Abstract
PDE3A (phosphodiesterase 3A) was identified as a phosphoprotein that co-immunoprecipitates with endogenous 14-3-3 proteins from HeLa cell extracts, and binds directly to 14-3-3 proteins in a phosphorylation-dependent manner. Among cellular stimuli tested, PMA promoted maximal binding of PDE3A to 14-3-3 proteins. While p42/p44 MAPK (mitogen-activated protein kinase), SAPK2 (stress-activated protein kinase 2)/p38 and PKC (protein kinase C) were all activated by PMA in HeLa cells, the PMA-induced binding of PDE3A to 14-3-3 proteins was inhibited by the non-specific PKC inhibitors Ro 318220 and H-7, but not by PD 184352, which inhibits MAPK activation, nor by SB 203580 and BIRB0796, which inhibit SAPK2 activation. Binding of PDE3A to 14-3-3 proteins was also blocked by the DNA replication inhibitors aphidicolin and mimosine, but the PDE3A–14-3-3 interaction was not cell-cycle-regulated. PDE3A isolated from cells was able to bind to 14-3-3 proteins after in vitro phosphorylation with PKC isoforms. Using MS/MS of IMAC (immobilized metal ion affinity chromatography)-enriched tryptic phosphopeptides and phosphospecific antibodies, at least five sites on PDE3A were found to be phosphorylated in vivo, of which Ser428 was selectively phosphorylated in response to PMA and dephosphorylated in cells treated with aphidicolin and mimosine. Phosphorylation of Ser428 therefore correlated with 14-3-3 binding to PDE3A. Ser312 of PDE3A was phosphorylated in an H-89-sensitive response to forskolin, indicative of phosphorylation by PKA (cAMP-dependent protein kinase), but phosphorylation at this site did not stimulate 14-3-3 binding. Thus 14-3-3 proteins can discriminate between sites in a region of multisite phosphorylation on PDE3A. An additional observation was that the cytoskeletal cross-linker protein plectin-1 coimmunoprecipitated with PDE3A independently of 14-3-3 binding.
Keywords: cAMP, cytoskeleton, 14-3-3 protein, phorbol ester, phosphodiesterase 3A (PDE3A), protein kinase C (PKC)
Abbreviations: DIG, digoxygenin; GFP, green fluorescent protein; HSL, hormone-sensitive lipase; IGF1, insulin-like growth factor 1; MALDI, matrix-assisted laser-desorption ionization; MAPK, mitogen-activated protein kinase; PDE, phosphodiesterase; PI3K, phosphoinositide 3-kinase; PKA, cAMP-dependent protein kinase; PKB, protein kinase B/Akt; PKC, protein kinase C; PKD, protein kinase D; SAPK, stress-activated protein kinase; TNF, tumour necrosis factor α; TOF, time-of-flight; VASP, vasodilator-stimulated phosphoprotein
INTRODUCTION
Cyclic nucleotide PDEs (phosphodiesterases) hydrolyse cAMP and cGMP, two important second messengers. There are at least 11 mammalian PDE gene families (PDEs 1–11), with multiple isoforms generated by expression from different genes, different transcription initiation sites and mRNA splice variants [1].
The PDE3 family contains variants of PDE3A and PDE3B [2,3]. These enzymes bind both cAMP and cGMP, although cGMP is a poor substrate and inhibits cAMP hydrolysis, hence the original name of ‘cGMP-inhibited cAMP phosphodiesterase’. The modes of binding of cAMP, cGMP and inhibitors have been deduced from the crystal structure of the PDE3B catalytic domain [4]. The full-length forms of PDE3A and PDE3B both have an N-terminal transmembrane domain that is thought to traverse the endoplasmic reticulum membrane (residues ∼1–300), followed by a cytoplasmic 50-residue hydrophobic sequence and a ‘regulatory’ region with phosphorylation sites (residues ∼301–690; see the Results section), catalytic domain (residues ∼690–974, including a 44-amino-acid sequence unique to the PDE3s and different in PDE3A and PDE3B) and a C-terminal tail (residues 974–1100) [3]. There may also be soluble forms of PDE3s that lack the transmembrane domain [3,5].
PDE3A and PDE3B have distinct, but overlapping, tissue distributions. PDE3A appears to predominate in heart, platelets, vascular smooth muscle and oocytes, while PDE3B is the major isoform in adipocytes, hepatocytes and developing spermatocytes [3].
The therapeutic potential of the PDE3 enzymes has been indicated by the fact that PDE3A-deficient mice are viable [6], and because PDE3 inhibitors have been found to have beneficial effects in some chronic heart failure patients [7,8]. In addition, the use of inhibitors, and the phenotypes of PDE3 gene-knockout mice, have implicated PDE3 enzymes in the regulation of diverse cellular processes that are relevant to health and disease, including lipolysis in adipocytes [9–11], glycogenolysis, myocardial contractility, smooth muscle relaxation, platelet activation and aggregation [12], meiotic maturation in oocytes [6,13], proliferation of rat mesangial cells, secretion of insulin from pancreatic β-cells [14,15], and renin secretion [16]. Best characterized is the regulation of HSL (hormone-sensitive lipase), the rate-limiting enzyme in the mobilization of triacylglycerols in adipose tissue. HSL is phosphorylated and activated by PKA (cAMP-dependent protein kinase) in response to adrenaline. This effect is transient, however, because PKA also activates PDE3B, lowering the concentration of cAMP and thereby inhibiting PKA. In addition, insulin antagonizes the adrenergic activation of HSL by the PKB (protein kinase B/Akt)-mediated phosphorylation and activation of PDE3B [9,10,17,18]. There is some uncertainty over the PKB sites, which have been reported to be pSer273 (pSer279 in rat; [10]) or pSer296 (pSer302 in rat; [19]) on mouse PDE3B, or possibly both sites [11]. The activating effects of phosphorylation at PKA and PKB on PDE3B were found to be additive. It has been discovered that PDE3B from insulin-stimulated adipocytes, and PDE3B that was phosphorylated by PKB in vitro, bind to 14-3-3 proteins [11]. The role of 14-3-3 proteins in the regulation of PDE3B by insulin is not yet clear. 14-3-3 proteins are ubiquitous eukaryotic proteins with central regulatory roles that they exert by binding to specific phosphorylated sites in diverse target proteins [20,21]. PDE3B also appears to be regulated by a pathway that involves protein interactions controlled by PI3Kγ (phosphoinositide 3-kinase γ) [22].
The regulation of PDE3A is less well characterized. In platelets, intracellular cAMP levels increase in response to stimuli such as adenosine and prostaglandin E1 that act via Gs/adenylate cyclase-coupled receptors. The elevated cAMP levels inhibit platelet activation (Ca2+ mobilization, secretion and integrin-dependent aggregation) in response to agonists that signal via Gi/z [23,24]. Resting cells appear to have a ‘basal’ level of cAMP that restricts activation, and PDE3 inhibitors cause cAMP levels to rise without exogenous stimulation of Gs/adenylate cyclase. These findings suggest that PDE3A keeps cAMP poised at a low threshold level, which is sufficient to maintain platelets in the resting state [12,25,26]. PDE3 inhibitors also stimulated phosphorylation of PKA sites (Ser157 and Ser239) on VASP (vasodilator-stimulated phosphoprotein), a regulator of actin dynamics, suggesting that PDE3A hydrolyses the cAMP pool that regulates PKA phosphorylation of VASP [27,28]. Little is known about the regulation of PDE3A activity, except for its inhibition by cGMP and the proposal of a negative-feedback loop in which phosphorylation of PDE3A by PKA increases its activity [29].
PDE3A was recently reported among 14-3-3 affinity-purified proteins [30]. In the present paper, we also identified PDE3A among human HeLa cell proteins that were isolated by 14-3-3-affinity chromatography. In contrast with PDE3B, the 14-3-3-binding site(s) on PDE3A were not phosphorylated by the PI3K/PKB pathway. Instead, PDE3A bound to 14-3-3 proteins when PKC (protein kinase C) was activated by PMA treatment of HeLa cells. Overall, at least five sites on PDE3A were found to be phosphorylated in vivo. In addition, PDE3A was found to bind to the cytoskeletal linker protein, plectin, by a mechanism that is independent of binding to 14-3-3 proteins.
EXPERIMENTAL
Materials
IGF1 (insulin-like growth factor 1) was from Biosource; dialysed foetal calf serum and other tissue culture reagents were from Invitrogen. LY 294002, H-89, SB 203580, rapamycin, the catalytic subunit of PKC from rat brain and recombinant human PKCα were from Calbiochem. DMSO, forskolin and H-7 were from Sigma. BIRB0796 was synthesized by Ms Natalia Shpiro and Dr Rodolfo Marguez, School of Life Sciences, University of Dundee. Pre-cast SDS/polyacrylamide gels and Colloidal Blue protein stain were from Invitrogen. All synthetic peptides used in the present study were synthesized by Dr Graham Bloomberg (University of Bristol, Bristol, U.K.). Microcystin-LR was from Professor Linda Lawton (The Robert Gordon University, Aberdeen, U.K.). Vivaspin concentrators were from Vivascience. Protease inhibitor mixture tablets and sequencing-grade trypsin were from Roche Molecular Biochemicals. All chromatographic columns, including [NHS (hydroxysuccinimide)-activated] CH-Sepharose, Protein G–Sepharose and glutathione–Sepharose were from Amersham Biosciences. All other chemicals were from BDH Chemicals or Sigma–Aldrich.
Antibodies
A PDE3A isoform-specific antibody was raised in sheep at Diagnostics Scotland (Penicuik, U.K.) against the synthetic peptide RLAGIENQSLDQTPQS, corresponding to residues 1095–1110 of human PDE3A. The peptide was coupled separately to BSA and keyhole-limpet haemocyanin before being mixed and injected into a sheep. The antibody was affinity-purified on CH-Sepharose coupled to the same peptide. The phosphospecific antibody recognizing PDE3A phosphorylated at Ser312 (anti-pSer312 antibody) was raised in sheep against the peptide SHRRTpS312LPCIP, corresponding to residues 307–317 of human PDE3A. The antibodies were affinity-purified on CH-Sepharose covalently coupled to the phosphorylated peptide, then passed through a column of CH-Sepharose coupled to the nonphosphorylated peptide. Similarly, phosphospecific antibodies were generated in sheep against the following phosphopeptides and affinity-purified: KRLRRpS428LPPGL (for anti-pSer428 antibody); LRRVpS438STWTT (for anti-pSer438 antibody); RDRS-TpS465IKLQEAP (for anti-pSer465 antibody); and HRALTYpT568QSAPDL (for anti-pThr568 antibody). Initially, we used the K-19 anti-14-3-3 antibody from Santa Cruz, until we raised a sheep polyclonal antibody that recognizes several or all of the isoforms of human 14-3-3 proteins by immunizing with the conjugated synthetic peptide KSELVQKAKLAEQAERYDD. The antibody against plectin was mouse monoclonal 9F8, an unpublished antibody generated by the last two authors of [31], which was a gift from Professor Birgit Lane, School of Life Sciences, University of Dundee.
Cell culture, cell-cycle synchronization and cell lysis
HeLa cells were cultured in DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% foetal bovine serum, 100 units/ml penicillin and 100 units/ml streptomycin. After stimulation, the medium was aspirated, and the cells were solubilized in 0.2 ml of ice-cold lysis buffer [50 mM Tris/acetate (pH 7.0), 1 mM EDTA, 1 mM EGTA, 1% (w/v) Triton X-100, 1 mM sodium orthovanadate, 10 mM sodium glycerophosphate, 50 mM NaF, 5 mM sodium pyrophosphate, 0.27 M sucrose, 2 μM microcystin-LR, 1 mM benzamidine, 0.1% (v/v) 2-mercaptoethanol and Complete™ proteinase inhibitor mixture (Roche; 1 tablet per 50 ml)]. The samples were snap-frozen in liquid nitrogen and stored at −80 °C until analysis. Protein concentrations were determined using the Bradford reagent (Perbio).
Immunoblotting and DIG (digoxygenin)–14-3-3 overlays
For blots of total cell lysates, 20 μg of protein was immunoblotted. Membranes were incubated in 50 mM Tris/HCl, pH 7.5, 0.15 M NaCl and 0.2% (v/v) Tween 20 containing 5% (w/v) BSA and then were immunoblotted at 4 °C for 16 h using the indicated antibodies (2 μg/ml for the sheep antibodies or 1000-fold dilution of commercial antibodies). Detection was performed using horseradish-peroxidase-conjugated secondary antibodies (Promega) and ECL® (enhanced chemiluminescence reagent; Amersham Biosciences).
DIG–14-3-3 overlays were performed using DIG-labelled 14-3-3 in place of primary antibody, followed by an anti-DIG–horseradish peroxidase secondary antibody [21].
14-3-3-affinity chromatography
HeLa cells (5×109 frozen cells from Computer Cell Culture Center, 14 Rue de la Marlette, 7180 Seneffe, Belgium) were thawed in 20 ml of lysis buffer containing 50 mM Tris/HCl, pH 7.5, 1 mM EDTA, 1 mM EGTA, 1% (w/v) Triton X-100, 1 mM sodium orthovanadate, 50 mM NaF, 10 mM sodium β-glycerophosphate, 0.27 M sucrose, 1 μM microcystin-LR, 0.1% (v/v) 2-mercaptoethanol and Complete™ proteinase inhibitor mixture (one tablet/50 ml). The lysed cells were centrifuged at 10000 g at 4 °C for 60 min, and the supernatant was diluted with 20 ml of buffer A [25 mM Tris/HCl, pH 7.5 (4 °C), 100 mM NaCl and 25 mM NaF]. The solution was clarified by filtration if necessary, and mixed end-over-end for 1 h at 4 °C with 6 ml of Sepharose linked to 6 mg each of BMH1/BMH2 (the Saccharomyces cerevisiae 14-3-3 isoforms) [21]. The mixture was poured into a column, and the flow-through was collected. The column was washed with 1–3 litres of 50 mM Hepes/NaOH, pH 7.5, 500 mM NaCl, 1 mM dithiothreitol, 1 μM microcystin-LR. (Microcystin-LR was not included from this point when preparations were to be used for dephosphorylation experiments.) The column was rinsed and mock-eluted with a synthetic phosphopeptide that does not bind 14-3-3 proteins (1 mM RSRTRTDpSYSAGQSV in buffer A for the experiments shown here), before proteins that bind to the phosphopeptide binding site of 14-3-3 proteins were eluted with 1 mM ARAApSAPA phosphopeptide.
Immunoprecipitations of PDE3A from HeLa cell extracts
Clarified protein extracts (1–5 mg) were mixed with 10 μg of the anti-C-terminal-PDE3A peptide antibody coupled to Protein G–Sepharose for 3 h at 4 °C. The beads were washed twice with lysis buffer containing 0.5 M NaCl and once in buffer A without NaCl, and the samples were eluted by boiling for 10 min in SDS sample buffer containing 100 mM dithiothreitol. Eluted proteins were resolved by SDS/PAGE, and immunoblot analyses were performed as described above.
Protein identification and phosphorylation site analysis
Proteins were identified from ‘in gel’ tryptic digestion of colloidal Coomassie-Blue-stained SDS/PAGE gel bands, by peptide mass fingerprinting combined with MALDI (matrix-assisted laser-desorption ionization)–MS/MS on a 4700 Proteomics Analyser (Applied Biosystems) as described previously [21]. Phosphopeptides were enriched from tryptic digests using 3 μl of PHOS-beads (Sigma P9740) previously equilibrated in 0.25 M acetic acid/30% acetonitrile (wash/bind buffer). Tryptic digests were diluted to 0.1 ml in wash/bind buffer and mixed with the PHOS-select beads by gentle vortex-mixing for 30 min. The beads were collected into a μC18 ZipTip (Millipore), washed by passing 3×25 μl of wash/bind buffer through the ZipTip and the peptides eluted with 2×15 μl of 0.4 M NH4OH. The eluted fraction was dried under a vacuum, reconstituted in 1% formic acid in water applied to a C18 StageTip (Proxeon), and the peptides were eluted with 50% acetonitrile/0.1% trifluoroacetic acid in water before MALDI–TOF (time-of-flight)/TOF analysis.
32P-labelled phosphopeptides were separated by reverse-phase HPLC and analysed by a combination of MS and solidphase Edman degradation as described previously [32].
RESULTS
Phosphorylated PDE3A extracted from HeLa cells binds directly to 14-3-3 proteins
PDE3A was identified by MALDI–TOF tryptic mass finger-printing of an approx. 105 kDa protein that was present in a pool of (phospho)proteins that were eluted from a 14-3-3 column with a 14-3-3-binding phosphopeptide, ARAApSAPA, run as described previously [21] (Figure 1A). The identity of PDE3A in the 14-3-3 column eluate was confirmed by Western blotting with an antibody (anti-C-terminal-PDE3A antibody) raised against the peptide RLAGIENQSLDQTPQS, a sequence that is not found in PDE3B (Figure 1A).
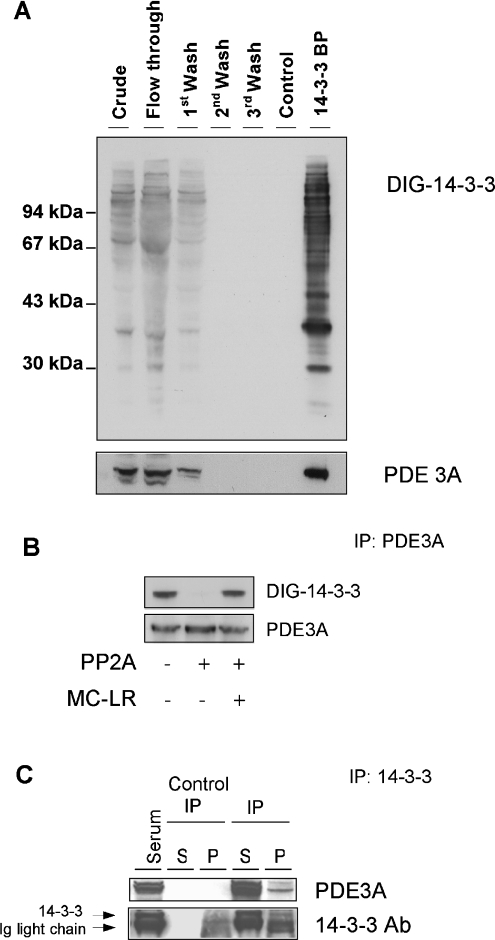
Figure 1. Phosphorylation-dependent binding of PDE3A to 14-3-3 proteins.
(A) Isolation of PDE3A by 14-3-3-affinity chromatography of HeLa cell extracts. The clarified HeLa cell extract was chromatographed on 14-3-3–Sepharose (see the Experimental section). The extract and column flow through were analysed without being concentrated, while 12 ml samples from the beginning, middle and end of the salt wash, a mock elution with control phosphopeptide (RSRTRTDpSYSAGQSV) and the ARAApSAPA elution pool were collected and concentrated in Vivaspin 6 concentrators (Vivascience) to 400 μl, of which 4 μl of each was run on SDS/PAGE using NuPage 10% Bis-Tris gels (Invitrogen) and transferred on to nitrocellulose membranes (Schleicher and Schuell). Amounts of protein run on SDS/PAGE were: extract, flow through and start of salt wash (40 μg of each); end of salt wash (protein undetectable); control phosphopeptide pool (<1 μg); and ARAApSAPA pool (2 μg; 14-3-3-binding proteins, labelled 14-3-3 BP). Blots of column fractions were analysed for proteins that bind directly to 14-3-3 proteins by DIG–14-3-3 overlay (upper panel), and Western blotting with anti-C-terminal-PDE3A antibody (lower panel). Molecular-mass sizes are indicated in kDa. (B) Direct binding of 14-3-3 proteins to phosphorylated PDE3A. PDE3A was immunoprecipitated (IP) from 1.5 mg HeLa cell extracts using 10 μg of the anti-C-terminal-PDE3A antibody. The purified protein was incubated for 30 min at 30 °C in the presence or absence of 50 m-units/ml protein phosphatase 2A (PP2A), with or without the inhibitor microcystin-LR (MC-LR; 5 μM), as indicated. The samples were run on SDS/PAGE, transferred on to nitrocellulose and probed for binding to anti-PDE3A (lower panel) and DIG–14-3-3 proteins in an overlay assay (upper panel). (C) Binding of endogenous 14-3-3 proteins to endogenous PDE3A. Protein from extract of serum-grown HeLa cells (3 mg) was immunoprecipitated (IP) with anti-14-3-3–Sepharose. The control contained no cell extract. After washing, the immunoprecipitates (P) and first supernatants (S) were run on SDS/PAGE using NuPage 10% Bis-Tris gels and transferred on to nitrocellulose. Crude cell extract (30 μg) was run in the first lane. Western blotting was used to identify the 14-3-3 protein, which ran just above the Ig light chain (lower panel) and co-immunoprecipitated PDE3A, which ran at approx. 105 kDa (anti-C-terminal-PDE3A antibody, upper panel). Ab, antibody.
The form of PDE3A isolated here does not appear to be the full-length PDE3A. Its molecular mass (∼105 kDa), the absence of peptides corresponding to the N-terminal transmembrane region, recovery in the soluble fraction on subcellular fractionation, and Km (results not shown) mean that it is similar to the previously reported Δ1-PDE3A variant protein that was found to lack the N-terminal transmembrane domain [5].
The anti-C-terminal-PDE3A antibody could immunoprecipitate the endogenous PDE3A protein, which was detected as a protein band of ∼105 kDa by immunoblotting with the same antibody (Figure 1B) and by tryptic mass fingerprinting (see results below). The immunoprecipitated PDE3A bound directly to DIG–14-3-3 proteins in an overlay assay (Figure 1B). The interaction was dependent on phosphorylation of the PDE3A because the 14-3-3 binding signal was abolished by dephosphorylation of the PDE3A with protein phosphatase 2A (Figure 1B).
Complexes of PDE3A and endogenous 14-3-3 proteins were identified in HeLa cell extracts by immunoprecipitating 14-3-3 proteins with an antibody that recognizes human 14-3-3 isoforms. When the washed immunoprecipitates were analysed by immunoblotting, PDE3A was found to have co-precipitated with the endogenous 14-3-3 proteins (Figure 1C).
PDE3A binds to 14-3-3 proteins in response to PMA in vivo and phosphorylation by PKC in vitro
The HeLa cells used for our 14-3-3-binding phosphoproteomics study had been cultured in medium containing serum, which contains a mixture of cell effectors. To pinpoint the cellular stimuli that are responsible for promoting the phosphorylation of PDE3A at site(s) that facilitate binding to 14-3-3 proteins, cells were deprived of serum and were stimulated in different ways. The endogenous PDE3A was then immunoprecipitated and its ability to bind to 14-3-3 proteins was assessed by incubating the blots with DIG-labelled 14-3-3.
Among the stimuli tested, the 14-3-3-binding signal was strongest for PDE3A from cells treated with the phorbol ester PMA (Figure 2A, and results not shown). In addition to its well-characterized ability to activate isoforms of PKC [33], PMA was found to stimulate the phosphorylation of both SAPK2a (stress-activated protein kinase 2a)/p38α and p42/p44 MAPK (mitogen-activated protein kinase) [ERK1 (extracellular-signal-regulated kinase 1)/ERK2] in HeLa cells (Figure 2A), and MAPKAPK2 (MAPK-activated protein kinase-2), a downstream target of SAPK2/p38 (results not shown). However, there are several reasons why neither SAPK2/p38 nor MAPK can be responsible for the PMA-induced binding of PDE3A to 14-3-3 proteins. First, UV and TNFα (tumour necrosis factor α) did not promote the 14-3-3 binding of PDE3A to the same level as did PMA, even though UV or TNFα both activated SAPK2a/p38α (Figure 2A). Neither was the PDE3A/14-3-3 interaction stimulated by EGF (epidermal growth factor), which promoted the phosphorylation and activation of p42/p44 MAPK (Figure 2A). Furthermore, the binding of PDE3A to 14-3-3 proteins in response to PMA was unaffected by the drugs SB 203580, which inhibited SAPK2/p38 ([34,35], and results not shown), BIRB0796, which also inhibited SAPK2/p38 isoforms [36] (Figure 2B) and/or PD 184352, which inhibited the phosphorylation of the p42 and p44 MAPKs [35] (Figure 2B). The response of PDE3A to PMA was, however, strongly inhibited by H-7 and Ro 318220 (Figure 2B), which inhibit a number of other protein kinases, including the PMA-activated PKC ([35], and http://www.emdbiosciences.com/product/371955 and references cited therein).
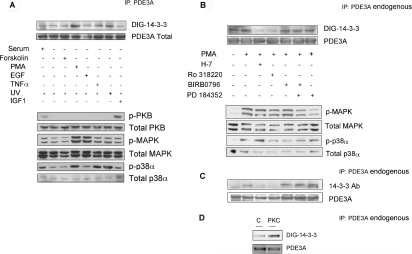
Figure 2. Binding of 14-3-3 proteins to PDE3A immunoprecipitated from PMA-stimulated HeLa cells.
(A) Cells growing in medium containing serum were harvested (serum), or serum-starved for 12 h, then stimulated in the following ways: forskolin (20 μM for 30 min), PMA (400 ng/ml for 15 min), epidermal growth factor (EGF; 50 ng/ml for 15 min), TNFα (10 μg/ml for 15 min), UV light (20 mJ/cm2 followed by 30 min incubation in serum-free medium), IGF1 (50 ng/ml for 20 min). PDE3A immunoprecipitated (IP) from 1.5 mg of cell extract was analysed by DIG–14-3-3 overlay and Western blotting with the anti-C-terminal-PDE3A antibody (top two panels). The six lower panels show the results of probing cell extracts (30 μg) with phosphospecific antibodies against pSer473 on PKBα (p-PKB), total PKB, the active phosphorylated forms of MAPK (p-MAPK), total MAPK, phosphorylated p38α (p-p38α) and total p38α. (B) Cells growing in medium containing serum were harvested (serum), or serum-starved for 12 h. Where indicated, cells were incubated with H-7 (100 μM), Ro 318220 (1 μM), SB 203580 (10 μM), PD 184352 (2 μM) or no inhibitor for 1 h, or BIRB0796 (10 μM) for 2 h, before stimulation with PMA (400 ng/ml) for a further 15 min. PDE3A immunoprecipitated (IP) from 3 mg of each cell extract was analysed by DIG–14-3-3 overlay (top panel) and Western blotting with anti-C-terminal-PDE3A antibody (second panel). The four lower panels show the results of probing cell extracts (30 μg) with phosphospecific antibodies against the active phosphorylated forms of MAPK (p-MAPK), total MAPK, phosphorylated p38α (p-p38α) and total p38α. (C) As for (B), except the anti-PDE3A immunoprecipitates (IP) were analysed by Western blotting to detect co-precipitating 14-3-3 proteins (using the sheep anti-14-3-3 antibody, see the Experimental section). (D) Direct binding of 14-3-3 proteins after phosphorylation of PDE3A with PKC. PDE3A was immunoprecipitated (IP) from cells that had been serum-starved for 12 h. The PDE3A was incubated in the presence of PKC (1 unit/ml rat brain PKC) with MgATP for 30 min at 30 °C, and analysed by DIG–14-3-3 overlays. The lane marked C is a control with no PKC. In other control experiments, PKC had no effect on PDE3A in the absence of Mg-ATP.
Consistent with the DIG–14-3-3 overlays (Figures 2A and 2B), the ability of PMA to induce the interaction between PDE3A and 14-3-3 proteins was confirmed by experiments in which PDE3A was immunoprecipitated, followed by immunoblotting to detect co-precipitated 14-3-3 proteins (Figure 2C). These experiments also confirmed that the PMA-induced PDE3A–14-3-3 interaction was selectively diminished when cells were treated with H-7 and Ro 318220 (Figure 2C).
In contrast with PDE3B [11], PFK-2 (fructose-2,6-bisphosphate kinase/phosphatase) [37] and other HeLa cell proteins whose binding to 14-3-3 proteins is promoted by PKB phosphorylation (K. M. Geraghty, J. Murphy, B. H. C. Wong and C. MacKintosh, unpublished work), 14-3-3 proteins did not bind well to PDE3A that was immunoprecipitated from extracts of cells stimulated with IGF1, in which PKB was phosphorylated and activated (Figure 2A, and results not shown for other time points after IGF1 stimulation).
Together, these findings suggested that PKC isoforms, or a kinase activated downstream of PKC, were responsible for phosphorylating at least one 14-3-3-binding site(s) on PDE3A, although the involvement of other protein kinases cannot be ruled out. Consistent with a role for PKC, when PDE3A from unstimulated cells was phosphorylated in vitro with PKC, the binding of the PDE to 14-3-3 proteins was markedly enhanced (Figure 2D). Similar results were obtained with the catalytic subunit of PKC from rat brain (Figure 2D) or recombinant human PKCα (results not shown).
Binding of PDE3A to 14-3-3 proteins is blocked by the DNA replication inhibitors aphidicolin and mimosine, but the PDE3A–14-3-3 interaction does not change during the cell cycle
cAMP regulates cell-cycle progression by multiple mechanisms, prompting us to investigate whether the phosphorylation and 14-3-3 binding of PDE3A was regulated in a cell-cycle-dependent manner. We found that the signal for 14-3-3 binding to PDE3A was lost when cells were treated with the DNA replication inhibitors, aphidicolin and mimosine, when assessed by either 14-3-3 overlay assays (Figure 3A), or co-immunoprecipitation of cellular 14-3-3 proteins with anti-PDE3A (Figure 3B). While these inhibitors block the cell cycle at S-phase, their effect on PDE3A does not seem to be related to cell-cycle regulation. We observed no change in the PDE3A–14-3-3 interaction when cells were synchronized in metaphase with nocodozole, and the synchronized cells were allowed to progress through S-phase and into G2 (results not shown). Thus aphidicolin and mimosine must act on PDE3A by some other mechanism. For example, these compounds may activate a PDE3A phosphatase.
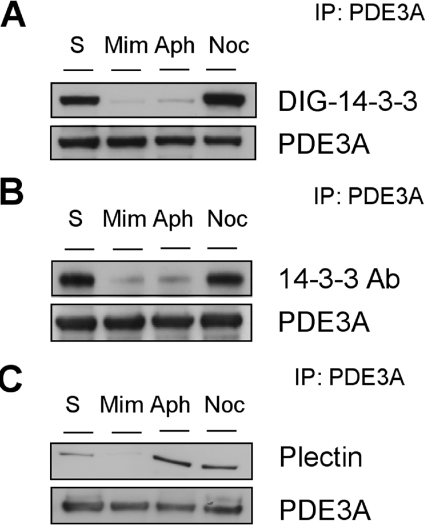
Figure 3. Effect of mimosine and aphidicolin on 14-3-3 binding to PDE3A.
(A) Cells growing in medium containing serum were harvested (S), incubated with mimosine (Mim; 0.2 mM for 16 h), aphidicolin (Aph; 10 μg/ml for 16 h) or nocodozole (Noc; 40 ng/ml for 16 h). PDE3A was immunoprecipitated (IP) from 3 mg of cell extract, and the immunoprecipitated PDE3A was analysed by DIG–14-3-3 overlay (upper panel) and detected with Western blotting with anti-PDE3A antibody (lower panel). (B) As for (A), except the anti-PDE3A immunoprecipitates were analysed by Western blotting to detect co-precipitating 14-3-3 proteins (using the sheep anti-14-3-3 antibody (Ab), see the Experimental section). (C) As for (A) and (B), except the SDS gels were 3–8% Tris/acetate and the anti-PDE3A immunoprecipitates were analysed by Western blotting to detect co-precipitating plectin. Note that, compared with other Figures, the 14-3-3 binding signals for PDE3A from serum-grown cells are stronger here because the blots were exposed in ECL® reagent for longer to highlight the contrast with the diminished signals caused by mimosine and aphidicolin.
Phosphorylation site mapping of PDE3A using IMAC (immobilized metal ion affinity chromatography) and MS
In larger-scale experiments, immunoprecipitated PDE3A from cell extracts was visible as a strong Coomassie-Blue-staining protein band (Figure 4A). When the immunoprecipitated PDE3A from cells stimulated with PMA was excised, digested with trypsin and analysed by MALDI–TOF-MS, no phosphopeptides were observed (Figure 4B, top panel). However, after IMAC of the tryptic digest, MALDI–TOF analysis revealed a number of new peaks whose masses corresponded to phosphorylated peptides (Figure 4B, bottom panel). Using MALDI–TOF/TOF (MS/MS), two of these peptides were identified as pS428LPPGLLR435 and A564LTYpTQSAPDLSPQILTPPVICSSCGR590 (where pT is pThr568).
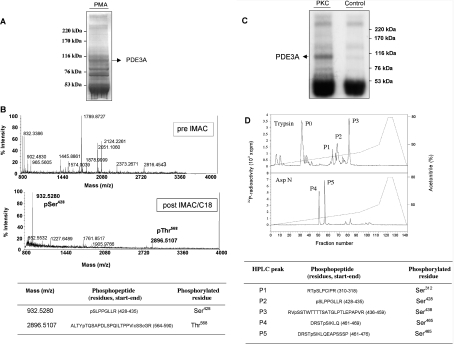
Figure 4. Sites on PDE3A that are phosphorylated in cells stimulated with PMA and by in vitro phosphorylation with PKC.
(A) PDE3A was immunoprecipitated from 30 mg of extract from cells that had been stimulated with PMA. The immunoprecipitate was run on SDS/PAGE using a 3–8% Tris/acetate gel. Molecular-mass sizes are indicated in kDa. (B) The immunoprecipitated PDE3A from (A) was excised from the gel and digested with trypsin, and the peptides were analysed by MALDI–TOF-MS. The mass spectra are shown before (upper spectrum) and after (lower spectrum) IMAC/C18 chromatography to enrich for phosphopeptides. In the sequences, c represents a half-cystine from an internal disulphide bond in this peptide. (C) Identification of the sites on PDE3A phosphorylated in vitro by PKC. PDE3A immunoprecipitated from serum-starved cells was incubated for 30 min at 30 °C with Mg[γ-32P]ATP in the presence of 1 unit/ml PKC (rat brain), and subjected to SDS/PAGE (3–8% Tris/acetate gel) and autoradiography. Molecular-mass sizes are indicated in kDa. (D) For the upper trace, the 32P-labelled PDE3A was excised from the gel, digested with trypsin and the peptides applied to a C18 HPLC column [Vydac 218TP5215 C18 column in 0.1% (v/v) trifluoroacetic acid in water]. The column was developed with an acetonitrile gradient (broken line), and 32P-radioactivity counted by an online Berthold LD509 detector is shown by the solid line. The phosphopeptides P0–P3 were eluted at 6.3% (P0), 12.6% (P1), 13.6% (P2) and 16% (P3) acetonitrile. P0 could not be identified, so, in the lower trace, another sample of 32P-labelled PDE3A was digested with the protease Asp-N, and two peaks of 32P-radioactivity were identified, termed P4 at 10% and P5 at 11% acetonitrile. The identities of phosphopeptides P1 to P5 are given in the Table. All residues were identified by a combination of MALDI–TOF, MS/MS and solid-phase Edman sequencing. Note that in the Asp-N protease digest, the pSer312, pSer428 and pSer438 sites are predicted to be within two long peptides (49 and 62 residues), and peptides containing these sites were not recovered from the reverse-phase HPLC column. It is possible that a tryptic peptide containing pSer465 had been present in P0 as STpSIK, as solid-phase Edman sequencing of P0 released counts in cycle 3.
PDE3A from serum-grown HeLa cells was immunoprecipitated and used as a substrate for in vitro phosphorylation by PKC using [γ-32P]ATP. Tryptic digests of the phosphorylated PDE3A were separated by HPLC and 32P-labelled peptides analysed by MALDI–TOF-MS and solid-phase Edman sequencing (Figures 4C and 4D). The HPLC profile showed four major radio-active phosphopeptides, P0–P3 (Figure 4D, top panel). The MALDI–TOF mass spectrum of P1 contained ions whose masses matched the mono-phosphorylated peptide R310TpSLPCIPR318 of PDE3A and those of the poorly resolved daughter ion characteristic of the loss of H3PO4 from the phosphopeptide in a post-source decay event. When subject to solid-phase Edman degradation, Cerenkov counting showed that [32P]Pi was released at cycle 3, which means that the residue corresponding to Ser312 was phosphorylated (Figure 4D). Using similar methods, P2 was identified as pS428LPPGLLR435 and P3 was V437pSSTWTTTTSATGLPTLEPAPVR459 (where pS is pSer438). The residues in PDE3A that precede each of these peptides are arginine or lysine, consistent with the specificity of trypsin. Phosphopeptide P0 was in a complex mixture of peptides and could not be identified. However, when the PKC-phosphorylated PDE3A was digested with the Asp-N protease, two radioactive peaks, P4 and P5, were identified by HPLC (Figure 4D). MALDI–TOF-MS and solid-phase sequencing showed that P4 represented the peptide D461RSTpSIKLQ469 (where pS is pSer465), while P5 was a longer version of the same phosphopeptide, D461RSTpSIKLQEAPSSSP476 (Figure 4D).
Overall, these data suggested that pSer428 was a potential 14-3-3-binding site because this residue was phosphorylated in both PDE3A from PMA-stimulated cells (Figures 4A and 4B) and PDE3A phosphorylated with PKC in vitro (Figures 4C and 4D), both conditions where 14-3-3 binding to PDE3A was enhanced (Figure 2). However, the IMAC and MALDI–MS analyses have limitations. The absence of a particular phosphopeptide does not necessarily mean that the corresponding site was not phosphorylated in the protein, because not all phosphopeptides bind to IMAC resins [38]. In addition, signals from phosphorylated peptides can be differentially suppressed in MALDI–MS, depending on the characteristics of the other phosphopeptides in the mixture, making it impossible to assess phosphopeptides quantitatively using IMAC and MALDI–MS analysis alone. For these reasons, we raised phosphospecific antibodies to assess how phosphorylation of the various sites on PDE3A changes in response to cell stimuli.
PMA promotes phosphorylation of Ser428 and binding of 14-3-3 proteins, while mimosine and aphidicolin promote dephosphorylation of sites including Ser428 and loss of 14-3-3 binding
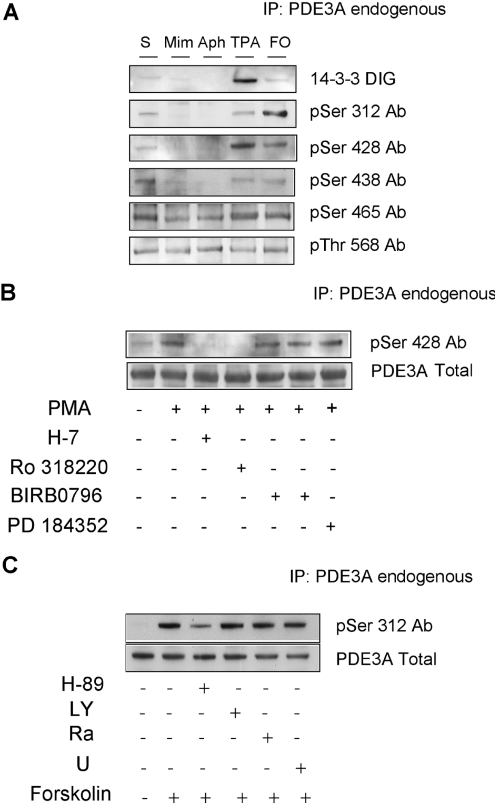
Phosphospecific antibodies were raised against five phosphopeptides that encompass the Ser312, Ser428, Ser438, Ser465 and Thr568 phosphorylation sites on PDE3A (see the Experimental section). The specificity of these antibodies was first established with phosphopeptides and unphosphorylated peptides spotted on to nitrocellulose. Each antibody was tested against each peptide. Where some cross-reactivity was observed, this could be eliminated by pre-incubating the antibodies with the cross-reacting peptides. Supplementary Figure 1 (http://www.BiochemJ.org/bj/392/bj3920163add.htm) shows blots using the optimal conditions (see the legend) where each antibody performed well in selectively detecting only its own immunogen phosphopeptide.
These antibodies were used to study the site-specific phosphorylation of PDE3A after stimulating HeLa cells in ways that had been found to cause no binding of PDE3A to 14-3-3 proteins (mimosine and aphidicolin), partial binding of PDE3A to 14-3-3 proteins (serum) and maximal binding of 14-3-3 proteins (PMA). PMA was found to markedly enhance the phosphorylation of Ser428, in parallel with increased 14-3-3 binding (Figure 5A), and both events increased in the first 20 min after PMA stimulation and were sustained for at least 2 h (results not shown). There was partial phosphorylation at this site and partial 14-3-3 binding of PDE3A from cells growing in serum, while mimosine and aphidicolin abolished both phosphorylation of Ser428 and the ability of PDE3A to bind to 14-3-3 proteins. The DNA replication blockers also diminished phosphorylation at Ser312, Ser438 and Ser465 (Figure 5A). In contrast with these regulated phosphorylations, the phosphorylation status of Thr568 was unchanged under all the conditions tested (Figure 5B, and results not shown).
Figure 5. Use of phosphospecific antibodies to map the sites on PDE3A that are phosphorylated in response to different cellular stimuli.
(A) Cells growing in medium containing serum were harvested (serum), or incubated with mimosine (Mim; 0.2 mM for 16 h), aphidicolin (Aph; 10 μg/ml for 16 h), or serum-starved for 12 h and incubated with PMA (TPA; 400 ng/ml) or forskolin (FO; 20 μM). PDE3A immunoprecipitated (IP) from 1.5 mg of cell extract was analysed by DIG–14-3-3 overlay and Western blotting with antibodies (Ab) that recognize the pSer312, pSer428, pSer438, pSer465 and pThr568 sites on PDE3A. The antibodies were incubated with the blots under the optimal conditions established in peptide dot blots (see Supplementary Figure 1 at http://www.BiochemJ.org/bj/392/bj3920163add.htm) as follows: the anti-pSer312 antibody was used in the absence of any competitor peptides; antipSer428 antibody was incubated in the presence of 20 μg/ml KRLRRSLPPGL; anti-pSer438 antibody was incubated in the presence of 20 μg/ml LRRVSSTWTT; anti-pSer465 antibody was incubated in the presence of 10 μg/ml each of RDRSTSIKLQEAP, KRLRRpSLPPGL and KRLRRSLPPGL; anti-pThr568 antibody was incubated in the presence of 20 μg/ml HRALTYTQSAPDL. (B) Cells growing in medium containing serum were serumstarved for 12 h. Where indicated, cells were incubated with H-7 (100 μM), Ro 318220 (1 μM), PD 184352 (2 μM) or no inhibitor for 1 h, or BIRB0796 (10 μM) for 2 h, before stimulation with PMA (400 ng/ml) for a further 15 min. PDE3A immunoprecipitated (IP) from 3 mg of each cell extract was analysed by Western blotting with anti-pSer428 antibody (Ab) (upper panel) and anti-C-terminal-PDE3A antibody (lower panel). (C) Cells were serum-starved for 12 h and stimulated with forskolin. Where indicated, cells were incubated with H-89 (10 μM), LY 294002 (LY; 100 μM), rapamycin (Ra; 100 nM) or UO126 (U; 10 μM) for 1 h, before stimulation with forskolin (20 μM) for a further 30 min. PDE3A was immunoprecipitated (IP) from 1.5 mg of each cell extract, and the immunoprecipitated PDE3A was analysed by Western blotting with the antibody that specifically recognizes phosphorylated Ser312 of PDE3A (upper panel) and Western blotting with anti-C-terminal-PDE3A antibody (lower panel).
Further experiments showed that phosphorylation of Ser428 was abolished when PMA-stimulated cells were incubated with H-7 or Ro 318220, while H-89, SB 203580, BIRB0796 and PD 184352 had no effect on PMA-induced phosphorylation of Ser428 (Figure 5B, and results not shown).
Together, these analyses support the hypothesis that pSer428 on PDE3A is the major phosphorylated site that is responsible for the PMA-induced increase in 14-3-3 binding to the enzyme. These findings are also consistent with Ser428 being phosphorylated in vivo by a PKC or PKC-activated protein kinase.
Phosphorylation of Ser312 of PDE3A in response to forskolin
Treatment of HeLa cells with forskolin activates adenylate cyclase activity and thus increases intracellular cAMP levels, leading to the activation of PKA. Forskolin did not promote the binding of 14-3-3 proteins to PDE3A (Figures 2A and 5A). However, using the pSer312-PDE3A antibodies, we found that forskolin stimulated the phosphorylation of PDE3A at Ser312 (Figures 5A and 5C). This residue lies within a sequence SHRRTS312LPCIP that conforms to a consensus site for PKA (RRXASXB, where, for optimal recognition, XA is not negatively charged and XB is hydrophobic). The forskolin-stimulated phosphorylation of Ser312 was inhibited by the PKA inhibitor H-89 (Figure 5C), and by mimosine and aphidicolin (Figure 5A), but not by inhibitors of PI3K (LY 294002), mTOR (mammalian target of rapamycin) (rapamycin) or the MAPK cascade (U0126) (Figure 5C; see [35] for inhibitor specificities), nor by H-7 (results not shown).
Plectin is co-immunoprecipitated with PDE3A, independently of 14-3-3 binding
In the larger-scale PDE3A immunoprecipitation experiments, additional higher-molecular-mass proteins were observed on Coomassie-Blue-stained gels, including a protein of approx. 500 kDa that was identified as the cytoskeletal linker protein, plectin (results not shown). Western blotting experiments with two different anti-plectin antibodies confirmed that plectin was co-immunoprecipitated from cell extracts with anti-PDE3A antibodies (Figure 3C). The binding of plectin to PDE3A appeared to occur independently of the PDE3A–14-3-3 interaction, because plectin was present in the anti-PDE3A immunoprecipitates from both cells treated with PMA, where PDE3A bound well to 14-3-3 proteins, and unstimulated cells, in which the PDE3A displayed little or no 14-3-3 binding (results not shown). However, the interaction between PDE3A and plectin seemed to be disrupted by incubation of cells with mimosine, although not with the other DNA replication inhibitor, aphidicolin (Figure 3C).
DISCUSSION
In the present study, the finding that 14-3-3 proteins bind to a phosphorylated ∼105 kDa form of PDE3A inside HeLa cells led to a full analysis of multisite phosphorylation of PDE3A in response to various cellular stimuli, using a battery of techniques including IMAC, MS, in vitro 32P-labelling coupled with solid-phase sequencing and the use of phosphospecific antibodies that recognize five phosphorylated sites on PDE3A.
In overlay assays, 14-3-3 proteins discriminated between PDE3A from mimosine- and aphidicolin-treated HeLa cells (no 14-3-3 binding), unstimulated serum-starved cells or cells stimulated with serum, UV, TNFα or forskolin (partial 14-3-3 binding), and cells stimulated with PMA (full 14-3-3 binding). The full 14-3-3 binding in response to PMA appears to be due to activation of PKC, as it was blocked by the non-specific inhibitors of PKC, H-7 and Ro 318220, but not by inhibitors of two other kinases that were also activated by PMA in HeLa cells, namely SAPK2/p38 and p42/p44 MAPK.
Ser428 was pinpointed as the PMA-regulated phosphorylation site that promotes maximal binding of 14-3-3 proteins to PDE3A (Figure 6). However, other phosphorylated sites may also contribute to 14-3-3 binding. 14-3-3 proteins function as dimers, and therefore have two sites with the potential to bind phosphorylated residues. It is possible that a 14-3-3 dimer can bind with low affinity to a site that is phosphorylated in cells growing in serum, but that the PMA-phosphorylated pSer428 site binds to the second site on the 14-3-3 dimer to generate a high-affinity interaction. In displaying this sort of graded binding response, 14-3-3 proteins could act as a ‘signal integrators’ that bind to their target most strongly when signals from two different kinases are detected. Multisite phosphorylation by one or more protein kinases is a widely recognized feature of cellular regulation [39], which recent advances in mass spectrometric techniques is making even more apparent. Our findings indicate the potential of 14-3-3 proteins in ‘interpreting’ different patterns of multisite phosphorylation.
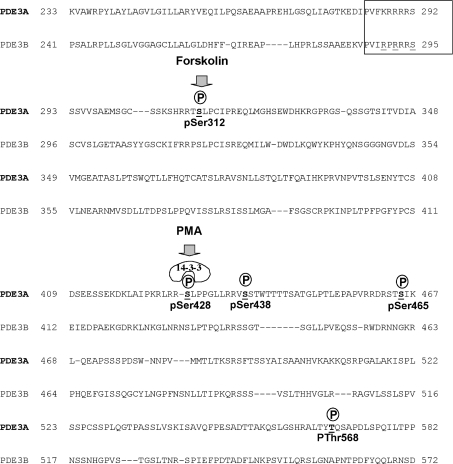
Figure 6. Amino acid sequences of human PDE3A and PDE3B within the regulatory phosphorylated and 14-3-3-binding domains.
The amino acid sequences of human PDE3A and PDE3B within the regulatory phosphorylated domain are shown. The boxed region highlights the comparison between PDE3A and PDE3B at a site analogous to a reported PKB phosphorylation site on PDE3B from rodents [10]. This sequence fits the canonical PKB consensus in PDE3B (RXRXXS), but not in PDE3A (KXRXXS). The phosphorylated sites on PDE3A that were identified in the present study are shown, including the forskolin-stimulated Ser312 phosphorylation site, and the PMA-induced phosphorylation of Ser428, which is linked to 14-3-3 binding.
While the combined use of phorbol ester and non-selective PKC inhibitors indicate PKC involvement, the PKC family comprises many isoforms [33], and our studies do not allow us to distinguish which members of this extensive kinase family may regulate PDE3A. Nevertheless, our findings of regulatory connections linking the PKCs and PDE3A are particularly intriguing because both activities have been implicated in promoting the activation and aggregation of platelets. Thus, when platelets were incubated with PDE3 inhibitors, intracellular cAMP levels rose, and platelet activation and aggregation were inhibited [12,25,26]. Many platelet receptor agonists activate PKC isoforms and downstream targets, including PKD (protein kinase D). These PKC-dependent events have been implicated as being functionally important in platelet activation [40,41]. It is therefore tempting to speculate that our findings could connect these observations if platelet agonists activate PKC leading to the phosphorylation and 14-3-3 binding of PDE3A, thereby activating PDE3A and keeping intracellular cAMP levels low, which would tend to promote platelet aggregation and activation. While only a hypothesis at the moment, our antibodies and 14-3-3-affinity reagents should be very useful for dissecting the regulation of PDE3A in platelets and other cell types, particularly combined with developments in more specific pharmacological probes and mutant mice to distinguish the actions of members of the PKC/PKD family [33].
In addition to PKC, p38/SAPK2 was strongly activated by PMA in HeLa cells, which may mean that some effects that have been attributed to PKCs on the basis of phorbol ester regulation may actually be mediated by SAPK2/p38.
We identified Ser312 as a site that becomes phosphorylated in cells in response to the adenylate cyclase activator forskolin, and this phosphorylation was prevented by H-89, indicating that PKA is the Ser312 kinase (Figure 6). However, phosphorylation of Ser312 did not stimulate 14-3-3 binding, even though this site looks like a classical mode I 14-3-3-binding phosphopeptide motif (http://scansite.mit.edu/motifsca_seq.phtml). We did not identify any 14-3-3 binding to PDE3A from cells in which PKB was activated by stimulation with IGF1, which we checked because PDE3B has been reported bind to 14-3-3 proteins when phosphorylated by PKB in insulin-stimulated cells [9–11]. One of the potential PKB sites on PDE3B is highlighted by the boxed sequence in Figure 6. This sequence conforms to a classical PKB site (RXRXXpS) in rat, mouse and human PDE3Bs, while, in the PDE3A from these species, a lysine residue takes the place of the first arginine (Figure 6), making this site unlikely to be phosphorylated with PKB, which is consistent with our findings. Our results do not exclude the possibility that PKB phosphorylates PDE3A at a site that does not promote binding to 14-3-3 proteins. Perhaps the discovery of multisite phosphorylation of PDE3A means that the regulatory phosphorylation sites on PDE3B warrant further analysis.
We performed several experiments to determine the functions of phosphorylation and 14-3-3 binding on PDE3A, including assays of enzyme activity and intracellular cAMP levels, but have not detected any changes thus far. While we were able to express active soluble forms (wild-type and phosphorylation site mutants) of PDE3A as GFP (green fluorescent protein) fusions in cells, we found that the transfected protein did not get phosphorylated or bind 14-3-3 proteins in the same way as the endogenous protein did in response to PMA. These findings precluded the use of these constructs to facilitate functional studies. It is possible that the GFP–PDE3A protein was not localized properly in the cells, and/or that phosphorylation/14-3-3 binding of PDE3A has very localized intracellular effects that cannot be seen with the isolated enzyme, or by looking at global cAMP levels. In this connection, we were interested to find that the cytoskeletal linker protein plectin-1 co-precipitates with PDE3A from extracts of HeLa cells. Plectin is a >500 kDa linker protein that can interact with all three intracellular fibrous networks (actin, microtubules and intermediate filaments), integrin β subunits (in competition with actin) and other membrane and signalling proteins [31,42]. Our finding is intriguing because PDE3A seems to regulate a sub-pool of cAMP that controls the PKA phosphorylation of proteins that regulate cytoskeletal dynamics. Thus inhibitors of PDE3, but not PDE2, promoted phosphorylation of the PKA sites on the actin-regulating protein VASP [25,27,28], and inhibitors of PDE3A inhibit platelet aggregation, a process involving regulation of integrin αIIbβ3 affinity [12]. Perhaps PDE3A hydrolyses a sub-pool of cAMP in a spatial localization that is influenced by plectin, and that connects PDE3A to its downstream cAMP/PKA-regulated targets in actin dynamics.
Online data
Acknowledgments
We thank the European Community programme ‘Quality of Life and Management of Living Resources’ for a Marie Curie Fellowship to M.P.R. under contract number QLK1-CT-2000-51184, and the U.K. Biotechnology and Biological Sciences Research Council (BBSRC) and U.K. Medical Research Council (MRC) for research grants. This study was supported by the Division of Signal Transduction Therapy (DSTT) at the University of Dundee, with funding from AstraZeneca, Boehringer Ingelheim, GlaxoSmithKline, Merck and Co., Merck KGaA and Pfizer. Thanks to the DSTT protein production and antibody purification teams co-ordinated by Hilary McLauchlan and James Hastie for affinity purification of antibodies, Leanne Brown and Colin Bell for tissue culture support, the Sequencing Service (School of Life Sciences, University of Dundee; http://www.dnaseq.co.uk) for DNA sequencing, and the Post Genomics and Molecular Interactions Centre for MS facilities. Thanks also to the laboratory group of Professor Pauline Schaap for help with cAMP assays.
References
- 1.Francis S. H., Turko I. V., Corbin J. D. Cyclic nucleotide phosphodiesterases: relating structure and function. Prog. Nucleic Acid Res. Mol. Biol. 2001;65:1–52. doi: 10.1016/s0079-6603(00)65001-8. [DOI] [PubMed] [Google Scholar]
- 2.Choi Y. H., Ekholm D., Krall J., Ahmad F., Degerman E., Manganiello V. C., Movsesian M. A. Identification of a novel isoform of the cyclic-nucleotide phosphodiesterase PDE3A expressed in vascular smooth-muscle myocytes. Biochem. J. 2001;353:41–50. [PMC free article] [PubMed] [Google Scholar]
- 3.Shakur Y., Takeda K., Kenan Y., Yu Z. X., Rena G., Brandt D., Houslay M. D., Degerman E., Ferrans V. J., Manganiello V. C. Membrane localization of cyclic nucleotide phosphodiesterase 3 (PDE3): two N-terminal domains are required for the efficient targeting to, and association of, PDE3 with endoplasmic reticulum. J. Biol. Chem. 2000;275:38749–38761. doi: 10.1074/jbc.M001734200. [DOI] [PubMed] [Google Scholar]
- 4.Scapin G., Patel S. B., Chung C., Varnerin J. P., Edmondson S. D., Mastracchio A., Parmee E. R., Singh S. B., Becker J. W., Van der Ploeg L. H., Tota M. R. Crystal structure of human phosphodiesterase 3B: atomic basis for substrate and inhibitor specificity. Biochemistry. 2004;43:6091–6100. doi: 10.1021/bi049868i. [DOI] [PubMed] [Google Scholar]
- 5.Shitsukawa K., Anderson C. B., Richard F. J., Horner A. K., Wiersma A., van Diun M., Conti M. Cloning and characterization of the cyclic guanosine monophosphate-inhibited phosphodiesterase PDE3A expressed in mouse oocyte. Biol. Reprod. 2001;65:188–196. doi: 10.1095/biolreprod65.1.188. [DOI] [PubMed] [Google Scholar]
- 6.Masciarelli S., Horner K., Liu C., Park S. H., Hinckley M., Hockman S., Nedachi T., Jin C., Conti M., Manganiello V. Cyclic nucleotide phosphodiesterase 3A-deficient mice as a model of female infertility. J. Clin. Invest. 2004;114:196–205. doi: 10.1172/JCI21804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kieback A. G., Iven H., Stolzenburg K., Eichner E., Ruckdeschel W., Baumann G. Pharmacokinetics and hemodynamic effects of the phosphodiesterase III inhibitor saterinone in patients with chronic heart failure. Int. J. Cardiol. 2003;91:201–208. doi: 10.1016/s0167-5273(03)00030-5. [DOI] [PubMed] [Google Scholar]
- 8.Movsesian M. A. PDE3 inhibition in dilated cardiomyopathy: reasons to reconsider. J. Card. Failure. 2003;9:475–480. doi: 10.1016/s1071-9164(03)00135-0. [DOI] [PubMed] [Google Scholar]
- 9.Ahmad F., Cong L.-N., Holst L. S., Wang L.-M., Landstrom T. R., Pierce J. H., Quon M. J., Degerman E., Manganiello V. C. Cyclic nucleotide phosphodiesterase 3B is a downstream target of protein kinase B and may be involved in regulation of effects of protein kinase B on thymidine incorporation in FDCP2 cells. J. Immunol. 2000;164:4678–4688. doi: 10.4049/jimmunol.164.9.4678. [DOI] [PubMed] [Google Scholar]
- 10.Kitamura T., Kitamura Y., Kuroda S., Hino Y., Ando M., Kotani K., Konishi H., Matsuzaki H., Kikkawa U., Ogawa W., Kasuga M. Insulin-induced phosphorylation and activation of cyclic nucleotide phosphodiesterase 3B by the serine-threonine kinase Akt. Mol. Cell. Biol. 1999;19:6286–6296. doi: 10.1128/mcb.19.9.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Onuma H., Osawa H., Yamada K., Ogura T., Tanabe F., Granner D. K., Makino H. Identification of the insulin-regulated interaction of phosphodiesterase 3B with 14-3-3β protein. Diabetes. 2002;51:3362–3367. doi: 10.2337/diabetes.51.12.3362. [DOI] [PubMed] [Google Scholar]
- 12.Feijge M. A., Ansink K., Vanschoonbeek K., Heemskerk J. W. Control of platelet activation by cyclic AMP turnover and cyclic nucleotide phosphodiesterase type-3. Biochem. Pharmacol. 2004;67:1559–1567. doi: 10.1016/j.bcp.2003.12.028. [DOI] [PubMed] [Google Scholar]
- 13.Bilodeau-Goeseels S. Effects of phosphodiesterase inhibitors on spontaneous nuclear maturation and cAMP concentrations in bovine oocytes. Theriogenology. 2003;60:1679–1690. doi: 10.1016/s0093-691x(03)00173-0. [DOI] [PubMed] [Google Scholar]
- 14.Degerman E., Manganiello V., Holst J. J., Ahren B. Milrinone efficiently potentiates insulin secretion induced by orally but not intravenously administered glucose in C57BL6J mice. Eur. J. Pharmacol. 2004;498:319–323. doi: 10.1016/j.ejphar.2004.07.096. [DOI] [PubMed] [Google Scholar]
- 15.Harndahl L., Wierup N., Enerback S., Mulder H., Manganiello V. C., Sundler F., Degerman E., Ahren B., Holst L. β-Cell-targeted overexpression of phosphodiesterase 3B in mice causes impaired insulin secretion, glucose intolerance, and deranged islet morphology. J. Biol. Chem. 2004;279:15214–15222. doi: 10.1074/jbc.M308952200. [DOI] [PubMed] [Google Scholar]
- 16.Friis U. G., Jensen B. L., Sethi S., Andreasen D., Hansen P. B., Skott O. Control of renin secretion from rat juxtaglomerular cells by cAMP-specific phosphodiesterases. Circ. Res. 2002;90:996–1003. doi: 10.1161/01.res.0000017622.25365.71. [DOI] [PubMed] [Google Scholar]
- 17.Wijkander J., Landstrom T. R., Manganiello V., Belfrage P., Degerman E. Insulin-induced phosphorylation and activation of phosphodiesterase 3B in rat adipocytes: possible role for protein kinase B but not mitogen-activated protein kinase or p70 S6 kinase. Endocrinology. 1998;139:219–227. doi: 10.1210/endo.139.1.5693. [DOI] [PubMed] [Google Scholar]
- 18.Zhao A. Z., Zhao H., Teague J., Fujimoto W., Beavo J. A. Attenuation of insulin secretion by insulin-like growth factor 1 is mediated through activation of phosphodiesterase 3B. Proc. Natl. Acad. Sci. U.S.A. 1997;94:3223–3228. doi: 10.1073/pnas.94.7.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rahn T., Ronnstrand L., Leroy M. J., Wernstedt C., Tornqvist H., Manganiello V. C., Belfage P., Degerman E. Identification of the site in the cGMP-inhibited phosphodiesterase phosphorylated in adipocytes in response to insulin and isoproterenol. J. Biol. Chem. 1996;271:11575–11580. doi: 10.1074/jbc.271.19.11575. [DOI] [PubMed] [Google Scholar]
- 20.MacKintosh C. Dynamic interactions between 14-3-3 proteins and phosphoproteins regulate diverse cellular processes. Biochem. J. 2004;381:329–342. doi: 10.1042/BJ20031332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pozuelo Rubio M. P., Geraghty K. M., Wong B. H., Wood N. T., Campbell D. G., Morrice N., MacKintosh C. 14-3-3-affinity purification of over 200 human phosphoproteins reveals new links to regulation of cellular metabolism, proliferation and trafficking. Biochem. J. 2004;379:395–408. doi: 10.1042/BJ20031797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patrucco E., Notte A., Barberis L., Selvetella G., Maffei A., Brancaccio M., Marengo S., Russo G., Azzolino O., Rybalkin S. D., et al. PI3Kγ modulates the cardiac response to chronic pressure overload by distinct kinase-dependent and -independent effects. Cell. 2004;118:375–387. doi: 10.1016/j.cell.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 23.Feoktistov I., Murray J. J., Biaggioni I. Positive modulation of intracellular Ca2+ levels by adenosine A2b receptors, prostacyclin, and prostaglandin E1 via a cholera toxin-sensitive mechanism in human erythroleukemia cells. Mol. Pharmacol. 1994;45:1160–1167. [PubMed] [Google Scholar]
- 24.Colman R. W. Platelet cyclic adenosine monophosphate phosphodiesterases: targets for regulating platelet-related thrombosis. Semin. Thromb. Hemostasis. 2004;30:451–460. doi: 10.1055/s-2004-833480. [DOI] [PubMed] [Google Scholar]
- 25.Manns J. M., Brenna K. J., Colman R. W., Sheth S. B. Differential regulation of human platelet responses by cGMP inhibited and stimulated cAMP phosphodiesterases. Thromb. Haemostasis. 2002;87:873–879. [PubMed] [Google Scholar]
- 26.Sim D. S., Merrill-Skoloff G., Furie B. C., Furie B., Flaumenhaft R. Initial accumulation of platelets during arterial thrombus formation in vivo is inhibited by elevation of basal cAMP levels. Blood. 2004;103:2127–2134. doi: 10.1182/blood-2003-04-1133. [DOI] [PubMed] [Google Scholar]
- 27.Sudo T., Ito H., Kimura Y. Phosphorylation of the vasodilator-stimulated phosphoprotein (VASP) by the anti-platelet drug, cilostazol, in platelets. Platelets. 2003;14:381–390. doi: 10.1080/09537100310001598819. [DOI] [PubMed] [Google Scholar]
- 28.Jensen B. O., Selheim F., Doskeland S. O., Gear A. R., Holmsen H. Protein kinase A mediates inhibition of the thrombin-induced platelet shape change by nitric oxide. Blood. 2004;104:2775–2782. doi: 10.1182/blood-2004-03-1058. [DOI] [PubMed] [Google Scholar]
- 29.Macphee C. H., Reifsnyder D. H., Moore T. A., Lerea K. M., Beavo J. A. Phosphorylation results in activation of a cAMP phosphodiesterase in human platelets. J. Biol. Chem. 1988;263:10353–10358. [PubMed] [Google Scholar]
- 30.Meek S. E., Lane W. S., Piwnica-Worms H. Comprehensive proteomic analysis of interphase and mitotic 14-3-3-binding proteins. J. Biol. Chem. 2004;279:32046–32054. doi: 10.1074/jbc.M403044200. [DOI] [PubMed] [Google Scholar]
- 31.Smith F. J., Eady R. A., Leigh I. M., McMillan J. R., Rugg E. L., Kelsell D. P., Bryant S. P., Spurr N. K., Geddes J. F., Kirtschig G., et al. Plectin deficiency results in muscular dystrophy with epidermolysis bullosa. Nat. Genet. 1996;13:450–457. doi: 10.1038/ng0896-450. [DOI] [PubMed] [Google Scholar]
- 32.Campbell D. G., Morrice N. A. Identification of protein phosphorylation sites by a combination of mass spectrometry and solid phase Edman sequencing. J. Biomol. Techniques. 2002;13:119–130. [PMC free article] [PubMed] [Google Scholar]
- 33.Parker P. J., Murray-Rust J. PKC at a glance. J. Cell Sci. 2004;117:131–132. doi: 10.1242/jcs.00982. [DOI] [PubMed] [Google Scholar]
- 34.Cuenda A., Rouse J., Doza Y. N., Meier R., Cohen P., Gallagher T. F., Young P. R., Lee J. C. SB 203580 is a specific inhibitor of a MAP kinase homologue which is stimulated by cellular stresses and interleukin-1. FEBS Lett. 1995;364:229–233. doi: 10.1016/0014-5793(95)00357-f. [DOI] [PubMed] [Google Scholar]
- 35.Davies S. P., Reddy H., Caivano M., Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuma Y., Sabio G., Bain J., Shpiro N., Marquez R., Cuenda A. BIRB796 inhibits all p38 MAPK isoforms in vitro and in vivo. J. Biol. Chem. 2005;280:19472–19479. doi: 10.1074/jbc.M414221200. [DOI] [PubMed] [Google Scholar]
- 37.Pozuelo Rubio M., Peggie M., Wong B. H., Morrice N., MacKintosh C. 14-3-3s regulate fructose-2,6-bisphosphate levels by binding to PKB-phosphorylated cardiac fructose-2,6-bisphosphate kinase/phosphatase. EMBO J. 2003;22:3514–3523. doi: 10.1093/emboj/cdg363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morrice N. Protein phosphorylation site analysis using PHOS-Select enrichment prior to mass spectrometry. Sigma-Origins. 2005;18:24–25. [Google Scholar]
- 39.Cohen P. The regulation of protein function by multisite phosphorylation – a 25 year update. Trends Biochem. Sci. 2000;25:596–601. doi: 10.1016/s0968-0004(00)01712-6. [DOI] [PubMed] [Google Scholar]
- 40.Stafford M. J., Watson S. P., Pears C. PKD: a new protein kinase C dependent pathway in platelets. Blood. 2003;101:1392–1399. doi: 10.1182/blood-2002-08-2384. [DOI] [PubMed] [Google Scholar]
- 41.Calcerrada M. C., Latorre E., Mora-Gil M. V., Catalan R. E., Miguel B. G., Martinez A. M. Selective translocation of protein kinase C isozymes by PAF in rabbit platelets. Prostaglandins Other Lipid Mediators. 2005;75:35–46. doi: 10.1016/j.prostaglandins.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 42.Leung C. L., Green K. J., Liem R. K. Plakins: a family of versatile cytolinker proteins. Trends Cell. Biol. 2002;12:37–45. doi: 10.1016/s0962-8924(01)02180-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.