Abstract
HSF1 (heat-shock factor 1) plays an essential role in mediating the appropriate cellular response to diverse forms of physiological stresses. However, it is not clear how HSF1 is regulated by interacting proteins under normal and stressful conditions. In the present study, Hsc70 (heat-shock cognate 70) was identified as a HSF1-interacting protein using the TAP (tandem affinity purification) system and MS. HSF1 can interact with Hsc70 in vivo and directly in vitro. Interestingly, Hsc70 is required for the regulation of HSF1 during heat stress and subsequent target gene expression in mammalian cells. Moreover, cells transfected with siRNAs (small interfering RNAs) targeted to Hsc70 showed greatly decreased HSF1 activation with expression of HSF1 target genes being dramatically reduced. Finally, loss of Hsc70 expression in cells resulted in an increase in stress-induced apoptosis. These results indicate that Hsc70 is a necessary and critical regulator of HSF1 activities.
Keywords: chaperone, heat-shock cognate 70 (Hsc70), heat-shock factor 1 (HSF1), heat-shock protein 70 (Hsp70), stress response
Abbreviations: EGS, ethylene glycol bis(succinimidyl succinate); EMSA, electrophoretic mobility-shift assay; GST, glutathione S-transferase; HEK, human embryonic kidney; Hsc70, heat-shock cognate 70; HSE, heat-shock element; HSF, heat-shock factor; HSF1 (FL), full-length HSF1; Hsp, heat-shock protein; MALDI, matrix-assisted laser-desorption ionization; mEF, mouse embryonic fibroblast; PSD, post-source decay; RNAi, RNA interference; siRNA, small interfering RNA; TAP, tandem affinity purification; TEV, tobacco etch virus; TOF, time of flight; WT, wild type
INTRODUCTION
Various physiological and cellular stresses can disrupt essential cellular signalling pathways and inhibit protein synthesis leading to dramatic increases in the levels of unfolded proteins, disruption of the cytoskeleton and loss of mitochondrial function [1,2]. The cellular stress response induces the expression of highly conserved Hsps (heat-shock proteins) that serve as molecular chaperones to accelerate refolding of damaged proteins and protect native proteins from unfolding under stressful conditions [3–5].
Stress-induced expression of Hsp genes is carried out by the actions of the HSFs (heat-shock factors), a family of transcription factors remarkably conserved from yeast to human. In baker's yeast Saccharomyces cerevisiae, a single HSF gene is essential for cell viability under all conditions and is required for both basal and stress induced transcription of Hsp genes [6,7]. Mammals have multiple distinct HSF genes, encoding isoforms denoted HSF1, HSF2 and HSF4. HSF1 is the predominant HSF isoform that responds to thermal and oxidative stress to activate the expression of Hsp genes [7,8]. In unstressed cells, HSF1 largely localizes in the cytoplasm as an inactive monomer. In response to cellular stress, HSF1 undergoes the transition from a monomer to a homotrimer form and localizes to the nucleus, where it acquires DNA-binding and transactivation activity [8,9].
Interacting proteins play a crucial role in the HSF1 regulatory pathway. Under normal physiological conditions, HSF1 activation is repressed by multichaperone complexes consisting of Hsps [10]. Recent reports demonstrated that HSF1 is negatively regulated by Hsp90 or Hsp90 multiprotein complexes [11–14]. In response to cellular stress, denatured proteins accumulate and compete with HSF1 for binding to the Hsp90 complex. In this situation, the Hsp90–HSF1 complex is interrupted, and free HSF1 is activated [11–14]. Hsp70 has also emerged as a key factor for HSF1 regulation. Several studies showed that Hsp70 interacts directly with HSF1 and attenuates heat-shock response [15–18]. Also, HSBP1 (heat-shock-factor-binding protein 1) has been reported to interact with the HSF1 trimer together with Hsp70 in order to suppress HSF1 transcriptional activity [19]. Clearly, there are a number of distinct steps in the HSF1 activation pathway that involve many molecular participants. Although several HSF1 regulatory proteins have been identified, the molecular mechanism of HSF1 activation remains to be elucidated.
In the present paper, we identified Hsc70 (heat-shock cognate 70) as an HSF1-interacting protein by the TAP (tandem affinity purification) system followed by MS. Although it has been reported that HSF1 and Hsc70 associate in high-molecular-mass complexes in the cytoplasm of unstressed cells, the functional significance of this interaction has not been characterized [20]. We demonstrate that the C-terminal region of HSF1 interacts directly with the substrate-binding domain of Hsc70. Interestingly, the complex of HSF1 and Hsc70 is co-localized into the nucleus after heat-shock treatment. We also show that Hsc70 is required for the trimerization of HSF1 and HSF1-mediated gene expression in response to heat shock. Collectively, the present study demonstrates a previously unreported functional role of Hsc70 for HSF1 activation.
MATERIALS AND METHODS
Cell culture, transfection and heat-shock treatment
HEK-293 (human embryonic kidney) and mEF (mouse embryonic fibroblast) cells were cultured at 37 °C in a humidified atmosphere with 5% CO2. DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% (v/v) foetal bovine serum was used as a growth medium for all cell lines. mEF cells derived from congenic WT (wild-type) mice and hsf1−/− mice were a gift from Dr Ivor Benjamin (Southwestern Medical Center, University of Texas, Dallas, TX, U.S.A.). Cells were transfected using the FuGENE 6 reagent (Roche Molecular Biochemicals) according to the manufacturer's instructions. For the heat-shock treatment, culture plates were wrapped with Parafilm™ and immersed in the water bath for the times and temperatures specified in the Figure legends.
Plasmids
Vectors for the expression of bacterial recombinant GST (glutathione S-transferase)-tagged full-length HSF1 [HSF1 (FL)] and C-terminal deleted HSF1 [HSF1-(1–290)] fusion proteins have been described in [21]. The open reading frame of Hsc70 (HSPA8, 1941 nucleotides) corresponding to GenBank® accession number BC016179 was amplified by PCR using human skeletal muscle cDNA as a template (Clontech). A vector for the expression of bacterial recombinant His6-tagged Hsc70 was created using the pET21a vector (Stratagene). The pET-Hsc70 vector was created by inserting a DNA fragment containing the Hsc70 open reading frame without a stop codon into the 5′ BamHI and 3′ XhoI sites of pET21a in-frame with the His6 tag. Mammalian expression vectors for C-terminally TAP-tagged HSF1 (FL) and C-terminal-deleted HSF1 (amino acids 1–290) proteins were created by inserting cDNA fragments into the 5′ KpnI and 3′ NotI sites of pCDNA3.1-TAP. Vectors for the expression of C-terminally TAP-tagged full-length Hsc70 and C-terminal-deleted Hsc70 proteins (amino acids 1–386 and 1–510) in mammalian cells were created by inserting cDNA fragments encoding each of these proteins into the 5′ KpnI and 3′ NotI sites of pCDNA3.1-TAP. Mammalian expression vector for N-terminally FLAG-tagged Hsc70 protein was created by inserting the cDNA fragment containing the complete open reading frame into the 5′ KpnI and 3′ XbaI sites of pCDNA3.1-NF.
TAP purification
HEK-293 cells were transfected with plasmids that expressed C-terminally TAP-tagged HSF1 (FL) or HSF1-(1–290) respectively. HSF1 (FL)–TAP and HSF1-(1–290)–TAP fusion proteins were purified according to the TAP-tagged protein purification procedure [22,23], with some modifications. Briefly, 36 h after transfection, cells were washed twice with cold PBS and lysed by sonication. Cell lysates were cleared by centrifugation at 10000 g for 10 min. The supernatants were allowed to bind IgG–Sepharose for 2 h at 4 °C using a Bio-Rad disposable column. The IgG–Sepharose column was washed with TNP buffer (10 mM Tris/HCl, pH 8.0, 150 mM NaCl and 0.1% Nonidet P40) lacking protease inhibitors. TEV (tobacco etch virus) cleavage was performed using rTEV protease (Amersham Biosciences) according to the manufacturer's instruction. TEV-cleaved proteins were transferred to the column containing calmodulin beads (Amersham Biosciences) and incubated for 2 h at 4 °C. After the beads were washed with calmodulin-binding buffer, the bound proteins were boiled in SDS/PAGE sample buffer.
MS
The gel-separated proteins of interest were excised and in-gel digested with trypsin as described in [24]. In-gel digestion was also performed on gel pieces that were excised from the similar mobility region from the untransfected control. The pool of tryptic peptides from the samples was analysed by MALDI (matrix-assisted laser-desorption ionization)–TOF (time-of-flight) MS in linear positive mode to generate a peptide mass map using α-cyano-4-hydroxycinnaminic acid (saturated solution in 50% acetonitrile with 0.1% trifluoroacetic acid) as the UV-absorbing matrix. MALDI–TOF MS was performed on a Voyager-DE Pro time-of-flight instrument (Applied Biosystems), equipped with a nitrogen laser operating at 337 nm. All mass spectra were externally calibrated with bradykinin and insulin and internally calibrated with trypsin autolysis peaks. The peptide mass values were used to search a non-redundant database (NCBInr) using the software tool MS-Digest. Selected peptides were subjected to PSD (post-source decay) analysis to obtain partial peptide sequencing (MS/MS) information. MALDI–PSD analysis was performed as described in [24].
Immunoblotting
HEK-293 cells were transfected with either pCDNA3.1-HSF1-TAP or pCDNA3.1-FLAG-Hsc70 mammalian expression vector. At 48 h after transfection, whole cell lysates were prepared as described previously [25]. Proteins were resolved by SDS/PAGE (12% gels) and immunoblotted using the following antibodies: anti-Hsp70 (Santa Cruz Biotechnology), anti-actin (C-11, Santa Cruz Biotechnology), anti-Hsc70 (sc-24, Santa Cruz Biotechnology), anti-Hsp27 (SPA-815, StressGen), and anti-HSF1 (a gift from Dr Carl Wu, National Institutes of Health, Bethesda, MD, U.S.A.).
In vitro HSF1 cross-linking analysis
For cross-linking experiments, whole-cell extracts were prepared by thawing frozen cell pellets in HEG buffer [20 mM Hepes, pH 7.9, 0.5 mM EDTA, 10% (v/v) glycerol containing 0.42 M NaCl, 1.5 mM MgCl2 and protease inhibitor cocktail (Roche Molecular Biochemicals)]. Cells were dispersed by repeated pipetting, and were incubated on ice for 15 min. Cell extracts were cleared by centrifugation at 12000 g for 15 min at 4 °C, and supernatants (30 μg of protein) were used for HSF1 trimerization studies. EGS [ethylene glycol bis(succinimidyl succinate)] cross-linking was carried out as described previously [26]. EGS was added to the final concentrations specified in Figure legends and incubated at room temperature (25 °C) for 30 min. After quenching the cross-linking reactions with excess 1 M Tris/HCl, pH 7.5, samples were resolved by SDS/PAGE (6% gels) and analysed by immunoblotting using anti-HSF1 antibody.
EMSA (electrophoretic mobility-shift assay)
Nuclear extracts were prepared from heat-shock-treated HEK-293 cells as described previously [25]. Extract (5 μg) was incubated with 32P-labelled HSE (heat-shock element) consensus sequence oligonucleotide for 15 min at room temperature in binding buffer [20 mM Hepes, pH 7.6, 5 mM EDTA, 1 mM dithiothreitol, 150 mM KCl, 50 mM (NH4)2SO4 and 1% (v/v) Tween-20]. Following native 5% PAGE, HSF1–HSE DNA complexes were visualized by autoradiography.
siRNA (small interfering RNA) experiment
siRNA was carried out as described previously [27,28]. A 19-nucleotide double-stranded siRNA was generated against Hsc70 using oligonucleotide (5′-CAGCACGGAAAAGTCGAGA-3′) and inserted into the 5′ XhoI and 3′ XbaI sites of the pSuppressorNeo vector (Imgenex). HEK-293 cells were transfected with RNAi (RNA interference) expression vector for Hsc70 (pSuppres-sorNeo-Hsc70) using FuGENE 6 reagent according to the manufacturer's instructions. Cells were harvested 48 h after transfection. Cell lysates were separated by SDS/PAGE (10% gels) and analysed by immunoblotting as described above.
Apoptosis measurement
Apoptotic cells were assayed using annexin V–FLUOS and propidium iodide (Roche Molecular Biochemicals) and visualized using a Nikon Eclipse E800 automated fluorescent microscope equipped with a digital camera or analysed by flow cytometry. Apoptosis was quantified using a cell death detection ELISA kit (Roche Molecular Biochemicals). Relative apoptosis correlates with absorption at 405 nm with a reference wavelength of 490 nm according to the manufacturer's instructions.
RESULTS
Identification of HSF1-interacting proteins
To investigate the regulatory mechanism of HSF1, we searched for potential proteins that interact with HSF1 using the TAP system. HEK-293 cells were transfected with C-terminally TAP-tagged HSF1 mammalian expression vector (pCDNA3.1-HSF1-TAP), and proteins associated with HSF1 were purified by the two-step affinity purification method as described in the Materials and methods section. The proteins present in the eluted fraction were concentrated and resolved on an SDS/12% polyacrylamide gel and visualized by Coomassie Blue staining. Interestingly, two pro-tein bands of approx. 70 kDa showed a strong interaction with HSF1 (Figure 1A). To identify these two proteins, MALDI–TOF MS was performed as described in the Materials and methods section. The corresponding mobility region of the vector control lane was also used as a negative control. Peptide values unique to the sample were used to search the NCBInr database. Also, MALDI–PSD was employed to obtain partial sequence information on prominent peptides that were detected in the MALDI–TOF spectrum. The peptide mass fingerprint data, along with the MALDI–PSD data, identified Hsc70 (Figure 1A, upper band) and Hsp70 (Figure 1A, lower band) as the proteins interacting specifically with HSF1. Because of the high sequence similarity between Hsc70 and Hsp70, the MALDI–PSD MS/MS data was essential to obtain unique sequence information for unambiguous identification (Figure 1B).
Figure 1. Identification of HSF1-interacting proteins.
(A) HEK-293 cells were transfected with either pCDNA3.1-TAP (C) or pCDNA3.1-HSF1-TAP (HSF1) expression vector. At 48 h after transfection, TAP-tagged HSF1 and its associated proteins were purified using the TAP system. Co-purified proteins were separated by SDS/PAGE, and the gel was stained with Coomassie Blue. Two bands excised for MALDI–TOF MS analysis are marked with an asterisk (*). Molecular-mass sizes are indicated in kDa. (B) MALDI–TOF MS of the tryptic digests of Hsc70. Shown in bold are peptide masses unique to p71 (upper band in A) that were sequenced by MALDI–PSD (labelled MS/MS). (C) HEK-293 cells were transfected with TAP-tagged HSF1-(1–290), TAP-tagged HSF1 (FL) expression vectors or empty vector control (C). At 48 h after transfection, cell extracts were subjected to immunoprecipitation using the TAP system, and endogenous Hsc70 was detected by Western blot (WB) analysis using anti-Hsc70 antibodies. (D) Purified GST-tagged bacterial recombinant HSF1-(1–290) or HSF1 (FL) (1 μg) were incubated with equimolar His6-tagged bacterial recombinant Hsc70. HSF1 was pulled-down using glutathione–agarose beads. Co-precipitated proteins were resolved by SDS/PAGE and detected by Western blot (WB) analysis using anti-His antibodies. C, empty vector control. (E) HEK-293 cells were transfected with TAP-tagged HSF1 (FL) expression vector. At 48 h after transfection, cells were untreated (−) or heat-shocked at 42 °C (+) for 1 h. Cell extracts were subjected to immunoprecipitation using the TAP system, and endogenous Hsc70 was detected by Western blot analysis using anti-Hsc70 antibodies.
Since the Hsp70 interaction with HSF1 has been well characterized, we focused on the interaction between HSF1 and Hsc70. To confirm the interaction between HSF1 and Hsc70, HEK-293 cells were transfected with an expression vector for TAP-tagged HSF1 (pCDNA3.1-HSF1-TAP). Western blot analysis using antibodies specific for Hsc70 confirmed the interaction between HSF1 and endogenous Hsc70 (Figure 1C, right-hand panel). Interestingly, HSF1-(1–290), encompassing the N-terminal DNA-binding domain and coiled-coil motif, did not interact with Hsc70. These results suggest that the interaction between HSF1 and Hsc70 is mediated by the C-terminal region of HSF1 which includes its transactivation domain.
To investigate whether the interaction between HSF1 and Hsc70 is direct, we examined the binding in vitro using GST pull-down assays. GST-tagged HSF1 (FL) and HSF1-(1–290) were expressed in bacteria and purified. Bacterial recombinant His6-tagged Hsc70 was also purified. As shown in Figure 1(D), Hsc70 specifically interacts with HSF1 (FL). However, HSF1-(1–290) did not show any interaction with Hsc70, confirming that the C-terminal domain of HSF1 is required for binding with Hsc70.
We next examined whether the interaction between HSF1 and Hsc70 is maintained during heat stress. HEK-293 cells were transfected with the TAP-tagged HSF1 expression vector (pCDNA3.1-HSF1-TAP). At 48 h after transfection, cells were heat shocked for 1 h at 42 °C. Western blot analysis using anti-Hsc70 antibody showed that the interaction between HSF1 and Hsc70 was not affected by heat stress (Figure 1E).
Hsc70 interacts with HSF1 through its C-terminal region
Hsc70 consists of three domains (Figure 2A): an N-terminal ATPase domain (amino acids 1–383), a proximal substrate-binding domain (amino acids 384–612), containing a coiled-coil domain, and a domain of unknown function (amino acids 613–646), which includes the binding site for the cofactor Hsp40. To identify the region of Hsc70 that is necessary for the interaction with HSF1, we constructed several TAP-tagged Hsc70 deletion mutants and examined their ability to interact with endogenous HSF1 in HEK-293 cells. As shown in Figure 2B (upper panel), HSF1 interacts with full-length Hsc70, but not with Hsc70 (amino acids 1–386) (which contains the ATPase domain only) and Hsc70 (amino acids 1–510) (which contains the ATPase domain and substrate-binding domain minus the coiled-coil domain). These results suggest that the C-terminal domain of the Hsc70 is necessary for its interaction with HSF1.
Figure 2. Hsc70 interacts with HSF1 through its C-terminal region.
(A) Schematic diagram of Hsc70 domain organization. Hsc70 is composed of an N-terminal ATPase domain, a central substrate-binding domain and a C-terminal domain of unknown function. The substrate-binding domain contains a coiled-coil domain that is likely to facilitate protein–protein interactions. Domain boundaries were obtained from the SMART and COIL programs. (B) HEK-293 cells were transfected with the indicated TAP-tagged expression vectors or empty vector control (C). At 48 h after transfection, TAP-tagged Hsc70 was purified, and co-precipitated endogenous HSF1 was analysed by Western blot analysis using anti-HSF1 antibodies (upper panel). The expression level of proteins in the transfected cells was monitored by Western blot analysis using anti-Hsc70 antibodies (lower panel). Molecular-mass sizes are given in kDa.
Hsc70 induces HSF1 trimerization and DNA-binding activity in vivo
Under non-stressful conditions, HSF1 is found predominantly as a cytoplasmic monomer that lacks specific DNA-binding activity. When cells are stressed, HSF1 homotrimerizes, acquires DNA-binding activity, translocates from the cytoplasm to the nucleus, is hyperphosphorylated and becomes transcriptionally competent. The finding that HSF1 interacts with Hsc70 raises the possibility that Hsc70 may regulate the activation of HSF1.
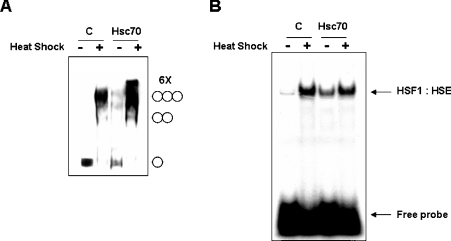
To examine whether the Hsc70 can modulate the HSF1 trimerization, cross-linking experiments were performed using whole-cell extracts prepared from HEK-293 cells that expressed FLAG-tagged Hsc70. As shown in Figure 3(A), heat shock induces HSF1 trimerization in control cells. Interestingly, overexpression of Hsc70 increased HSF1 trimerization further under the heat shock treatment (Figure 3A). Furthermore, while HSF1 exists as a monomer in non-heat-shocked cells, HSF1 trimerization can be moderately induced in the absence of heat shock when Hsc70 was overexpressed (Figure 3A). We next examined the potential role of Hsc70 in stress-inducible HSF1 DNA-binding activity. HEK-293 cells were transfected with FLAG-tagged Hsc70 expression vector (pCDNA3.1-FLAG-Hsc70), and the DNA-binding activity of HSF1 was examined by EMSA. As shown in Figure 3(B), overexpression of Hsc70 results in a significant induction of HSF1 DNA-binding activity in the absence of heat shock. Collectively, these results demonstrate that the Hsc70–HSF1 interaction may positively regulate HSF1 activation.
Figure 3. Hsc70 affects HSF1-trimerization and DNA-binding activities.
HEK-293 cells were transfected with either pCDNA3.1 (C) or pCDNA3.1-FLAG-Hsc70 mammalian expression vector. At 48 h after transfection, cells were untreated (−) or heat shocked at 42 °C (+) for 1 h. (A) Whole-cell extracts (20 μg) were subjected to in vitro cross-linking experiment with 2 mM EGS. HSF1 was detected by Western blot analysis using anti-HSF1 antibodies. The positions of HSF1 monomers, dimers, trimers and putative hexamers (6×) are shown on the right. (B) Nuclear extracts were prepared, and HSF1 DNA-binding activity was measured by EMSA using 32P-labelled HSF1 DNA-binding fragments known as HSEs. The position of the HSF1–HSE complex and the free HSE DNA fragments are shown on the right. C, empty vector control.
Hsc70 is localized to the nucleus upon heat shock
The distribution of endogenous Hsc70 was examined using WT and hsf1−/− mEF cells. To assess the distribution of Hsc70, cellular lysates were fractionated, and Western blot analysis was performed before and after exposure to heat shock. Under the non-stress conditions, both HSF1 and Hsc70 were predominantly localized in the cytoplasm of WT mEF cells (Figure 4A). Upon heat shock, endogenous HSF1 and Hsc70 redistributed to the nucleus (Figure 4A). Immunoblotting using the same extracts assured that the c-fos localized in the nucleus, whereas actin remained largely in the cytoplasm under both stressed and non-stressed conditions (Figure 4A). In contrast, Hsc70 remained largely in the cytoplasm upon heat shock in hsf1−/− mEF cells, suggesting that Hsc70 co-localizes with HSF1 to the nucleus under heat-stressed conditions (Figure 4B). We examined further the subcellular distribution of HSF1 and Hsc70 using fluorescence microscopy. hsf1−/− mEF cells were co-transfected with pEGFP-HSF1 and pCDNA3.1-FLAG-Hsc70 mammalian expression vectors. At 48 h after transfection, cells were heat shocked for 1 h at 42 °C, and indirect immunofluorescence analysis was performed. As shown in Figure 4(C), HSF1 and Hsc70 were found predominately in the cytoplasm under non-stressed conditions and translocated to the nucleus upon heat stress.
Figure 4. HSF1 and Hsc70 are co-localized in the nucleus after heat shock.
(A) WT and (B) hsf1−/− mEF cells were heat-shocked at 42 °C for 1 h. Cytosolic (C) and nuclear (N) fractions were prepared and resolved by SDS/PAGE. HSF1, Hsc70, actin and c-fos were detected by Western blot analysis. (C) hsf1−/− mEF cells were co-transfected with pEGFP-HSF1 and pCDNA3.1-FLAG-Hsc70 mammalian expression vectors. At 48 h after transfection, cells were untreated (Control) or heat-shocked at 42 °C for 1 h. Cells were fixed and immunostained using anti-FLAG antibodies. Nuclei were stained with DAPI (4,6-diamidino-2-phenylindole).
Hsc70 regulates heat-induced HSF1 activation
The role of Hsc70 in the activation of HSF1 in vivo was evaluated using specific siRNAs to knockdown the expression of Hsc70. Transfection of the RNAi expression vector for Hsc70 (pSuppressorNeo-Hsc70) led to a marked inhibition of endogenous Hsc70 expression in HEK-293 cells (Figure 5A). As shown in Figure 5(B), transfected cells with Hsc70 siRNA vector did not significantly prevent HSF1–HSE complex formation under non-stressed conditions. In fact, a slight increase in HSF1 DNA binding was observed, suggesting a possible compensatory pathway is activated in response to knocking down Hsc70 expression. However, under heat-shock conditions, knocking down Hsc70 expression strongly prevented HSF1 DNA-binding activity, suggesting that Hsc70 is required for HSF1 activation in response to heat shock.
Figure 5. Depletion of endogenous Hsc70 by RNAi inhibits heat-stress-mediated HSF1 activation.
HEK-293 cells were transiently transfected with a pSuppressorNeo (C) or pSuppressorNeo-Hsc70 RNAi expression vector. (A) At 48 h after transfection, total cell extracts were prepared, and Hsc70 and actin protein levels were detected by Western blot analysis. (B) At 48 h after transfection, cells were untreated (−) or heat-shocked at 42 °C (+) for 1 h. Nuclear extracts were prepared, and HSF1 DNA-binding activity was measured by EMSA using 32P-labelled HSF1 DNA-binding fragments known as HSEs. The position of the HSF1–HSE complex and the free HSE DNA fragments are shown on the right. (C) At 48 h after transfection, cells were untreated (−) or heat-shocked at 42 °C (+) for 1 h, followed by recovery at 37 °C for 24 or 48 h. Total cell extracts were prepared, and Hsp70, Hsp27, HSF1 and actin protein levels were detected by Western blot analysis.
Previous reports have clearly demonstrated the important role of mammalian HSF1 in stress-induced Hsp gene expression. To examine the role of Hsc70 on HSF1 target gene expression, HEK-293 cells were transfected with Hsc70 siRNA vector or an empty siRNA vector. At 48 h after transfection, cells were heat shocked for 1 h and recovered for the indicated time periods to activate the expression of Hsp70 and Hsp27. As shown in Figure 5(C), the typical expression of Hsp70 and Hsp27 was induced upon heat-shock treatment in control cells. Interestingly, under non-stressed conditions, Hsc70 knockdown cells expressed a low level of Hsp70 compared with WT cells, again demonstrating a possible compensatory response to the loss of Hsc70 expression. However, cells transfected with Hsc70 siRNA vector showed a severe reduction of target gene expression upon heat stress (Figure 5C), clearly indicating Hsc70 involvement in the transcriptional activation function of HSF1 during cellular insults.
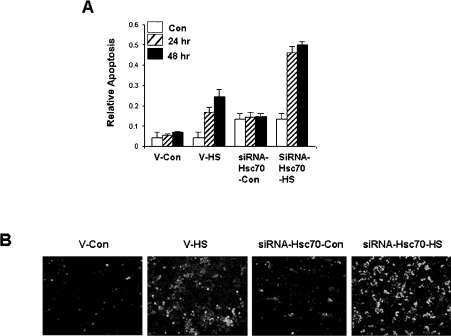
We next examined whether knocking down Hsc70 protein expression affected stress-induced apoptosis using cell death detection ELISA analysis. Control cells allowed recovering for 24 or 48 h after heat-shock treatment showed a low level of cell death (Figure 6A). However, Hsc70-siRNA-vector-transfected cells showed an increased susceptibility to programmed cell death by heat stress (Figure 6A). Annexin V–FLUOS analysis also revealed an elevated number of apoptotic cells in Hsc70-siRNA-vector-transfected cells compared with empty-vector-transfected control cells following heat stress (Figure 6B).
Figure 6. Depletion of endogenous Hsc70 by RNAi leads to heat-stress-induced apoptosis.
HEK-293 cells were transiently transfected with a pSuppressorNeo (V) or pSuppressorNeo-Hsc70 RNAi expression vector. (A) At 48 h after transfection, cells were untreated (Con) or heat-shocked at 44 °C (HS) for 1 h, followed by recovery at 37 °C for 24 or 48 h. Relative cell death was measured as a ‘relative apoptosis’ index, see the Materials and methods section for details. (B) At 48 h after transfection, cells were untreated (Con) or heat-shocked at 44 °C (HS) for 1 h and then cultured for a further 24 h at 37 °C. Apoptosis was detected by annexin V–FLUOS and propidium iodide staining.
DISCUSSION
Many biochemical and genetic studies have demonstrated that the mammalian HSF1, and the corresponding Drosophila HSF respond to stress to activate target gene transcription [7,8,26]. The activation of mammalian HSF1 is a multi-step process that involves the conversion of a cytoplasmically localized monomer, which binds to HSEs with low affinity, into a homotrimer, followed by nuclear translocation, high-affinity binding to HSEs and target gene activation [9]. Consistent with the transient nature of Hsp target gene activation in the stress response, trimerized HSF1 is ultimately converted back into the cytosolic monomer [11,12].
In the present study, we characterized a novel mechanism of HSF1 activation. Hsc70 was identified as a strong HSF1-interacting protein using a strenuous TAP system followed by MS. We showed that Hsc70 is required for HSF1 to become activated and target expression of appropriate genes during heat stress.
Hsc70 is best known as a molecular chaperone because of its major role in protein folding [1,3,4,29]. Two members of the Hsp70 family, Hsc70 and Hsp70, have a high degree of sequence homology. Several groups have recently reported that the stress-inducible Hsp70 binds with the HSF1 transactivation domain as a part of a down-regulation of the heat-shock response [15–18]. Since HSF1 induces Hsp70 gene expression in response to heat stress, it has been suggested that Hsp70 may function in a negative-feedback mechanism to return HSF1 to its inactive monomeric state [18]. Although it has been reported that HSF1 and Hsc70 associate in a high-molecular-mass complex in the cytoplasm of unstressed cells [20], the role of Hsc70 in HSF1 regulation is unknown. Although both Hsc70 and Hsp70 interact tightly with HSF1, the function of their respective interactions with HSF1 is probably different [15,20]. First, Hsc70 and Hsp70 have different expression patterns; for example, Hsp70 expression is induced by stress, whereas Hsc70 is constitutively expressed in cells. Secondly, although both Hsc70 and Hsp70 have been shown to interact directly with lipid bilayers and promote membrane protein folding and polypeptide translocation, Hsc70 has a more dramatic effect than Hsp70 [30]. Also, Hsc70 and Hsp70 differently acquire immunogenic peptides during oxidative stress. Hsp70 associates with peptides more quantitatively than Hsc70 under oxidative conditions [30]. Moreover, the secondary structure of Hsp70 is more dramatically changed than Hsc70 under oxidative conditions [31]. Although the overall structures of these two proteins are very similar, they have considerable differences in their C-terminal domains. This domain may therefore mediate substrate specificity and specific biological functions [32–34].
We observed that HSF1 interacts directly with Hsc70 under both unstressed and heat-stressed conditions. Heat stress did not affect the level of HSF1–Hsc70 interaction (Figure 1E). Why the overexpression of Hsc70 did not fully activate HSF1 in unstressed cells (Figure 3) is likely to be due to the involvement of additional regulatory associating proteins. Hsp90 has emerged as a key factor for the regulation of HSF1. Many studies provide evidence that the transcriptional activity of trimeric HSF1 is repressed by multichaperone complexes consisting of Hsp90 and its co-chaperones (Hip, Hop, p23, Hsp70 and Cyp-40) [13,14]. Hsc70 co-chaperones may also regulate the HSF1–Hsc70 stress-sensing mechanism by positively regulating the availability of a functional Hsc70.
Several interesting observations were made when Hsc70 expression was repressed using siRNA (Figure 5). Under non-stressed conditions, the DNA-binding activity of HSF1 was slightly elevated (Figure 5B), and a low level of Hsp70 target gene expression was detected (Figure 5C) in Hsc70 knockdown cells compared with control cells. The observed low levels may represent an insignificant difference between samples, but, more likely, the results suggest that the cells, using a currently unknown mechanism, were attempting to compensate for the loss of Hsc70. Nevertheless, the most striking observation was when the cells were challenged with heat shock. Knocking down the expression of Hsc70 greatly diminished HSF1 activities, indicating that Hsc70 is a required component of the HSF1-mediated heat-shock-response mechanism.
In the present study, we showed that the overexpression of Hsc70 induced HSF1 activation, including HSF1 multimerization and induction of DNA-binding activity in vivo. Furthermore, under heat-shock conditions, RNAi disruption of endogenous Hsc70 expression resulted in a marked decrease in HSF1 DNA-binding activity and decreased its ability to induce expression of necessary stress-response proteins, such as Hsp70 and Hsp27. A consequence of this inhibition is an increase in cellular apoptosis. These studies clearly implicate Hsc70 as a critical facilitator of HSF1-mediated cell survival in response to cellular insults. Future genetic and biochemical experiments will focus on the mechanistic details of this activation.
Acknowledgments
We thank Dr Dennis Thiele for helpful suggestions.
References
- 1.Parsell D. A., Lindquist S. The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu. Rev. Genet. 1993;27:437–496. doi: 10.1146/annurev.ge.27.120193.002253. [DOI] [PubMed] [Google Scholar]
- 2.Zugel U., Kaufmann S. H. Role of heat shock proteins in protection from and pathogenesis of infectious diseases. Clin. Microbiol. Rev. 1999;12:19–39. doi: 10.1128/cmr.12.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morimoto R. I., Tissieres A., Georgopoulos C. Cold Spring Harbor Monograph Series, vol. 26. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1994. The biology of the heat shock proteins and molecular chaperones; p. 610. [Google Scholar]
- 4.Fink A. L. Chaperone-mediated protein folding. Physiol. Rev. 1999;79:425–449. doi: 10.1152/physrev.1999.79.2.425. [DOI] [PubMed] [Google Scholar]
- 5.Kaufman R. J. Molecular chaperones and the heat shock response. Sponsored by Cold Spring Harbor Laboratory, 6–10 May 1998. Biochim. Biophys. Acta. 1999;1423:R13–R27. doi: 10.1016/s0304-419x(98)00029-8. [DOI] [PubMed] [Google Scholar]
- 6.Liu X. D., Liu P. C., Santoro N., Thiele D. J. Conservation of a stress response: human heat shock transcription factors functionally substitute for yeast HSF. EMBO J. 1997;16:6466–6477. doi: 10.1093/emboj/16.21.6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morano K. A., Thiele D. J. Heat shock factor function and regulation in response to cellular stress, growth, and differentiation signals. Gene Expression. 1999;7:271–282. [PMC free article] [PubMed] [Google Scholar]
- 8.Wu C. Heat shock transcription factors: structure and regulation. Annu. Rev. Cell Dev. Biol. 1995;11:441–469. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- 9.Zuo J., Baler R., Dahl G., Voellmy R. Activation of the DNA-binding ability of human heat shock transcription factor 1 may involve the transition from an intramolecular to an intermolecular triple-stranded coiled-coil structure. Mol. Cell. Biol. 1994;14:7557–7568. doi: 10.1128/mcb.14.11.7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo Y., Guettouche T., Fenna M., Boellmann F., Pratt W. B., Toft D. O., Smith D. F., Voellmy R. Evidence for a mechanism of repression of heat shock factor 1 transcriptional activity by a multichaperone complex. J. Biol. Chem. 2001;276:45791–45799. doi: 10.1074/jbc.M105931200. [DOI] [PubMed] [Google Scholar]
- 11.Morimoto R. I., Kroeger P. E., Cotto J. J. The transcriptional regulation of heat shock genes: a plethora of heat shock factors and regulatory conditions. In: Feige U., Morimoto R. I., Yahara I., Polla B. S., editors. Stress-Inducible Cellular Responses. Basel: Birkhhaeuser Verlag; 1996. pp. 139–163. [DOI] [PubMed] [Google Scholar]
- 12.Voellmy R. Sensing and responding to stress. In: Feige U., Morimoto R. I., Yahara I., Polla B. S., editors. Stress-Inducible Cellular Responses. Basel: Birkhhaeuser Verlag; 1996. pp. 121–137. [Google Scholar]
- 13.Zuo J., Guo Y., Guettouche T., Smith D. F., Voellmy R. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell. 1998;94:471–480. doi: 10.1016/s0092-8674(00)81588-3. [DOI] [PubMed] [Google Scholar]
- 14.Bharadwaj S., Ali A., Ovsenek N. Multiple components of the HSP90 chaperone complex function in regulation of heat shock factor 1 in vivo. Mol. Cell. Biol. 1999;19:8033–8041. doi: 10.1128/mcb.19.12.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abravaya K., Myers M. P., Murphy S. P., Morimoto R. I. The human heat shock protein hsp70 interacts with HSF, the transcription factor that regulates heat shock gene expression. Genes Dev. 1992;6:1153–1164. doi: 10.1101/gad.6.7.1153. [DOI] [PubMed] [Google Scholar]
- 16.Mosser D. D., Duchaine J., Massie B. The DNA-binding activity of the human heat shock transcription factor is regulated in vivo by hsp70. Mol. Cell. Biol. 1993;13:5427–5438. doi: 10.1128/mcb.13.9.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rabindran S. K., Wisniewski J., Li L., Li G. C., Wu C. Interaction between heat shock factor and hsp70 is insufficient to suppress induction of DNA-binding activity in vivo. Mol. Cell. Biol. 1994;14:6552–6560. doi: 10.1128/mcb.14.10.6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi Y., Mosser D. D., Morimoto R. I. Molecular chaperones as HSF1-specific transcriptional repressors. Genes Dev. 1998;12:654–666. doi: 10.1101/gad.12.5.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Satyal S. H., Chen D., Fox S. G., Kramer J. M., Morimoto R. I. Negative regulation of the heat shock transcriptional response by HSBP1. Genes Dev. 1998;12:1962–1974. doi: 10.1101/gad.12.13.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nunes S. L., Calderwood S. K. Heat shock factor-1 and the heat shock cognate 70 protein associate in high molecular weight complexes in the cytoplasm of NIH-3T3 cells. Biochem. Biophys. Res. Commun. 1995;213:1–6. doi: 10.1006/bbrc.1995.2090. [DOI] [PubMed] [Google Scholar]
- 21.Ahn S. G., Thiele D. J. Redox regulation of mammalian heat shock factor 1 is essential for Hsp gene activation and protection from stress. Genes Dev. 2003;17:516–528. doi: 10.1101/gad.1044503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 23.Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Seraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 24.Vacratsis P. O., Phinney B. S., Gage D. A., Gallo K. A. Identification of in vivo phosphorylation sites of MLK3 by mass spectrometry and phosphopeptide mapping. Biochemistry. 2002;41:5613–5624. doi: 10.1021/bi016075c. [DOI] [PubMed] [Google Scholar]
- 25.Ahn S. G., Liu P. C., Klyachko K., Morimoto R. I., Thiele D. J. The loop domain of heat shock transcription factor 1 dictates DNA-binding specificity and responses to heat stress. Genes Dev. 2001;15:2134–2145. doi: 10.1101/gad.894801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarge K. D., Murphy S. P., Morimoto R. I. Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA-binding activity, and nuclear localization and can occur in the absence of stress. Mol. Cell. Biol. 1993;13:1392–1407. doi: 10.1128/mcb.13.3.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brummelkamp T. R., Bernards R., Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 28.Paul C. P., Good P. D., Winer I., Engelke D. R. Effective expression of small interfering RNA in human cells. Nat. Biotechnol. 2002;20:505–508. doi: 10.1038/nbt0502-505. [DOI] [PubMed] [Google Scholar]
- 29.Chirico W. J., Waters M. G., Blobel G. 70K heat shock related proteins stimulate protein translocation into microsomes. Nature (London) 1988;332:805–810. doi: 10.1038/332805a0. [DOI] [PubMed] [Google Scholar]
- 30.Arispe N., Doh M., De Maio A. Lipid interaction differentiates the constitutive and stress-induced heat shock proteins Hsc70 and Hsp70. Cell Stress Chaperones. 2002;7:330–338. doi: 10.1379/1466-1268(2002)007<0330:lidtca>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Callahan M. K., Chaillot D., Jacquin C., Clark P. R., Menoret A. Differential acquisition of antigenic peptides by Hsp70 and Hsc70 under oxidative conditions. J. Biol. Chem. 2002;277:33604–33609. doi: 10.1074/jbc.M202890200. [DOI] [PubMed] [Google Scholar]
- 32.Hu S. M., Wang C. Involvement of the 10-kDa C-terminal fragment of hsc70 in complexing with unfolded protein. Arch. Biochem. Biophys. 1996;332:163–169. doi: 10.1006/abbi.1996.0328. [DOI] [PubMed] [Google Scholar]
- 33.Bertelsen E. B., Zhou H., Lowry D. F., Flynn G. C., Dahlquist F. W. Topology and dynamics of the 10 kDa C-terminal domain of DnaK in solution. Protein Sci. 1999;8:343–354. doi: 10.1110/ps.8.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morshauser R. C., Hu W., Wang H., Pang Y., Flynn G. C., Zuiderweg E. R. High-resolution solution structure of the 18 kDa substrate-binding domain of the mammalian chaperone protein Hsc70. J. Mol. Biol. 1999;289:1387–1403. doi: 10.1006/jmbi.1999.2776. [DOI] [PubMed] [Google Scholar]