMART/PFAM databases failed to reliably identify the PLD and the PHY domains (also known as P2 and P4 respectively [7]) in the collection of Phys. As a consequence, the outer limits of these domains were identified by sequence alignments and refined by hand analyses. The predicted PLD and PHY domains in this final alignment with the lowest SMART or PFAM E-values were used for subsequent analyses. Environmental sequences (EnvSeq) [29,30] predicted to be part of the Phy superfamily were identified by BLAST searches with representative PLD, GAF and PHY sequences based on the E-value cut-off of 0.01. All of the EnvSeq sequences were predicted to be partial clones based on SMART with the exception of EnvSeq-1, -2 and -12.
Alignment and phylogenetic analysis
Amino acid sequences were aligned using CLUSTALX MAC v1.81 (http://www.embl.de/~chenna/clustal/darwin/). The unrooted phylogenetic tree based on the GAF domain was generated in CLUSTALX by the neighbour-joining method and a 1000 times bootstrap replicate, and displayed using Treeview PC v1.5.2 (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html). Only those EnvSeqs predicted to have a complete GAF domain were included. Amino acid sequence alignments were calculated and displayed using MACBOXSHADE v2.15 (Institute of Animal Health, Pirbright, Sussex, U.K.). (It should be noted that most DNA sequences obtained from the various genomic databases were not reconfirmed here. Consequently, it is likely that some derived protein sequences contain errors that could alter their alignments and the extent of sequence conservation.) Additional domains were identified from the SMART and PFAM (http://www.sanger.ac.uk/Software/Pfam/) databases, BLAST searches, and sequence alignments. Accession numbers for the Sargasso Sea EnvSeq-1 to -11 sequences are: 1 (EAI29822), 2 (EAJ02301), 3 (EAG56416), 4 (EAF18794), 5 (EAG25097), 6 (EAE74053), 7 (EAK02112), 8 (EAG45313), 9 (AACY01244739), 10 (EAD68975) and 11 (AACY01305952) [29]. Two additional accessions, EAK71409 and EAK71410, overlapped with the sequence of EnvSeq-1. The accession number for EnvSeq-12 from the mine biofilm is AADL01000497 [30].
Chromophore binding and spectroscopic analyses
Deinococcus radiodurans (Dr) BphP(N321) was PCR amplified from the full-length cDNA described previously [5]. The coding regions for Agrobacterium tumefaciens (At) BphP1 and BphP2, BphP1(N504) and -(N320), and BphP2(N505) and -(N314) were PCR-amplified from strain C58 genomic DNA by using primers designed to introduce NdeI and XhoI sites before and after the designated length of coding region, respectively. The NdeI/XhoI-digested (Promega) PCR products were cloned into pET21b (Novagen), which was similarly digested, resulting in the addition of codons for a His6 tag before a stop codon. All site-directed mutations were introduced by the PCR-based QuikChange method (Stratagene, La Jolla, CA, U.S.A.). Each coding region was completely sequenced by the dideoxy method to confirm introduction of the appropriate mutation. Recombinant proteins were expressed in Escherichia coli strain BL21-Codon Plus (DE3)-RIL (Stratagene) and purified by nickel-chelate affinity chromatography (Novagen) as described in [5]. After purification, the buffer was exchanged for 70 mM Tris/HCl (pH 8.0)/1 mM Na4EDTA by ultrafiltration using a Centriprep YM-10 column.
For chromoprotein assembly, crude soluble extracts from E. coli expressing the recombinant apoproteins were incubated in darkness for 1 h, in at least a 10-fold molar excess of bilin, prior to affinity purification. BV IXα was obtained from Porphyrin Products (Logan, UT, U.S.A.), PCB from Dr Pill-Soon Song (Kumho Life and Environmental Science Laboratory, Gwangju, Korea), BV IIIα, BV XIIIα and mesoBV XIIIα from Dr Tony McDonagh (Division of Gastroenterology and the Liver Center, University of California-San Francisco, San Francisco, CA, U.S.A.), and 18-Et (ethyl)-BV IXα, 3-Et-BV IXα and 3-18-Et-BV IXα from Dr Katsuhiko Inomata (Division of Material Science, Kanazawa University, Ishikawa, Japan) as described in [26]. Absorption spectra of the assembled Phys were determined after overnight incubations in the dark or after saturating irradiations with light at 690 nm (R) and 775 nm (FR) provided by interference filters. Covalent attachment of bilin to the Phy polypeptides was monitored by zinc-induced fluorescence of the chromoproteins subjected to SDS/PAGE [5].
RESULTS
Phylogenetic analysis of the Phy superfamily
The GAF domain is an approx. 150-amino-acid motif, with very early evolutionary origins, that can be found in a number of cyclic nucleotide-binding proteins, phosphodiesterases and in a diverse set of proteins involved in phototransduction [31]. Phys, in particular, contain a GAF domain that is clearly distinct from those in other protein types, as determined by protein sequence alignments [8]. In many cases, it can also be distinguished biochemically by its ability to direct the autocatalytic attachment of bilins, and then associate with these chromophores, to help generate the unique spectral characteristics of this photoreceptor class (e.g. [5,8,22]).
Using the GAF sequences from representative Phys as queries, we exhaustively searched through the available genomic databases for related proteins by BLASTP. This search identified over 70 proteins containing a Phy-type GAF domain in 30 known microbial species and in two collections of prokaryotic species that remain to be classified. This list included previously described proteins from various cyanobacteria, α- and γ-proteobacteria, and fungi [5,16,17,19,22–25,32–34]. New to this list were GAF-containing sequences from the actinobacterium Kineococcus radiotolerans, the cyanobacteria (Anabena variabilis, Crocosphaera watsonii, Cytophaga hutchinsonii, Gleobacter violaceus and Thermosynechococcus elongatus), the proteobacteria (Magnetospirillum magnetotacticum, Rhodospirilum rubrum, Xanthomonas axonopodis and Xanthomonas campestris) and the fungi (Aspergillus nidulans, Gibberella zeae and Ustilago maydis). The K. radiotolerans sequence was significant because it provided the first example of a potential Phy-type protein in actinobacteria. Phy-type GAF domains were also detected in the archaeon Methanosarcina acetivorans, possibly extending the distribution of Phys to Archaea.
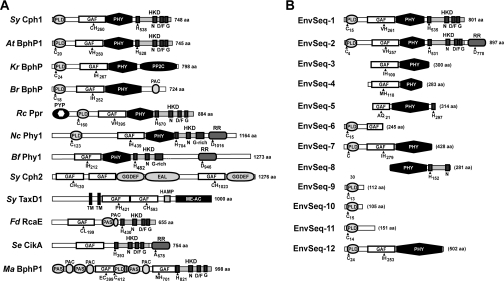
The protein organizations of representative Phys described previously and new species discovered here are shown in Figure 1(A). In several cases, more than one GAF domain was predicted by SMART. Two and three GAF domains were found in Synechocystis PCC6803 TaxD1 (also known as PixJ1) and Cph2 respectively, in accord with previous reports [7,19], and two GAF domains were evident in M. acetivorans BphP. Most of the microbial Phys had a HK domain downstream of the GAF domain, strongly suggesting that they function as TC-HKs. Some of the bacterial Phys (e.g. Synechococcus elongatus CikA [34] and Agrobacterium tumefaciens BphP2 [25]) and all of the fungal Phys {e.g. Neurospora crassa PHY-2 and Botryontinia fuckeliana PHY1 ([5,23] and the present study)} also contain an RR domain appended to the C-terminal end of the HK domain (Figure 1A), thus placing them in the hybrid kinase subfamily. However, the phosphotransferase activity of CikA is in question given the absence of the phosphoacceptor Asp [34]. For a limited number of Phys, other motifs were predicted C-terminal to the bilin-binding region, indicating that alternative sensory transduction chains are possible. These alternative forms include: Synechocystis TaxD1 that has a methyl-accepting domain, K. radiotolerans BphP that has a PP2C (phosphatase-2C) domain, and Synechocystis Cph2, Rhodobacter sphaeroides BphP and Thermosynechococcus elongatus Cph that have Gly-Gly-Asp-Glu-Phe (GGDEF) and Glu-Ala-Leu (EAL) motifs (Figure 1A, and results not shown), which have been connected in other protein contexts with diguanylate cyclase and diguanylate phosphodiesterase activities respectively [35].
Figure 1. Domain structures of representative members from the BphP, Fph, Cph and Phy-like clades (A) and predicted members of the Phy superfamily (EnvSeq) present in a random collection of DNA sequences from a marine environment (EnvSeq-1 to -11 [29]) and an acidophilic mine biofilm (EnvSeq-12 [30]) (B).
Species designations can be found in the legend of Figure 3. The amino acid (aa) length of each protein is indicated on the right; parentheses indicate partial sequence. Abbreviations not otherwise defined in the main text: EAL, motif bearing a consensus Glu/Ala/Leu sequence; GGDEF, motif bearing a consensus Gly/Gly/Asp/Glu/Phe sequence; HAMP, HK/adenylate cyclase/methyl-binding protein/phosphatase domain. HKD, HK domain (the H, N, D/F and G boxes are indicated); HKRD, HK-related domain. Me-Ac, methyl-accepting chemotaxis protein domain. PAC, PAS-like domain C-terminal to PAS; PYP, photoactive yellow protein domain; TM, transmembrane. The positions of signature amino acids in the PLD, GAF, HKD and RR domains are indicated.
In addition to the known species, we discovered eight new Phy-type proteins (designated EnvSeq) containing the signature GAF-domain in the recently released shotgun sequence databases of micro-organisms living in either marine or highly acidic environments (Figure 1B). For EnvSeq-1 to -7, the corresponding DNAs were isolated from unknown micro-organisms present in the Sargasso Sea [29]. The DNA for EnvSeq-12 was isolated from an acidophilic biofilm enriched in Leptospirillum group II proteobacteria and Ferroplasma type II archaebacteria [30]. With the exception of EnvSeq-6, for which insufficient sequence data were available, a PHY domain was apparent C-terminal to the GAF domain. Three entries with sufficient sequence information (EnvSeq-1, -2, and -5) also contained all or part of a C-terminal HK domain, strongly suggesting that they participate in TC-HK signalling, with EnvSeq-2 also containing a C-terminal RR (Figure 1B).
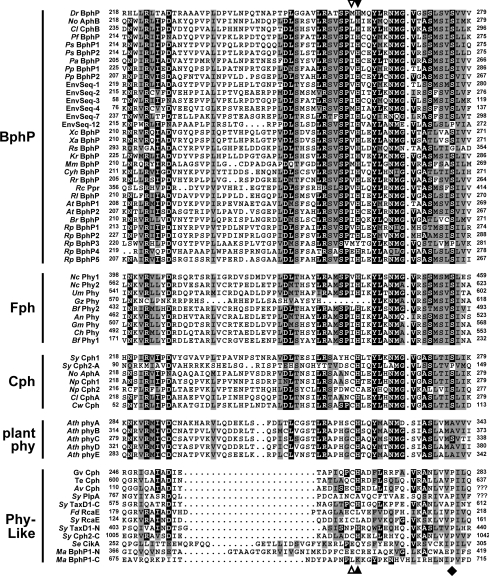
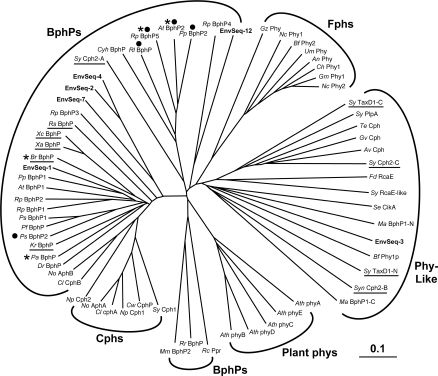
Alignment of our total collection of microbial Phy-type GAF sequences using BLASTP showed that they contained many of the signature residues that define the GAF domain in this group (Figure 2 and Supplemental Figure 1 at http://www.BiochemJ.org/bj/392/bj3920103add.htm). These include an Asp/Arg pair and a Glu (residues 171/172 and 189 respectively, in Synechocystis Cph1) that are essential for the bilin attachment [8] and a Tyr recently shown to be critical for maintaining the native photochemical properties of Synechocystis Cph1 (Tyr-176 [36]). Sequence alignments (Supplemental Figures 1–3 at http://www.BiochemJ.org/bj/392/bj3920103add.htm) and phylogenetic analysis (Figure 3) of these microbial GAF domains together with representatives from a higher plant (AthphyA-E from Ar. thaliana) roughly divided the microbial collection into four major Phy subgroups, which we have designated as Cphs (cyanobacterial Phys), BphPs (bacteriophytochromes), Fphs (fungal Phys) and a collection of Phy-like sequences. Consistent with their names, these families appear distinct from plant phys and show a strong preference for their respective microbial kingdoms. However, it should be emphasized that the clustering was not phyletically strict. For example, Calothrix PCC 7601 contains both a Cph-type (CphA) and a BphP-type (CphB) sequence [37], whereas Synechococcus contains two Cph-like (Cph1 and 2) and three Phy-like (TaxD1, PlpA and RcaE-like) sequences ([17–20] and the present study).
Figure 2. Amino-acid-sequence alignment of a portion of the GAF domain from members of the Phy superfamily.
Alignment of the entire domain can be found in Supplemental Figure 1 (http://www.BiochemJ.org/bj/392/bj3920103add.htm). The sequences are grouped by their proposed inclusion in the BphP, Fph, Cph, plant phy and Phy-like families. The numbers indicate the amino acid positions within each GAF sequence. Black and grey boxes denote identical and similar residues respectively. The Cys and His residues important for bilin binding are identified by the open and closed arrowheads respectively (see Figure 6). The diamond locates the Pro conserved among Phy-like proteins. Species and sequence designations can be found in the legend of Figure 3.
Figure 3. Phylogenetic analysis of the Phy superfamily based on an alignment of the GAF domain.
The BphP, Fph, Cph, plant phy and Phy-like families are indicated. The sequences were aligned by CLUSTALX and the unrooted phylogenetic tree was generated by the neighbour-joining method and displayed using Treeview. Underline indicates proteins without an obvious HK domain. Asterisks indicate known members of the bathyBphP subfamily. Closed circles denote HK proteins that belong to the HWE group [40]. Genus/species abbreviations shown in the Figure: Agrobacterium tumefaciens (At), Anabaena variabilis (Av), Arabidopsis thaliana (Ath), Aspergillus nidulans (An), Botryotinia fuckeliana (Bf), Bradyrhizobium ORS278 (Br), Calorthrix PCC7601 (Cl), Cochliobolus heterostrophus (Ch), Crocosphaera watsonii (Cw), Cytophaga hutchinsonii (Cyh), Deinococcus radiodurans (Dr), Fremyella diplosiphon (Fd), Gibberella moniliformis (Gm), Gibberella zeae (Gz), Gleobacter violaceus (Gv), Kineococcus radiotolerans (Kr), Methanosarcina acetivorans (Ma), Neurospora crassa (Nc), Nostoc PCC 7120 (No), Nostoc punctiforme (Np), Pseudomonas aeruginosa (Pa), Pseudomonas putida (Pp), Pseudomonas syringae (Ps), Rhizobium leguminosarium (Rl), Rhodospirillum centenum (Rc), Rhodopseudomonas palustris (Rp), Rhodospirilum rubrum (Rr), Synechocystis PCC6803 (Sy), Thermosynechococcus elongatus (Te), Ustilago maydis (Um), Xanthomonas axonopodis (Xa) and Xanthomonas campestris (Xc). EnvSeq-1, -2, -3, -4 and -7 were identified in a collection of genomic fragments randomly sequenced from a marine environmental sequence database [29]. EnvSeq-12 is a random DNA sequence isolated from an acidophilic mine biofilm [30]. Both the N-terminal (N) and C-terminal (C) GAF domains in Synechocystis TaxD1 and M. acetivorans BphP1, and two of the GAF domains (A and C) in Synechocystis Cph2, were included in the comparison.
Closer examination of the GAF sequence for important residues strengthened support for the subgroups based on phylogenetic clustering (Figure 2 and Supplemental Figure 1 at http://www.BiochemJ.org/bj/392/bj3920103add.htm). As reported earlier for a smaller collection [5], members of the Cph, BphP and Fph families almost all have a positionally conserved His (His-260 in D. radiodurans BphP) that is important for bilin binding (see below and [5,38]). All the classified Cph sequences, like higher plant phys, have a Cys immediately proximal to this His that serves as the actual bilin attachment site for these apoproteins [8,20,37]. This Cys is invariably absent from the BphP and Fph families and often is replaced by a small hydrophobic residue {Ile, Leu, Met or Val ([5] and Figure 2)}. As predicted from the phylogenetic tree, the Phy-like sequences displayed a much less robust alignment for their GAF sequences. A significant number of residues, which are nearly invariant within the Cph, BphP and Fph collections, are poorly conserved in this group (Figure 2 and Supplemental Figure 1 at http://www.BiochemJ.org/bj/392/bj3920103add.htm). For example, these Phy-like polypeptides are sometimes missing either or both residues of the Cys/His pair (e.g. S. elongates CikA and F. diplosiphon RcaE) and typically have a substantial deletion proximal to this site. Some are also missing the Tyr that helps maintain the native spectral properties of the chromophore [36]. Of potential significance is the substitution of a Ser and Met/Ile/Leu pair conserved in almost all other Phys with a conserved Pro and Asp/Glu pair (Pro-160 and Glu-192 in F. diplosiphon RcaE; Figure 2 and Supplemental Figure 1 at http://www.BiochemJ.org/bj/392/bj3920103add.htm).
A subfamily of Phys was recently discovered, designated the bathyPhys, that has the unique attribute of preferring the Pfr and not the Pr form as the ground state [25,32]. Studies with two representatives, Agrobacterium tumefaciens BphP2 (or Apg2) and Pseudomonas aeruginosa BphP, showed that they bind BV IXα as Pr but then rapidly convert into Pfr in the dark, thus needing FR and not R for the initial photoconversion [25,39]. Since this photochemical distinction could reflect unique interactions between the bilin and the GAF domain, we expected that the four known bathyPhys would cluster together on a tree based on this motif. Although Ag. tumefaciens BphP2 and Rhodopseudomonas palustris BphP5 were on the same branch, Bradyrhizobium BphP and Ps. aeruginosa BphP were on another (Figure 3), suggesting that sequences outside of the GAF domain influence this unusual spectral property. We observed some clustering of BphPs bearing the distinctive HWE-HK domain [40]. In contrast, those Phys without an obvious HK domain were distributed throughout the tree (Figure 3). Whether such a disperse distribution was generated by independent evolutionary events or by gene transfer from a common progenitor is unknown.
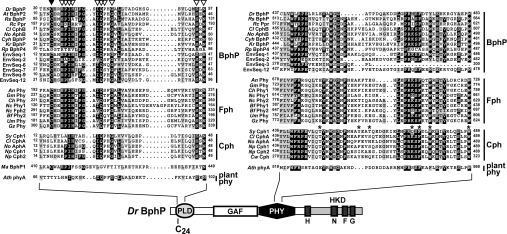
Mutagenic analyses of various Phys have shown that sequences upstream and downstream of the GAF domain are also critical for the spectral integrity of the photoreceptor. The PHY domain is particularly important for proper photoconversion of Pr to Pfr and the stability of Pfr [7–10,36]. Using a more robust alignment that included all 70 representatives, we refined the PHY domain to an approx. 130-amino-acid region which was evident in all members of the plant phy, Cph, BphP and Fph families but was not apparent in any members of Phy-like group (Figure 4 and Supplemental Figure 2 at http://www.BiochemJ.org/bj/392/bj3920103add.htm). It contains a number of islands with strong sequence conservation, especially within the Cph and Fph families, that are likely to participate in domain folding. Random mutagenesis of Synechocystis Cph1 identified several amino acids in the PHY region that influence R/FR photochemistry [36]. Although some mutations affected residues highly conserved within the Phy superfamily (e.g. Phe-475, Trp-478 and Trp-489 in SyCph1), implying that these sites have direct roles, other mutations affected variable positions (e.g. Glu-385, Asn-397 and Thr-461 in SyCph1; see Supplemental Figure 2 at http://www.BiochemJ.org/bj/392/bj3920103add.htm), suggesting that these residues have secondary effects.
Figure 4. Amino acid sequence alignment of portions of PLD (left) and PHY (right) from members of the BphP, Fph, Cph and plant phy clades.
Alignments of the entire domains can be found in Supplemental Figures 2 and 3 (http://www.BiochemJ.org/bj/392/bj3920103add.htm). The sequences were grouped by their proposed inclusion into BphP, Fph, Cph and plant phy families. The position of each domain in the linear Phy sequence of D. radiodurans BphP is shown. Black and grey boxes denote identical and similar residues respectively. The numbers identify the amino acid positions for each sequence. The cysteine in the PLD important for bilin binding is identified by the closed arrowhead (Cys-24 in DrBphP). The open arrowheads identify conserved residues in AtBphP2 that were tested for their importance in BV IXα ligation (see Figure 7). The asterisks locate amino acids in the PHY domain identified as being important for the spectral properties of Synechocystis Cph1 [36]. Species and sequence designations can be found in the legend of Figure 3.
Phylogenetic analysis with the PHY domain generated a similar clustering of the plant phy, Cph, BphP and Fph families to that seen with the GAF domain, supporting further our proposed groupings (results not shown). Given its functional significance, we used the PHY domain as an additional query to search the various microbial databases. Although no new Phys were detected in the databases of known micro-organisms, one additional Phy was found in the shotgun sequence collection from the Sargasso Sea [29]. The partial sequence for EnvSeq-8 harboured a full PHY domain followed by the N-terminal portion of a HK domain, suggesting that it also acts as a photoreceptor in a TC-HK phospho-relay (Figure 1B).
Lamparter and co-workers [26,27] recently described a conserved PLD upstream of the GAF domain in many microbial Phys that participates in bilin binding. Using the PLD as a query, we detected a related sequence in all the BphP and Fph sequences identified above (Figure 4 and Supplemental Figure 3 at http://www.BiochemJ.org/bj/392/bj3920103add.htm). Importantly, all but one also has the positionally conserved Cys (the exception was R. palustris BphP4, whose sequence has not yet been verified). Even R. centenum Ppr, which contains an appended N-terminal photoactive yellow protein motif that binds the chromophore p-hydroxycinnamic acid, has a PLD upstream of its GAF domain (Figure 1A). An obvious but less well-conserved PLD was evident in members of the Cph and plant phy families but this motif was notably missing the conserved Cys (Figure 4 and Supplemental Figure 3 http://www.BiochemJ.org/bj/392/bj3920103add.htm). Nothing resembling a PLD could be detected in most members of the Phy-like clade. The only exception was the possible Phy from the archaebacterium M. acetivorans (MaBphP1), where a Cys-containing PLD was detected upstream of the second GAF domain (Figures 1A and 4). Searches of the environmental databases with a consensus PLD sequence identified three additional Phy-type proteins [EnvSeq-9 to -11 (Figures 1B and 4)]. All three were partial sequences containing only the PLD, which prevented us from unequivocally assigning them to one of the four microbial families. However, the positionally conserved Cys was evident, suggesting that EnvSeq-9 to -11 are related to the BphP/Fph types.
Organization of the Phy operons
Like other sensory systems [41], several elements within the Phy sensory transduction chain are often connected genetically. For example, all the Fphs identified to date have an RR domain appended to the C-terminal end, supporting further their participation in TC-HK cascades ([5,23,25] and the present study). For the bacterial Phys, the gene encoding the photoreceptor apoprotein is often coupled in an operon to other open reading frames encoding factors important for TC-HK signal transmission. For Cphs and BphPs, these include separate RRs and sometimes other HKs (e.g. the Ag. tumefaciens BphP1 operon [25]) that may be part of the phospho-relay. For some BphP operons, an HO gene needed to synthesize BV IXα from haem is included or nearby [5,25,28].
For several of the new Phy sequences reported here, similar genetic connections were evident (Supplemental Figure 4 at http://www.BiochemJ.org/bj/392/bj3920103add.htm). For example, the BphP operon in K. radiotolerans includes the coding region for an HO gene (BphO) downstream of that for the BphP. For the EnvSeq sequences where enough surrounding genomic sequence was available, similar linkages of RR and/or HO genes were observed, with the latter arrangement further supporting their designation as a Phy. The EnvSeq-1 BphP gene is followed by an RR gene (BphR) in the same orientation and nearby is a HK gene in the opposite orientation. Both the EnvSeq-9 and -12 genes are downstream of a likely HO gene (BphO). For EnvSeq-9, the BphP and BphO coding sequences overlap by 4 bp, whereas for EnvSeq-12, the two are immediately adjacent with no intervening DNA sequence. The M. acetivorans Phy-like gene is in a cluster containing a putative transcription factor.
Role of the PHY domain in stabilizing Pfr
Alignments of the GAF, PHY and PLD sequences from plants and a wide array of micro-organisms clearly show that these domains have been conserved through evolution and identify a number of amino acids that may be important for the unique spectral properties and/or folding of this photoreceptor family. To examine these domains further biochemically, we analysed three representative BphPs by deletion or site-directed mutagenesis coupled with bilin-binding and spectroscopic assays. D. radiodurans BphP1 and Ag. tumefaciens BphP1 were chosen to help resolve the site for BV IXα attachment among the BphPs [22,26,27], and Ag. tumefaciens BphP2 was chosen to identify possible differences that discriminate the bathyPhys from the more photochemically conventional forms. Of particular interest was the role of the PHY domain in stabilizing the Pfr form of AtBphP2, given that it assumes this form without light [25]. Wild-type and mutant apoproteins were expressed with C-terminal His6 tags in E. coli, incubated with the bilin, and purified by nickel-chelate affinity chromatography. The recombinant proteins were then assayed for bilin attachment by zinc-induced fluorescence of the holoprotein following SDS/PAGE and for the distinctive R/FR photochromic absorption spectra characteristic of Phy-type pigments.
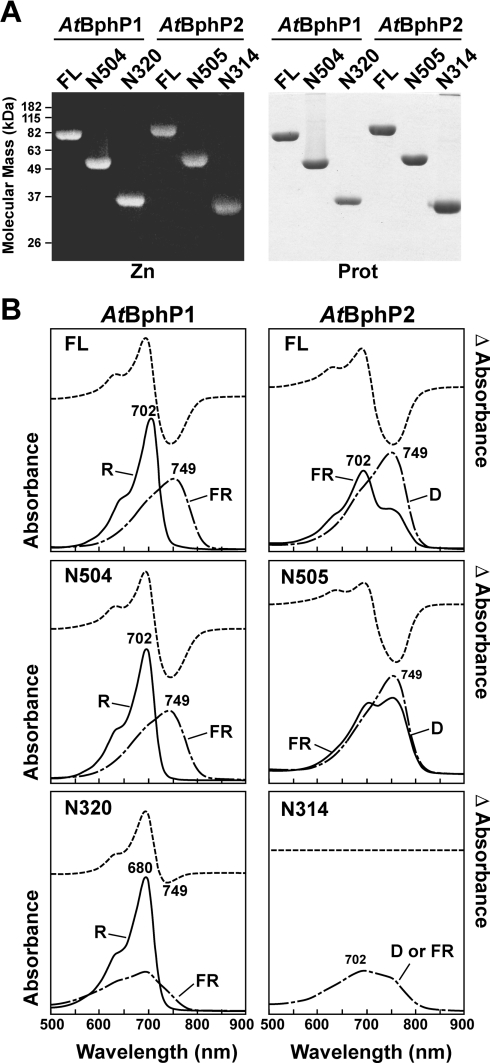
To examine further the role of the PHY domain in stabilizing the Pfr chromophore [8–10,36], we tested a pair of C-terminal truncations bracketing the PHY domain of Ag. tumefaciens BphP1 and -2. The longer set was missing only the C-terminal HK domain [AtBphP1(N504) and AtBphP2(N505), where the numbers indicate the final amino acid of the truncated polypeptide], whereas the shorter set was missing both the HK and the PHY domains [AtBphP1(N320) and AtBphP2(N314)]. As shown in Figure 5(A), both sets of truncations efficiently bound BV IXα (as judged by zinc-induced fluorescence of the bound chromophore following SDS/PAGE), indicating that neither the PHY domain nor the downstream HK domain has a substantial impact on the bilin lyase activity of the apoproteins.
Figure 5. Assembly and spectral properties of full-length (FL) and C-terminally truncated forms of Ag. tumefaciens BphP1 and BphP2.
The numbers indicate the last amino acid of the truncated polypeptides. (A) Covalent binding of BV IXα. The wild-type (WT) and mutant apoproteins were incubated with BV IXα for 1 h, purified by nickel-chelate affinity chromatography and then subjected to SDS/PAGE and either assayed for the bound bilin by zinc-induced fluorescence (Zn) or stained for protein with Coomassie Blue (Prot). (B) Absorption and R/FR (AtBphP1) or FR/D (AtBphP2) difference spectra of samples from (A). The absorption maxima of the Pr and Pfr forms are indicated. The spectra were adjusted for an equal amount of protein as determined by absorbance at 280 nm.
The PHY domain was essential for the photochromicity of both AtBphP1 and AtBphP2 (Figure 5B). For AtBphP1, the R/FR spectral properties of the N504 truncation were indistinguishable from the full-length holoprotein. In contrast, the N320 truncation generated a normal Pr, but saturating R irradiations created a ‘Pfr-like’ bleached state that rapidly reverted to Pr in the dark. A similar effect was seen for an equivalent PHY deletion of D. radiodurans BphP [DrBphP(N321) (see Figure 6)]. For AtBphP2, assembly of a spectrally normal Pfr was possible upon removal of the HK domain (Figure 5B). However, the AtBphP2(N505) truncation inefficiently photoconverted into a highly unstable Pr form in FR. In fact, thermal reversion back to Pfr was so rapid that it prevented accurate measurement of the Pr absorption spectrum, even after saturating FR irradiations (results not shown). Upon removal of the PHY domain, AtBphP2(N314) failed to assume the expected Pfr ground state upon BV IXα binding [25], but instead generated a bleached and red-shifted ‘Pr-like’ form in the dark. This bili-protein was not photochromic with either R or FR. Collectively, the results demonstrate that the PHY domain, although not essential for bilin binding, is important for stabilizing the Pfr form and proper photochromicity, even for a bathyPhy that uses Pfr as the ground state.
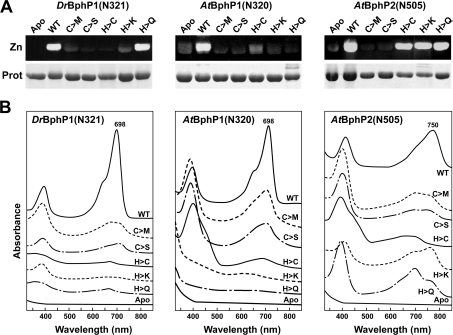
Figure 6. Requirements of the PLD Cys and the GAF His for bilin binding by D. radiodurans BphP, and BphP1 and BphP2 from Ag. tumefaciens.
Codons for the Cys and His residues were converted into those for the indicated amino acids by site-directed mutagenesis of DrBphP(N321), AtBphP1(N320) and AtBphP2(N505). The recombinant wild-type (WT) and mutant polypeptides were incubated with BV IXα for 1 h and purified by nickel-chelate chromatography. (A) Samples were subjected to SDS/PAGE and either assayed for the bound bilin by zinc-induced fluorescence (Zn) or stained for protein with Coomassie Blue (Prot). Apo, apoprotein prior to BV IXα incubation. (B) Absorption spectra of the samples following an extended incubation in darkness. The absorption maxima of the Pr (DrBphP and AtBphP1) and Pfr (AtBphP2) forms are indicated. The absorption spectra were adjusted for an equal amount of protein as determined by absorbance at 280 nm.
Roles of the GAF and PLD regions in chromophore binding
Whereas studies with Ag. tumefaciens BphP1 and Ps. aeruginosa BphP showed that the conserved Cys within the PLD is the bilin attachment site [27,39], a contradictory study by us with D. radiodurans BphP implicated the invariant His within the GAF domain [22]. To help resolve this discrepancy, we examined bilin assembly for a common set of Cys and His mutants in the three BphPs, DrBphP(N321), AtBphP1(N320) and AtBphP2(N505). All three wild-type sequences readily bound BV IXα (as judged by zinc-induced fluorescence of the holoprotein) and generated the expected Pr and Pfr states following a dark incubation (Figures 5, 6A and 6B). When the PLD Cys substitution mutants were tested similarly, none of the three apoproteins effectively assembled with BV IXα (Figure 6A). These Cys→Met or Cys→Ser mutants also displayed little absorption in the R/FR region (Figure 6B), with much of the residual absorption possibly reflecting non-covalent association of the BV IXα [26,42].
Some but not all of the His mutants exhibited a similar defect in BV IXα attachment and photochromicity but these effects were clearly influenced by the apoprotein and the nature of the His substitution (Figures 6A and 6B). For DrBphP(N321), the His→Cys and His→Lys mutants failed to bind BV IXα, whereas the His→Gln mutant effectively bound this bilin. All three His mutants displayed little or no absorption in the R region, an effect that was unexpected for His→Gln variant given its ability to covalently bind BV IXα. The three His mutants of AtBphP1(N320) failed to bind BV IXα well, with none displaying strong absorption in the R region. In contrast, all three His mutants of AtBphP2(N505) efficiently attached BV IXα, but the resulting bili-proteins displayed unusual absorption spectra after assembly. In particular, the spectrum of the His→Lys mutant prior to irradiations more closely resembled that of Pfr, while the spectrum of the His→Gln mutant more closely resembled that of Pr, suggesting that the preferred Pfr ground state of the holoprotein was shifted to Pr by the Gln substitution. Taken together, the data support the proposal by Lamparter et al. [26,27] that the PLD is the bilin-binding site. However, the GAF His also appears to be important, especially in those Phys that prefer Pr as the ground state. The unique ability of AtBphP2(N505) to tolerate various His substitutions, at least with respect to BV IXα ligation, suggests that it could represent another distinguishing feature of the bathyPhy subfamily.
Importance of the PLD
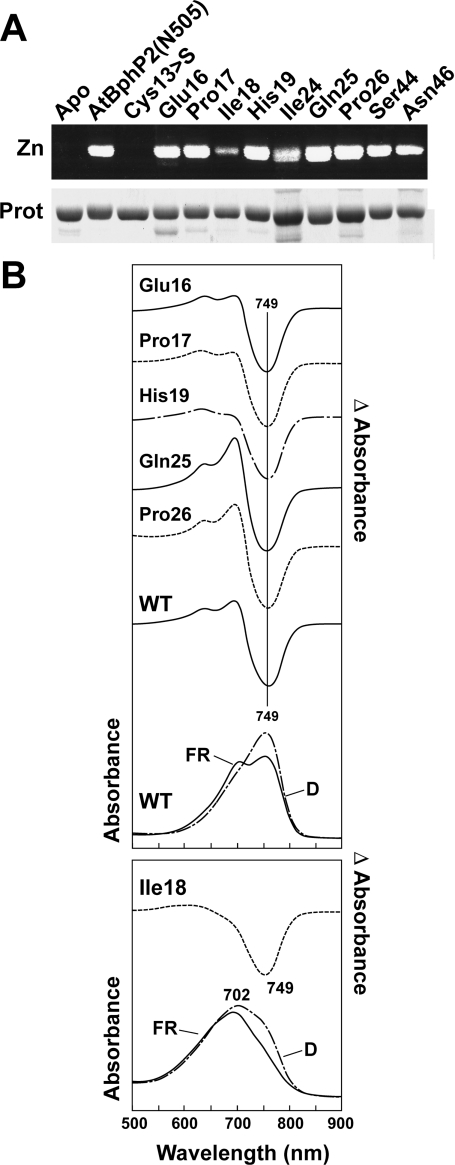
The alignment of the PLD revealed a number of other highly conserved residues besides the Cys that may be important for bilin attachment, R/FR photochromicity, and/or protein folding. In support, recent studies by Zhao et al. [43] showed through deletion analysis that one stretch (IQPHGV; residues 26–31) is essential for the bilin lyase activity of the Nostoc AphA, a member of the Cph family. The Ile in this motif was also identified by Bhoo et al. [38] as being critical for both bilin attachment and photochromicity in pea phyA. In the present paper, we describe the individual testing of the importance of some of these amino acids (Glu-16, Pro-17, Ile-18, His-19, Ile-24, Gln-25, Pro-26, Ser-44 and Asn-46) by alanine substitution mutagenesis of AtBphP2(N505). Most of these residues are conserved not only in the BphP and Fph families, but also in the plant phy and Cph families that do not attach bilins via this domain (Figure 4 and Supplemental Figure 3 at http://www.BiochemJ.org/bj/392/bj3920103add.htm). Many of the PLD mutants were readily expressed in soluble forms. The exceptions were the Ile-24, Ser-44 and Asn-46 mutants; the recombinant apoproteins accumulated mainly in the insoluble fraction. We obtained a small amount of these three mutants for BV IXα-binding assays, but the amounts of holoprotein were insufficient for spectroscopic analyses.
Surprisingly, although the PLD Cys→Ser mutant of AtBphP2(N505) strongly abrogated BV IXα attachment in vitro, only one of the nine other PLD mutants appreciably attenuated covalent binding [Ile-18 (Figure 7A)]. Most of the mutant chromoproteins also displayed relatively normal Pr and Pfr absorption and R/FR difference spectra. This indicates that the association of the bilin to its binding pocket and its photochromicity were not appreciably affected (Figure 7B and results not shown). We observed small changes in the height of the R maximum of the difference spectrum for the His-19, Gln-25, and Pro-26 mutants, which appeared to reflect an increase and decrease, respectively, in the Pr to Pfr dark reversion rate relative to the unmodified chromoprotein. The most significant change was for the Ile-18 mutant, which assumed a mixture of Pr and Pfr at equilibrium in the dark instead of solely being Pfr (Figure 7B). Taken together, the mild consequences of most PLD mutation imply that this domain outside of the bilin-binding Cys has only a minor impact on bilin ligation and Pr/Pfr photochemistry.
Figure 7. Importance of conserved residues within the PLD for BV IXα binding and the spectral properties of Ag. tumefaciens BphP2.
(A) Covalent binding of BV IXα to the wild-type and mutant versions of AtBphP2(N505). The recombinant polypeptide was incubated with BV for 1 h, purified by nickel-chelate affinity chromatography, and then subjected to SDS/PAGE and either assayed for the bound bilin by zinc-induced fluorescence (Zn) or stained for protein with Coomassie Blue (Prot). Apo, apoprotein prior to BV IXα incubation. (B) Absorption and FR/D difference spectra of samples from (A) following an overnight incubation in darkness (D) or saturating irradiation with FR. The absorption maxima of the Pfr forms are indicated.
Importance of the bilin A ring vinyl side chain for binding to Phys
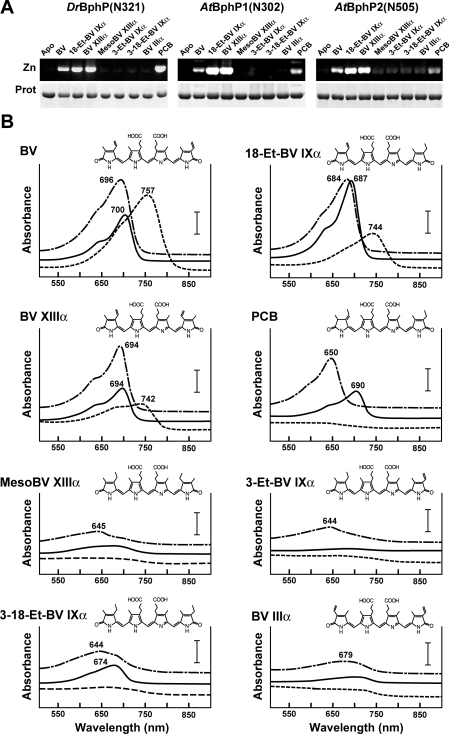
Previous studies showed that plant phys and Ag. tumefaciens BphP1 bind their respective bilins, PΦB and BV IXα, by the ethylidene/vinyl side chains attached to the A-ring C-3 position [26,44,45]. However, both the distinct photochemical properties of bathyBphP AtBphP2 [25] and the discriminating nature of the His mutants in DrBphP1 and AtBphP1 (Figure 7A) suggested that alternative binding sites in the BV IXα were possible, in particular the vinyl side chain of the D-ring. To address this issue, we examined the assembly of DrBphP(N321), AtBphP1(N320) and AtBphP2(N505) with a library of BV IXα derivatives altered at either or both of the A- and D-ring vinyl side chains. The derivatives included: (i) BV XIIIα and 18-Et-BV IXα, which have an A-ring vinyl group at the C-3 position but have the D-ring vinyl at position C-18 exchanged for ethyl and methyl groups respectively; (ii) PCB, which has the A-ring vinyl exchanged for an ethylidene group and an ethyl group connected to the D-ring; (iii) 3-Et-BV IXα and 3-18-Et-BV IXα, which have ethyl instead of vinyl groups at the C-3 or both the C-3 and C-18 positions respectively; (iv) meso-BV XIIIα, which has ethyl groups at the C-3 and C-17 positions; and (iv) BV IIIα, in which the vinyl group of the A-ring is switched to the C-2 position (see Figure 8B for structures).
Figure 8. Assembly and spectral properties of D. radiodurans BphP and Ag. tumefaciens BphP1 and BphP2 following incubation with BV IXα, PCB and various BV IXα derivatives.
The structure of each bilin is shown in (B). (A) Covalent binding of bilins to the apoproteins. The recombinant polypeptides [DrBphP(N321), AtBphP1(N320) and AtBphP2(N505)] were incubated with the bilin for 1 h, purified by nickel-chelate affinity chromatography, and then subjected to SDS/PAGE and either assayed for the bound bilin by zinc-induced fluorescence (Zn) or stained for protein with Coomassie Blue (Prot). Apo, apoprotein without BV IXα incubation. (B) Absorption spectra of the DrBphP(N321) (long dashed lines), AtBphP1(N320) (solid lines) and AtBphP2(N505) (short dashed lines) from (A) following an extended incubation in darkness. The absorbance maxima are indicated. The bar represents 0.18 A, 0.13 A and 0.05 A for DrBphP(N321), AtBphP1(N320) and AtBphP2(N505) respectively.
When incubated with the recombinant apoproteins, we found that the two bilin variants with a vinyl group at the A-ring C-3 position, BV XIIIα and 18-Et-BV IXα, covalently assembled with all three apoproteins like BV IXα, as judged by zinc-induced fluorescence of the resulting holoproteins (Figure 8A). PCB, which has an ethylidene group at this position also bound to the three apoproteins, but the assembly was clearly more robust for DrBphP(N321) as compared with both Ag. tumefaciens apoproteins. In most cases, strong R/FR-absorbing photochromic holoproteins were also generated. For the BV XIIIα- and 18-Et-BV IXα-containing species, the Pr and Pfr absorption spectra resembled those assembled with BV IXα (Figure 8B and results not shown). The PCB-containing DrBphP(N321) and AtBphP1(N320) holoproteins also exhibited the expected Pr and Pfr absorption spectra (with blue-shifted maxima as compared with BV IXα, given the less conjugated π-electron system). However, the PCB-AtBphP2(N505) adduct had little absorbance in the R or FR regions and displayed little or no change in absorption following sequential R/FR irradiations (Figure 8B and results not shown). The remaining four bilins without these C-3 vinyl/ethylidene groups (meso-BV XIIIα, 3-Et-BV IXα, 3-18-Et-BV IXα and BV IIIα) failed to bind covalently with any of the three apoproteins and, as expected, generated little or no R/FR absorption (Figure 8A and B). Together with the common use of a PLD Cys, the common need for the A-ring vinyl side chain among the three BphPs suggests that other members of the BphP family use a similar method to form the thioether linkage with BV IXα.
DISCUSSION
Organization of the Phy superfamily
Analysis of the expanding collection of Phy-type sequences performed by us and others shows that the Phy superfamily includes, in addition to those in plants, a diverse set of photoreceptors present in a wide array of proteobacteria, cyanobacteria and fungi. We can now include over 70 members from 30 known microbial species, with more likely to be found as additional genomic sequencing projects progress. New to the list is a Phy from K. radiotolerans, thus expanding the distribution of Phys to actinobacteria, and a potential Phy-related sequence in M. acetivorans, which may extend Phys to Archaea. Sequence alignments of the Phy collection identified a number of conserved motifs/residues surrounding the bilin-binding pocket. These presumably have important functions in chromophore attachment and the unique photochromic spectral properties of this photoreceptor class. Included is a signature GAF domain that is often preceded by a PLD and followed by a PHY domain, the combination of which appears to be specific for the Phy superfamily. Given the remarkable conservation of the PLD/GAF/PHY domains in species widely separated during evolution, they should be extremely useful in identifying new members of the Phy superfamily. As examples, we discovered 12 potential Phys in two microbial shotgun sequence databases. Taken together, this collection also shows that Phy-type photoreceptors radiated more widely than previously appreciated.
The structural organization of microbial Phys indicates that many, but not all, participate either directly or indirectly in TC-HK phospho-relays. This action is further supported by the presence of an RR domain, either translationally linked to the HK domain of the Phy to create a hybrid HK, or expressed as a separate protein genetically linked to the Phy within a common operon. Instead of initiating one or more TC-HK cascades, some Phys may dampen such phospho-relays. For example, the presence of a PP2C domain in K. radiotolerans BphP could attenuate one or more kinase cascades by releasing the phosphate signal in a light-dependent manner. The involvement of the PP2C-like protein (CpeR) in the complementary chromatic adaptation response of F. diplosiphon suggests that a similar dampening by dephosphorylation acts downstream of the Phy-like protein RcaE [46].
In several cases (Synechocystis Cph2, Rh. sphaeroides BphP and T. elongatus Cph), GGDEF and EAL motifs (both named for a consensus set of amino acids) are downstream of the bilin-binding site instead of a HK domain. These motifs, which are proposed to have diguanylate cyclase and diguanylate phosphodiesterase activities respectively [35], have been also found in a several non-Phy proteins required for motility (e.g. Pseudomonas aeruginosa FimX, Vibrio cholerae RocS, and Caulobacter cresentus PleD [47–49]). PleD for example, appears to directly regulate the flagellum motor through a HK cascade [47]. Consequently, the presence of the GGDEF and EAL motifs raises the possibility that some prokaryotic Phys photoregulate bacterial movement. In support, Synechocystis Cph2 has been shown to modulate taxis towards blue light [19].
Classification of the Phy superfamily into distinct families
Using phylogenetic analysis of the GAF domain coupled with a grouping based on specific biochemical characteristics (e.g. identity of the bilin and its linkage site, and spectral characteristics), we propose the formation of four major microbial Phy subgroups (BphPs, Cphs, Fphs and a collection of Phy-like sequences) that reflect their distribution within the bacterial and fungal kingdoms and their photobiological properties. The Cph family is most similar to plant phys. Like their plant relatives, members of the Cph family appear to bind bilins through the positionally conserved Cys in the GAF domain [17]. Although they have a discernible PLD, the absence of the Cys residue implies a priori that this motif is not directly involved. The BphP and Fph families are most closely related to each other. Members of these families are clearly distinguished from plant phys and Cphs by the absence of the Cys residue within the GAF domain, the use of BV IXα as the chromophore [5], and the probable use of a positionally conserved Cys with the PLD for bilin binding ([27,39] and the present study). Although the data from several representative BphPs support the PLD Cys, we note that a similar role in members of the Fph family awaits testing. Plant phys and probably Cphs bind their PΦB and PCB chromophores through the A-ring ethylidene group [17,50]. Based on our and others' analyses of several BV IXα derivatives, it is likely that members of the BphP and Fph families also bind their bilin via the vinyl group attached to the same A-ring ([26,27,39] and the present study).
Although our first analysis of a BphP implicated a GAF-domain His as the site for covalently attaching the bilin chromophores [22], more recent studies identified a Cys within the PLD [26,27,39]. Here, we provide further support for the PLD Cys using a suite of site-directed mutants affecting both His and Cys residues in three different BphPs. Whereas none of the PLD Cys mutants could bind bilins, some of the GAF His mutants remained active. The difference was especially pronounced for Ag. tumefaciens BphP2, for which three mutants that converted the His into either Cys, Lys or Gln were without effect. In the contexts of D. radiodurans BphP and Ag. tumefaciens BphP1 apoproteins, the GAF domain His is important, suggesting that it may in some situations participate in chromophore ligation. Other than the PLD Cys, most of the other conserved residues near this amino acid of Ag. tumefaciens BphP2 were non-essential. The only apparent exception was Ile-18. Ile-24 was previously identified as critical for chromophore attachment in pea phyA [38,43]; it did not appear to alter the BV IXα-binding ability of AtBphP2(N505) but did impair the stability of the recombinant apoprotein. It is intriguing that plant phy/Cph families and the BphP/Fph families bind spatially close sites in their respective chromophores but use Cys residues located in two different domains (GAF versus PLD). One likely scenario is that the respective PLD and GAF Cys acceptor sites are very close in the three-dimensional structures of BphPs, Fphs, Cphs and plant phys despite being widely separated in the primary sequence.
At present, the molecular explanation for the unusual photochemical properties of the bathyPhys is unknown. Sequence comparisons of their PLD, GAF and PHY domains with those of other BphPs failed to identify motifs/amino acids that could generate the preference for the Pfr ground state. Deletion analyses of AtBphP2 as compared with AtBphP1 and DrBphP showed that this bathyPhy also requires the PHY domain to help stabilize the Pfr form. However, several subtle differences were detected here that could be important distinctions, including the ability of His-250 mutants in AtBphP2 to bind bilins and the failure of AtBphP2 to assemble efficiently with PCB and generate a photochromic photoreceptor. Clearly, additional comparative studies with other bathyBphPs are needed to determine whether these effects are more general.
In addition to the more typical Phys, a collection of Phy-like proteins was evident from the alignments and phylogenetic analyses. Although members of the Phy-like group have the signature Phy-type GAF domain, other features are often missing, including the PHY motif, the PLD and/or the Cys/His residues within the GAF domain. Another discriminator is the presence of an approx. 25-residue deletion just upstream of the Cys in the GAF domain when aligned with members of the BphP, Cph and Fph families. Some of the Phy-like sequences have retained the GAF Cys, suggesting that they have the capacity to bind bilins through this domain. Synechocystis RcaE, for example, can bind bilins covalently but fails to become R/FR-photochromic [51]. While it remains possible that these Phy-like proteins are not photoreceptors, intriguing scenarios are that they (1) act as a bilin sensors and not as light sensors [7], (2) associate with non-bilin chromophores, and/or (3) bind bilin-type pigments but generate markedly different spectral properties. In support of the latter possibility, Yoshihara et al. [52] recently reported that Synechocystis TaxD1 (PixJ1) expressed in vivo associates with a bilin-type chromophore through its GAF Cys, but generates a blue/green-light photoreversible chromoprotein. The substantial blue shift in absorbance of the holoprotein could reflect either the use of a novel less-π-conjugated bilin or distinct interactions between the apoprotein and the bilin within the binding pocket. Given that TaxD1 and at least two other members of the Phy-like clade (PlpA and Cph2) control blue-light photobiology in Synechocystis [18,52,53], it is tempting to speculate that members of the Phy-like clade actually represent a unique class of blue/green light-absorbing bili-proteins.
Predictive value of the Phy families
The proposed Phy families should have value in predicting the behaviour of new, yet-to-be-characterized species. For example, our analysis of N. crassa Phy2 recently confirmed the expectation that this Fph covalently binds BV IXα and generates an R/FR chromoprotein (B. Noh and R. D. Vierstra, unpublished work). Similar to the BphPs studied here from D. radiodurans and Ag. tumefaciens (Figures 5 and 6), its PHY domain is also essential for Pfr stability, such that its removal generates a biliprotein spectroscopically similar to DrBphP(N321) and AtBphP1(N302). Likewise, we predict that the Phy sequence from the actinobacterium K. radiotolerans will have photochemical properties similar to members of the BphP family, given its similar PLD–GAF–PHY domain organization and phylogenetic clustering with other BphPs. We also detected a Phy-type protein in the genome of the archaebacterium M. acetivorans that awaits biochemical characterization. It does not have an obvious PHY domain and neither of its two GAF domains appear to be related to those in the BphP, Fph and Cph families. However, the more C-terminal GAF domain is preceded by a Cys-containing PLD, suggesting that it could bind bilins by a mechanism similar to BphPs/Fphs.
For the EnvSeqs identified here, the presence of a Cys-containing PLD in most (the exception is EnvSeq-3) and their phylogenetic clustering with other BphPs predicts that they are members of the BphP/Fph families. For EnvSeq-9 and -12, this designation is supported further by the coupling of an HO (BphO) gene to the coding region for the apoprotein. It is interesting to note that the Sargasso Sea collection contained almost 800 unique rhodopsin-like photoreceptors [29] but only 11 Phy-type receptors (the present study). Consequently, it appears that retinal-based signalling systems are much more prevalent than Phys among marine micro-organisms.
Evolution of the Phy superfamily
Close analysis of the microbial Phys now provides a potential route for the evolution of the Phy superfamily. Given their widespread presence in a number of photosynthetic and non-photosynthetic proteobacteria, the BphP family probably represents the progenitor for all of the bilin-containing photochromic pigments. The use of BV IXα as the chromophore also offers the simplest way to obtain a linear bilin, requiring just one enzymatic step from the cyclic haem precursor [5]. The BphP precursor may have been an ancient GAF-containing protein, related to FixL [54], that modified its haem-binding pocket to accommodate linear bilins and added a PAS derivative (i.e. PLD) for covalent attachment. An evolving PHY region contributed to the ability to become R/FR-photochromic by helping stabilize the Pfr conformer, while the addition of the HK domain provided a facile way to connect light perception to appropriate signalling networks.
Relatives of the BphPs then emerged during the evolution of the cyanobacterial, actinobacterial and fungal clades. Members of the Fph family probably evolved directly from BphPs and preserved the use of BV IXα as the chromophore. Like other fungal TC-HKs, the Fphs appended the target RR directly to the photoreceptor, presumably to facilitate phospho-relay transmission. At present, Fphs have been conclusively found only in filamentous fungi ([5,23] and the present study), with a preliminary action spectrum for light-induced sporulation suggesting that one is present in the slime mould Physarum polycephalum [15]. Their absence in unicellular fungi suggests that only some fungal phyla selectively retained these photoreceptors or acquired them from bacteria by gene transfer.
For the Cph family, PCB was adopted as the chromophore, which was readily available given its prime use as a photosynthetic accessory pigment [6]. Its blue-shifted Pr absorption spectrum more closely overlaps with chlorophyll thus providing a more accurate measure of photosynthetic wavelengths under competitive conditions. Clearly, the use of PCB as the chromophore would require a way to discriminate PCB from its precursor BV IXα. Both problems may have been overcome by the use of the GAF Cys to attach the chromophore covalently, as opposed to the PLD Cys. One possibility is that the position of the GAF Cys in the three-dimensional structure restricts binding to the ethylidene side chain of PCB instead of the vinyl side chain of BV IXα. Phy-like sequences have only been found in cyanobacteria, suggesting that they evolved from the Cphs through loss of the PLD and PHY motifs and changes within the GAF domain. The variable protein organization of these members implies widely diverse functions for the Phy-like group. The evolution of plant phys remains unclear given the absence of prokaryotic versions with a similar C-terminal architecture (e.g. tandem internal PAS domains), although their derivation from a Cph obtained during chloroplast endosymbiosis is most likely. Plant phys then became nuclear encoded and employed PΦB instead of PCB as the chromophore. The two internal PAS domains were added to help with signal output. The HK domain was also modified to alter or delete its HK activity; in fact, this domain may have been retained solely to promote homodimerization, a common characteristic among bona fide Phy photoreceptors [2,4].
Conclusions
The expanding database of Phy and Phy-like proteins now provides the opportunity to identify common amino acids that help define these unique photochromic photoreceptors. Using this strategy, we identified a number of residues within the PLD, GAF and PHY domains that must be important based on this evolutionary retention. For example, the Tyr residue identified by Fischer and Lagarias [36] as essential for the primary photoprocess of Synechocystis Cph1 is invariant among the BphP, Cph and Fph families and present in many members of the Phy-like clade. The PHY domain in particular appears essential for spectral properties and stability of the Pfr form (the present study, and [7–10,36]). Even for the bathyPhy from Ag. tumefaciens (BphP2) that prefers Pfr as the ground state, deletion of this domain generated a bili-protein that had lost its capacity to generate Pfr and remain in this form following photoconversion by R. Clearly, three-dimensional structures of representative Phys are now needed to confirm the importance of each of the conserved domains and residues identified here.
Online data
Acknowledgments
We thank Drs P.-S. Song, T. MacDonagh, T. Lamparter (Department of Plant Physiology, Freie Universitat, Berlin, Germany) and K. Inomata for supplying the various BV IXα derivatives, and Dr S.-H. Bhoo (Plant Metabolism Research Center, Kyung Hee University, Suwon, Korea) for providing the DrBphP gene. This work was supported by grants from the National Science Foundation (MCB-0424062) and the Research Division of the UW-College of Agriculture and Life Sciences (Hatch) to R.D.V., and a US-Israel BARD postdoctoral fellowship to B.K.
References
- 1.Briggs W. R., Spudich J. L. Weinheim, Germany: Wiley-VCH Verlag GmbH & Co; 2005. Handbook of Photosensory Systems. [Google Scholar]
- 2.Quail P. H. Phytochrome photosensory signalling networks. Nat. Rev. Mol. Cell Biol. 2002;3:85–93. doi: 10.1038/nrm728. [DOI] [PubMed] [Google Scholar]
- 3.Smith H. Phytochromes and light signal perception by plants – an emerging synthesis. Nature (London) 2000;407:585–591. doi: 10.1038/35036500. [DOI] [PubMed] [Google Scholar]
- 4.Vierstra R. D. Cyanophytochromes, bacteriophytochromes, and plant phytochromes: light-regulated kinases related to bacterial two-component regulators. In: Inouya M., Dutta R., editors. Histidine Kinases in Signal Transduction. New York: Academic Press; 2002. pp. 273–295. [Google Scholar]
- 5.Bhoo S. H., Davis S. J., Walker J., Karniol B., Vierstra R. D. Bacteriophytochromes are photochromic histidine kinases using a biliverdin chromophore. Nature (London) 2001;414:776–779. doi: 10.1038/414776a. [DOI] [PubMed] [Google Scholar]
- 6.Frankenberg N., Mukougawa K., Kohchi T., Lagarias J. C. Functional genomic analysis of the HY2 family of ferredoxin-dependent bilin reductases from oxygenic photosynthetic organisms. Plant Cell. 2001;13:965–978. doi: 10.1105/tpc.13.4.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Montgomery B. L., Lagarias J. C. Phytochrome ancestry: sensors of bilins and light. Trends Plant Sci. 2002;7:357–366. doi: 10.1016/s1360-1385(02)02304-x. [DOI] [PubMed] [Google Scholar]
- 8.Wu S. H., Lagarias J. C. Defining the bilin lyase domain: lessons from the extended phytochrome superfamily. Biochemistry. 2000;39:13487–13495. doi: 10.1021/bi001123z. [DOI] [PubMed] [Google Scholar]
- 9.Cherry J. R., Hondred D., Walker J. M., Keller J. M., Hershey H. P., Vierstra R. D. Carboxy-terminal deletion analysis of oat phytochrome A reveals the presence of separate domains required for structure and biological activity. Plant Cell. 1993;5:565–575. doi: 10.1105/tpc.5.5.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oka Y., Matsushita T., Mochizuki N., Suzuki T., Tokutomi S., Nagatani A. Functional analysis of a 450-amino acid N-terminal fragment of phytochrome B in Arabidopsis. Plant Cell. 2004;16:2104–2116. doi: 10.1105/tpc.104.022350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeh K. C., Lagarias J. C. Eukaryotic phytochromes – light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc. Natl. Acad. Sci. U.S.A. 1998;95:13976–13981. doi: 10.1073/pnas.95.23.13976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schneider-Poetsch H. A. Signal transduction by phytochrome: phytochromes have a module related to the transmitter modules of bacterial sensor proteins. Photochem. Photobiol. 1992;56:839–846. doi: 10.1111/j.1751-1097.1992.tb02241.x. [DOI] [PubMed] [Google Scholar]
- 13.Krall L., Reed J. W. The histidine kinase-related domain participates in phytochrome B function but is dispensable. Proc. Natl. Acad. Sci. U.S.A. 2000;97:8169–8174. doi: 10.1073/pnas.140520097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsushita T., Mochizuki N., Nagatani A. Dimers of the N-terminal domain of phytochrome B are functional in the nucleus. Nature (London) 2003;424:571–574. doi: 10.1038/nature01837. [DOI] [PubMed] [Google Scholar]
- 15.Marwan W., Starostzik C. The sequence of regulatory events in the sporulation control network of Physarum polycephalum analysed by time-resolved somatic complementation of mutants. Protist. 2002;153:391–400. doi: 10.1078/14344610260450127. [DOI] [PubMed] [Google Scholar]
- 16.Kehoe D. M., Grossman A. R. Similarity of a chromatic adaptation sensor to phytochrome and ethylene receptors. Science (Washington, D.C.) 1996;273:1409–1412. doi: 10.1126/science.273.5280.1409. [DOI] [PubMed] [Google Scholar]
- 17.Yeh K. C., Wu S. H., Murphy J. T., Lagarias J. C. A cyanobacterial phytochrome two-component light sensory system. Science (Washington, D.C.) 1997;277:1505–1508. doi: 10.1126/science.277.5331.1505. [DOI] [PubMed] [Google Scholar]
- 18.Wilde A., Churin Y., Schubert H., Borner T. Disruption of a Synechocystis sp. PCC 6803 gene with partial similarity to phytochrome genes alters growth under changing light qualities. FEBS Lett. 1997;406:89–92. doi: 10.1016/s0014-5793(97)00248-2. [DOI] [PubMed] [Google Scholar]
- 19.Bhaya D., Takahashi A., Grossman A. R. Light regulation of type IV pilus-dependent motility by chemosensor-like elements in Synechocystis PCC6803. Proc. Natl. Acad. Sci. U.S.A. 2001;98:7540–7545. doi: 10.1073/pnas.131201098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park C. M., Kim J. I., Yang S. S., Kang J. G., Kang J. H., Shim J. Y., Chung Y. H., Park Y. M., Song P. S. A second photochromic bacteriophytochrome from Synechocystis sp PCC 6803: spectral analysis and down-regulation by light. Biochemistry. 2000;39:10840–10847. doi: 10.1021/bi992831r. [DOI] [PubMed] [Google Scholar]
- 21.Hughes J., Lamparter T., Mittmann F., Hartmann E., Gartner W., Wilde A., Borner T. A prokaryotic phytochrome. Nature (London) 1997;386:663. doi: 10.1038/386663a0. [DOI] [PubMed] [Google Scholar]
- 22.Davis S. J., Vener A. V., Vierstra R. D. Bacteriophytochromes: phytochrome-like photoreceptors from nonphotosynthetic eubacteria. Science (Washington, D.C.) 1999;286:2517–2520. doi: 10.1126/science.286.5449.2517. [DOI] [PubMed] [Google Scholar]
- 23.Catlett N. L., Yoder O. C., Turgeon B. G. Whole-genome analysis of two-component signal transduction genes in fungal pathogens. Eukaryotic Cell. 2003;2:1151–1161. doi: 10.1128/EC.2.6.1151-1161.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamparter T., Michael N., Mittmann F., Esteban B. Phytochrome from Agrobacterium tumefaciens has unusual spectral properties and reveals an N-terminal chromophore attachment site. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11628–11633. doi: 10.1073/pnas.152263999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karniol B., Vierstra R. D. The pair of bacteriophytochromes from Agrobacterium tumefaciens are histidine kinases with opposing photobiological properties. Proc. Natl. Acad. Sci. U.S.A. 2003;100:2807–2812. doi: 10.1073/pnas.0437914100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamparter T., Michael N., Caspani O., Miyata T., Shirai K., Inomata K. Biliverdin binds covalently to Agrobacterium phytochrome Agp1 via its ring A vinyl side chain. J. Biol. Chem. 2003;278:33786–33792. doi: 10.1074/jbc.M305563200. [DOI] [PubMed] [Google Scholar]
- 27.Lamparter T., Carrascal M., Michael N., Martinez E., Rottwinkel G., Abian J. The biliverdin chromophore binds covalently to a conserved cysteine residue in the N-terminus of Agrobacterium phytochrome Agp1. Biochemistry. 2004;43:3659–3669. doi: 10.1021/bi035693l. [DOI] [PubMed] [Google Scholar]
- 28.Wegele R., Tasler R., Zeng Y. H., Rivera M., Frankenberg-Dinkel N. The heme oxygenase(s)-phytochrome system of Pseudomonas aeruginosa. J. Biol. Chem. 2004;279:45791–45802. doi: 10.1074/jbc.M408303200. [DOI] [PubMed] [Google Scholar]
- 29.Venter J. C., Remington K., Heidelberg J. F., Halpern A. L., Rusch D., Eisen J. A., Wu D. Y., Paulsen I., Nelson K. E., Nelson W., et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science (Washington, D.C.) 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 30.Tyson G. W., Chapman J., Hugenholtz P., Allen E. E., Ram R. J., Richardson P. M., Solovyev V. V., Rubin E. M., Rokhsar D. S., Banfield J. F. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature (London) 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- 31.Aravind L., Ponting C. P. The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem. Sci. 1997;22:458–459. doi: 10.1016/s0968-0004(97)01148-1. [DOI] [PubMed] [Google Scholar]
- 32.Giraud E., Fardoux J., Fourrier N., Hannibal L., Genty B., Bouyer P., Dreyfus B., Vermeglio A. Bacteriophytochrome controls photosystem synthesis in anoxygenic bacteria. Nature (London) 2002;417:202–205. doi: 10.1038/417202a. [DOI] [PubMed] [Google Scholar]
- 33.Herdman M., Coursin T., Rippka R., Houmard J., Tandeau de Marsac N. A new appraisal of the prokaryotic origin of eukaryotic phytochromes. J. Mol. Evol. 2000;51:205–213. doi: 10.1007/s002390010082. [DOI] [PubMed] [Google Scholar]
- 34.Schmitz O., Katayama M., Williams S. B., Kondo T., Golden S. S. CikA, a bacteriophytochrome that resets the cyanobacterial circadian clock. Science (Washington, D.C.) 2000;289:765–768. doi: 10.1126/science.289.5480.765. [DOI] [PubMed] [Google Scholar]
- 35.Galperin M. Y., Nikolskaya A. N., Koonin E. V. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 2001;203:11–21. doi: 10.1111/j.1574-6968.2001.tb10814.x. [DOI] [PubMed] [Google Scholar]
- 36.Fischer A. J., Lagarias J. C. Harnessing phytochrome's glowing potential. Proc. Natl. Acad. Sci. U.S.A. 2004;101:17334–17339. doi: 10.1073/pnas.0407645101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jorissen H., Quest B., Remberg A., Coursin T., Braslavsky S. E., Schaffner K., de Marsac N. T., Gartner W. Two independent, light-sensing two-component systems in a filamentous cyanobacterium. Eur. J. Biochem. 2002;269:2662–2671. doi: 10.1046/j.1432-1033.2002.02928.x. [DOI] [PubMed] [Google Scholar]
- 38.Bhoo S. H., Hirano T., Jeong H. Y., Lee J. G., Furuya M., Song P. S. Phytochrome photochromism probed by site-directed mutations and chromophore esterification. J. Am. Chem. Soc. 1997;119:11717–11718. [Google Scholar]
- 39.Talser R., Moises T., Frankenberg-Dinkel N. Biochemcial and spectroscopic characterization of the bacterial phytochrome of Pseudomonas aeroginosa. FEBS J. 2005;272:1927–1936. doi: 10.1111/j.1742-4658.2005.04623.x. [DOI] [PubMed] [Google Scholar]
- 40.Karniol B., Vierstra R. D. The HWE histidine kinases, a new family of bacterial two-component sensor kinases with potentially diverse roles in environmental signaling. J. Bacteriol. 2004;186:445–453. doi: 10.1128/JB.186.2.445-453.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Inouye M., Dutta R. New York: Academic Press; 2003. Histidine Kinases in Signal Transmission. [Google Scholar]
- 42.Jorissen H., Quest B., Lindner I., de Marsac N. T., Gartner W. Phytochromes with noncovalently bound chromophores: The ability of apophytochromes to direct tetrapyrrole photoisomerization. Photochem. Photobiol. 2002;75:554–559. doi: 10.1562/0031-8655(2002)075<0554:pwnbct>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 43.Zhao K. H., Ran Y., Li M., Sun Y. N., Zhou M., Storf M., Kupka M., Bohm S., Bubenzer C., Scheer H. Photochromic biliproteins from the cyanobacterium Anabaena sp PCC 7120: Lyase activities, chromophore exchange, and photochromism in phytochrome AphA. Biochemistry. 2004;43:11576–11588. doi: 10.1021/bi0491548. [DOI] [PubMed] [Google Scholar]
- 44.Hanzawa H., Shinomura T., Inomata K., Kakiuchi T., Kinoshita H., Wada K., Furuya M. Structural requirement of bilin chromophore for the photosensory specificity of phytochromes A and B. Proc. Natl. Acad. Sci. U.S.A. 2002;99:4725–4729. doi: 10.1073/pnas.062713399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hanzawa H., Inomata K., Kinoshita H., Kakiuchi T., Jayasundera K. P., Sawamoto D., Ohta A., Uchida K., Wada K., Furuya M. In vitro assembly of phytochrome B apoprotein with synthetic analogs of the phytochrome chromophore. Proc. Natl. Acad. Sci. U.S.A. 2001;98:3612–3617. doi: 10.1073/pnas.051629698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seib L. O., Kehoe D. M. A turquoise mutant genetically separates expression of genes encoding phycoerythrin and its associated linker peptides. J. Bacteriol. 2002;184:962–970. doi: 10.1128/jb.184.4.962-970.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aldridge P., Paul R., Goymer P., Rainey P., Jenal U. Role of the GGDEF regulator PleD in polar development of Caulobacter crescentus. Mol. Microbiol. 2003;47:1695–1708. doi: 10.1046/j.1365-2958.2003.03401.x. [DOI] [PubMed] [Google Scholar]
- 48.Huang B. X., Whitchurch C. B., Mattick J. S. FimX, a multidomain protein connecting environmental signals to twitching motility in Pseudomonas aeruginosa. J. Bacteriol. 2003;185:7068–7076. doi: 10.1128/JB.185.24.7068-7076.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rashid M. H., Rajanna C., All A., Karaolis D. K. R. Identification of genes involved in the switch between the smooth and rugose phenotypes of Vibrio cholerae. FEMS Microbiol. Lett. 2003;227:113–119. doi: 10.1016/S0378-1097(03)00657-8. [DOI] [PubMed] [Google Scholar]
- 50.Lagarias J. C., Rapoport H. Chromopeptides from phytochrome. The structure and linkage of the Pr form of the phytochrome chromophore. J. Am. Chem. Soc. 1980;102:4821–4829. [Google Scholar]
- 51.Terauchi K., Montgomery B. L., Grossman A. R., Lagarias J. C., Kehoe D. M. RcaE is a complementary chromatic adaptation photoreceptor required for green and red light responsiveness. Mol. Microbiol. 2004;51:567–577. doi: 10.1046/j.1365-2958.2003.03853.x. [DOI] [PubMed] [Google Scholar]
- 52.Yoshihara S., Katayama M., Geng X. X., Ikeuchi M. Cyanobacterial phytochrome-like PixJ1 holoprotein shows novel reversible photoconversion between blue- and green-absorbing forms. Plant Cell Physiol. 2004;45:1729–1737. doi: 10.1093/pcp/pch214. [DOI] [PubMed] [Google Scholar]
- 53.Wilde A., Fiedler B., Borner T. The cyanobacterial phytochrome Cph2 inhibits phototaxis towards blue light. Mol. Microbiol. 2002;44:981–988. doi: 10.1046/j.1365-2958.2002.02923.x. [DOI] [PubMed] [Google Scholar]
- 54.Gong W. M., Hao B., Mansy S. S., Gonzalez G., Gilles-Gonzalez M. A., Chan M. K. Structure of a biological oxygen sensor: A new mechanism for heme-driven signal transduction. Proc. Natl. Acad. Sci. U.S.A. 1998;95:15177–15182. doi: 10.1073/pnas.95.26.15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.