Abstract
Haem is essential for the health and function of nearly all cells. 5-Aminolaevulinic acid synthase-1 (ALAS-1) catalyses the first and rate-controlling step of haem biosynthesis. ALAS-1 is repressed by haem and is induced strongly by lipophilic drugs that also induce CYP (cytochrome P450) proteins. We investigated the effects on the avian ALAS-1 gene promoter of a phenobarbital-like chemical, Glut (glutethimide), and a haem synthesis inhibitor, DHA (4,6-dioxoheptanoic acid), using a reporter gene assay in transiently transfected LMH (Leghorn male hepatoma) hepatoma cells. A 9.1 kb cALAS-1 (chicken ALAS-1) promoter-luciferase-reporter construct, was poorly induced by Glut and not by DHA alone, but was synergistically induced by the combination. In contrast, a 3.5 kb promoter ALAS-1 construct was induced by Glut alone, without any further effect of DHA. In addition, exogenous haem (20 μM) repressed the basal and Glut- and DHA-induced activity of luciferase reporter constructs containing 9.1 and 6.3 kb of ALAS-1 5′-flanking region but not the construct containing the first 3.5 kb of promoter sequence. This effect of haem was subsequently shown to be dependent on the −6.3 to −3.5 kb region of the 5′-flanking region of cALAS-1 and requires the native orientation of the region. Two deletion constructs of this approx. 2.8 kb haem-repressive region (1.7 and 1.1 kb constructs) retained haem-dependent repression of basal and drug inductions, suggesting that more than one cis-acting elements are responsible for this haem-dependent repression of ALAS-1. These results demonstrate that there are regulatory regions in the 5′-flanking region of the cALAS-1 gene that respond to haem and provide a basis for further investigations of the molecular mechanisms by which haem down-regulates expression of the ALAS-1 gene.
Keywords: 5-aminolaevulinic acid synthase-1 (ALAS-1); 4,6-dioxoheptanoic acid (DHA); drug induction; glutethimide; haem; LMH cell line
Abbreviations: ALA, aminolaevulinic acid; ALAS, 5-aminolaevulinic acid synthase; cALAS-1, chicken ALAS-1; β-gal, β-galactosidase; DHA, 4,6-dioxoheptanoic acid; DR, hexamer half-site direct repeat; DRES, drug-responsive enhancer sequence; Glut, glutethimide; HO-1, haem oxygenase-1; LMH, Leghorn male hepatoma; 5′-UTR, 5′-untranslated region
INTRODUCTION
Haem biosynthesis occurs by a multi-step pathway that is present in virtually all cell types. In higher organisms, erythroid cells and hepatocytes are the major sites of haem biosynthesis [1,2]. 5-Aminolaevulinic acid synthase (ALAS) is the first and rate-controlling enzyme of haem biosynthesis in animals [3]. ALAS catalyses the condensation of glycine and succinyl-CoA to form ALA (5-aminolaevulinic acid) [4]. In higher organisms, including mammals and birds, ALAS is encoded by two genes: ALAS-1, which is the housekeeping form that is expressed ubiquitously, and ALAS-2, which is expressed only in erythrocytes and erythroid precursors [3,5,6].
Because haem is essential for the normal function of nearly all cells, defects in haem synthesis have far-reaching pathological and biochemical effects. For example, the acute porphyrias are a family of disorders characterized by defects in the production or activity of one of the enzymes in the haem biosynthesis pathway. In acute attacks, neuropsychiatric and visceral symptoms are precipitated by drugs, and appear to be associated with uncontrolled up-regulation of hepatic ALAS-1 [2,4,7]. Additionally, there are a number of acquired disorders of haem synthesis, including poisoning with heavy metals such as lead, in which similar symptoms and biochemical abnormalities occur [2,4].
Haem synthesis and degradation within the cell are tightly controlled so that the amount of available haem closely matches the cell's requirements. In hepatocytes, a major control point in haem synthesis occurs via loosely bound, unassigned haem in a regulatory haem pool, which may repress ALAS-1 by a number of negative feedback mechanisms, including reduction of transcription [7–10], reduction of mRNA stability [11–13] and inhibition of the transport of ALAS-1 into mitochondria [14–17]. Direct inhibition of hepatic ALAS activity by haem appears unlikely at physiological haem concentrations [18]. However, the molecular mechanism by which haem regulates ALAS-1 mRNA levels remains poorly characterized and, in particular, the down-regulation of ALAS-1 transcription by haem has remained controversial [8,10,12]. An important reason for this is that it has proven difficult to establish continuous cell-culture models that retain levels of mRNA, protein and activity of ALAS-1 as occurs in intact organisms and also retain inductive or repressive responses to chemicals. For example, virtually all mammalian hepatoma cell lines and cells from livers of tumour-bearing rats exhibit very low levels of ALAS-1, and these levels are only weakly inducible (1.5–2.0-fold induction), if at all [19,20]. Furthermore, mammalian hepatocytes in primary culture show rapid decreases in ALAS activities [21], and maintaining this activity has required costly [22–24] or complicated conditions of culture [24,25] or have been subject to unpredictable responses [26]. In addition, HuH-7 cells, a human hepatoma cell line, do not show accumulations of porphyrin in response to porphyrogenic drugs such as Glut (glutethimide) and deferoxamine (H. L. Bonkovsky, unpublished work). We were the first to demonstrate that LMH (Leghorn male hepatoma) cells, a chicken hepatoma cell line, simply cultured on 0.1% (w/v) gelatin, retain robust (>10-fold) inducibility of ALAS-1 and HO-1 (haem oxygenase-1) mRNA, and activity, and accumulate porphyrins in response to various treatments [27,28]. These cells have proven to be a useful, relevant model for study of porphyrin and haem metabolism and for testing porphyrogenicity of drugs and chemicals [27,29,30].
In contrast with haem, there is clear experimental evidence that porphyrogenic drugs directly increase hepatic ALAS-1 gene expression [12,31–34]. For example, using the avian LMH hepatoma cell line, transcriptional activation of ALAS-1 by porphyrogenic drugs has been shown to be mediated, at least in part, by two DRESs (drug-responsive enhancer sequences) located far upstream of the transcriptional start site [35]. These DRESs work independent of each other and contain functional DR4- and DR5-type recognition sequences (where DR stands for hexamer half-site direct repeat) for nuclear receptors and act as binding sites for the CXR (chicken xenobiotic-sensing receptor), a chick homologue of the CAR (constitutive androstane receptor) [35,36]. The existence of these DRESs does not exclude a role for other promoter or enhancer elements to up-regulate drug-dependent induction of ALAS-1, nor for haem to down-regulate the extent to which drugs can induce ALAS-1 gene expression.
In the present study, the issue of haem-dependent down-regulation of basal and drug-induced ALAS-1 gene transcriptions was approached by investigating, in LMH cells, the effects of the porphyrogenic drug, Glut, in combination with the inhibitor of haem synthesis, DHA (4,6-dioxoheptanoic acid; also called succinyl acetone), and/or haem on avian ALAS-1 promoter-reporter (luciferase) constructs. Using this system, we have identified separate haem- and drug-responsive regions in the cALAS-1 (chicken ALAS-1) promoter. In addition, we present data that the haem-responsive regions can suppress the drug-mediated promoter up-regulation by the DRES, leading to a model in which cellular haem levels may act at both transcriptional and post-transcriptional sites to limit induction of ALAS-1 following challenge by drugs.
MATERIALS AND METHODS
Materials
Haem (haemin chloride) and protoporphyrin IX were from Porphyrin Products (Logan, UT, U.S.A.). DMSO (Me2SO), DHA, ferric chloride and Glut were obtained from Sigma (St. Louis, MO, U.S.A.). Gelatin was from J. T. Baker (Phillipsburg, NJ, U.S.A.). All chemicals were of the highest purity available. LMH cells were a gift from D. L. Williams (Department of Pharmaceutical Sciences, SUNY-Stony Brook, Stony Brook, NY, U.S.A.). The pPGK-β-gal plasmid was a gift from P. Dobner (Department of Molecular Genetics and Microbiology, University of Massachusetts Medical School, Worcester, MA, U.S.A.). The pGL3-Basic, pGL3-Promoter construct, primer RV primer3 and primerGL2 were purchased from Promega (Madison, WI, U.S.A.).
Cell culture and treatment
LMH cells were maintained in Waymouth's MB 752/1 complete medium (containing 10%, v/v, fetal bovine serum with 100 units/ml penicillin and 100 μg/ml streptomycin) on 0.1% gelatin-coated flasks and passaged routinely once or twice a week [37]. Preparation and treatment of LMH cells with chemicals were exactly as described previously [38]. All chemicals were freshly prepared on the day of treatment. After 16 h of treatment with chemicals, cells were harvested, and luciferase and β-gal (β-galactosidase) activities and protein content were determined, as described below.
Plasmid construction
cALAS-1 promoter constructs (Figure 1)
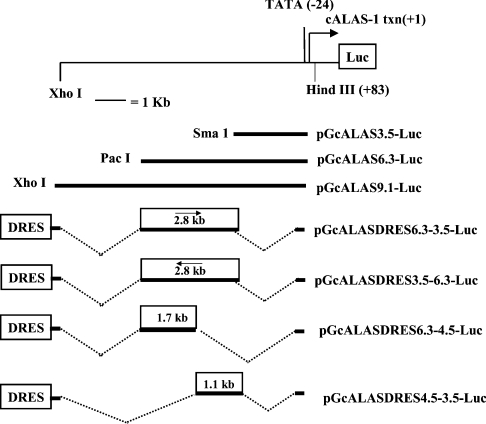
To generate cALAS-1 promoter constructs, the early promoter region and 5′-UTR (5′-untranslated region) of cALAS-1 (−299 to +82 bp) were amplified from the λ clone (cALAS-1) [39] with Pfu DNA polymerase (Stratagene, La Jolla, CA, U.S.A.) using the primers 5′-AATGAGAAGCTTTGAGATTTGTG-3′ and 5′-CCGCCTCCAAGCTTCCTCCTG-3, digested with HindIII (the HindIII site is underlined) and cloned into the HindIII site of a modified pGL3-Basic plasmid vector in which the NotI site at 4651 bp was destroyed to make pGcALAS0.3-Luc (where Luc stands for luciferase). To construct pGcALAS3.5-Luc, a SmaI fragment (−3447 to −165 bp) isolated from λ cALAS-1 clone was inserted into the SmaI site (−165 bp) of pGcALAS0.3-Luc. To generate pGcALAS9.1-Luc, an approx. 9.1 kb XhoI–NotI restriction fragment representing −9067 to +82 bp of the cALAS-1 promoter was cloned into the XhoI and NotI sites of pGcALAS0.3-Luc. To generate pGcALAS6.3-Luc, pGcALAS9.1-Luc was digested with XhoI (+9.1 kb) and PacI (+6.3 kb), end-filled with T4 DNA polymerase and re-ligated.
Heterologous promoter constructs containing DRES
To generate pGcALASDRES6.3-3.5-Luc and pGcALASDR-ES3.5-6.3-Luc (Figure 1), the cALAS-1 5′-flanking region containing the approx. 2.8 kb haem-repressive region (−6286 to −3475 bp) was amplified from pGcALAS9.1-Luc DNA using primers: 5′-ccgctcgagGGTAAGTAGGTCACAGTTATG-3′ and 5′-ctcgagcggTTCAGACTGGGAGCAGTACC-3′ (XhoI sites are underlined). The resulting PCR product was cloned into the XhoI site of the heterologous promoter construct, pGL3-Promoter (pGL3-Prom-Luc). The cALAS-1 5′-flanking region containing the 167 bp DRES (−13793 to −13271 bp) [35] was then amplified from chick genomic DNA using primers: 5′-cgacgcgtAGTACAGCTATGGATCTGTT-3′ and 5′-acgcgtcgGTCTTGATAGGGCTAACTTT-3′ (MluI sites are underlined). The resulting 538 bp PCR product was cloned into the MluI site of pGL3-Prom-Luc, pGcALAS6.3-3.5-Luc and pGcALAS3.5-6.3-Luc, to generate pGL3DRES-Luc, pGcALASDRES6.3-3.5-Luc and pGcALAS-DRES3.5-6.3-Luc respectively.
Figure 1. Partial restriction map of the 5′-UTR and 5′-flanking region of the cALAS-1 gene, and structures of cALAS-1 constructs.
Numbering starts at the transcription start point (+1). Recognition sites for restriction endonucleases XhoI, PacI, SmaI and HindIII used for plasmid construction are shown. The portions of the 5′-flanking region and 5′-UTR cloned into the reporter (Luc) gene plasmids (pGL3-Basic or pGL3-Promoter) are indicated, as are the abbreviations used to denote these constructs. A shorter construct, composed of the first 299 bp upstream of the transcription starting point, linked to the luciferase gene, was also studied. It showed no effect of up- or down-regulation.
To generate pGcALASDRES6.3-4.5-Luc and pGcALASDR-ES4.5-3.5-Luc (Figure 1), the 1.7 kb NheI (+6.3 kb) to BstEII (+4.5 kb) and 1.1 kb BstEII (+4.5 kb) to BglII (+3.5 kb) restriction fragments were isolated from pGcALAS6.3-Luc, end-filled and cloned into the SmaI site of pGL3-Prom-Luc. The PCR product containing the cALAS-1 DRES was then cloned upstream of these sequences to generate pGcALASDRES6.3-4.5-Luc and pGcALASDRES4.5-3.5-Luc respectively.
Transient transfections
LMH cells were plated on to 0.1%-gelatin-coated 6-well plates at a density of 1.5×105 cells/well. After 24 h, LMH cells were cotransfected with ALAS-1 promoter/reporter constructs (0.5 μg) and pPGK-β-gal (0.5 μg) using Lipofectamine™ Plus (0.33 μg/μl of reagent), according to the manufacturer's instructions. Total DNA transfected was kept constant by adding pBLUESCRIPT KS II+ plasmid DNA.
Assessment of reporter gene activity
Reporter gene expression was assessed by quantification of luciferase activity, normalized to β-gal activity and protein content. Transfected cells were washed twice with PBS and harvested in 250 μl of glycylglycine harvest buffer (25 mM glycylglycine, pH 7.8, 15 mM potassium phosphate, pH 7.8, 15 mM MgSO4, 4 mM EGTA and 1 mM dithiothreitol, added just prior to use). Cells were lysed by freeze–thawing (three cycles of 3 min in liquid nitrogen and 3 min at 37 °C) and cellular debris was removed by centrifugation (10–15 min, 14000 g and 4 °C). Luciferase activity was measured with a Monolight 2010® Luminometer (Analytical Luminescence Labs, Ann Arbor, MI, U.S.A.) and Luciferase Assay Reagent (Promega). Relative light units (RLU) produced in 10 s were measured as above and normalized to β-gal activities and protein content. β-Gal activity was measured by a colorimetric assay [40], and protein concentration by the BCA (bicinchoninic acid) method, using BSA as the standard [41].
Statistical analysis of data
In all experiments, each condition included at least triplicate samples. All experiments were repeated three to seven times with consistent results. Statistical analyses were performed using JMP 3.0.2 software (SAS Institute, Cary, NC, U.S.A.). Initial descriptive statistics showed that the results for continuous variables were distributed normally. Therefore the differences in mean values were assessed by ANOVA, with the Tukey–Kramer correction for multiple pairwise comparisons. P<0.05 was considered significant.
RESULTS
Glut and DHA can synergistically activate the cALAS-1 promoter
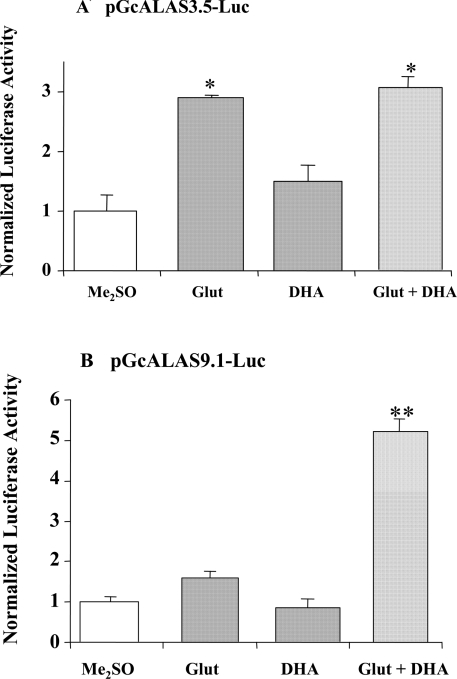
Previously, Glut and DHA have been shown to act synergistically to induce ALAS enzyme activity in primary cultures of chick liver cells [29] and LMH cells [27,29]. To determine if this synergistic effect occurred at the transcriptional level, LMH cells transiently transfected with the cALAS-1 reporter constructs pGcALAS9.1-Luc, pGcALAS3.5-Luc and pGcALAS0.3-Luc were treated with Glut alone, DHA alone, or a combination of Glut and DHA. Glut alone (50 μM) led to a 3-fold increase in luciferase activity in cells transfected with pGcALAS3.5-Luc (Figure 2A) and a lesser increase (1.5-fold) in cells transfected with pGcALAS9.1-Luc (Figure 2B). Whereas the shortest construct (pGcALAS0.3-Luc) was not responsive to Glut±DHA, 50 μM Glut led to a maximal up-regulation of the 3.5- or 9.1-ALAS-1 promoter constructs (results not shown). These results indicate that the cALAS-1 gene contains a drug-responsive region located in the first 3.5 kb of promoter, in addition to the DRES identified previously at −13 kb [27]. However, in cells transfected with pGcALAS9.1-Luc, the combination of 50 μM Glut and 250 μM DHA produced a synergistic induction in luciferase activity, compared with Glut or DHA alone (P<0.001; Figure 2B). In contrast, the combination of Glut and DHA did not significantly increase luciferase activity compared with Glut alone in cells transfected with pGcALAS3.5-Luc (Figure 2A). Increasing concentrations of DHA, up to 1 mM, had no effect on the Glut-induced reporter gene expression from pGcALAS3.5-Luc, indicating that pGcALAS3.5-Luc was unable to be synergistically induced by DHA (results not shown).
Figure 2. Effects of Glut and DHA on pGcALAS3.5-Luc (A) or pGcALAS9.1-Luc (B) transiently transfected into LMH cells.
Following transfection, LMH cells were treated with vehicle (Me2SO), Glut (50 μM), or DHA (250 μM), alone or in combination, for 16 h prior to harvest. Luciferase and β-gal activities and protein concentrations were assayed as described in the Materials and methods section. Luciferase activities are expressed as fold induction compared with vehicle-treated control, normalized to β-gal activity and protein concentrations. Results are means±S.E.M. for triplicate determinations, and representative of six independent experiments. *Differs from Me2SO or DHA alone, P<0.01. In (A), the two groups treated with Glut did not differ significantly from one another. **Differs from all others, P<0.001.
The synergistic induction by Glut and DHA with pGcALAS9.1-Luc, but not with pGcALAS3.5-Luc, suggested that there is a DHA-responsive region located within the −9.1 to −3.5 kb region of cALAS-1 that is distinct from the Glut-responsive element. Because DHA is a specific inhibitor of haem synthesis, it was likely that DHA was acting indirectly on the ALAS-1 promoter through lowering haem levels. As described further below, the −9.1 to −3.5 kb region of cALAS-1 5′-flanking region contains haem-responsive negative element(s) that repress Glut induction in the absence of DHA (normal cellular haem levels).
Effects of Glut, DHA and haem on reporter gene activity and deletion analysis of the −9.1 to −3.5 kb cALAS-1 region
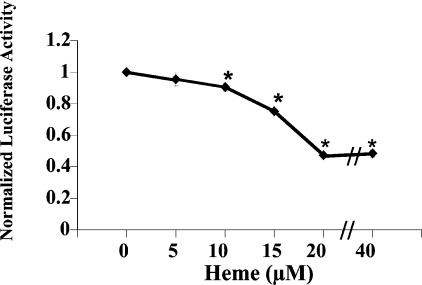
In order to determine if the cALAS-1 promoter can respond to increased haem levels, LMH cells transiently transfected with pGcALAS9.1-Luc were treated with increasing amounts of exogenous haem. Increasing concentrations of haem from 10 to 40 μM reduced luciferase activity in a dose-dependent manner when compared with Me2SO-treated control (Figure 3). Significant down-regulation by haem was noted at 10 and 15 μM (P<0.001), with maximal repression achieved at 20 μM (54% repression of luciferase activity; Figure 3).
Figure 3. Effect of exogenous haem on pGcALAS9.1-Luc activity.
LMH cells were transiently transfected with pGcALAS9.1-Luc and treated with increasing concentrations of haem (‘Heme’; 0, 5, 10, 15, 20 and 40 μM) dissolved in Me2SO. After 16 h, the cells were harvested and lysates were used to measure luciferase, β-gal and protein levels, as described in the Materials and methods section. Results are means±S.E.M. for triplicate determinations, and are representative of four independent experiments. Luciferase activities are expressed as fold induction compared with vehicle-treated control, normalized to β-gal and protein levels. The amount of Me2SO added per well was kept constant and was not greater than 2 μl/ml medium, a volume that was shown in previous studies to not affect luciferase activity [26,28,45,46]. *Differs from cells transfected with pGcALAS9.1-Luc and treated with 0 μM haem, P<0.001.
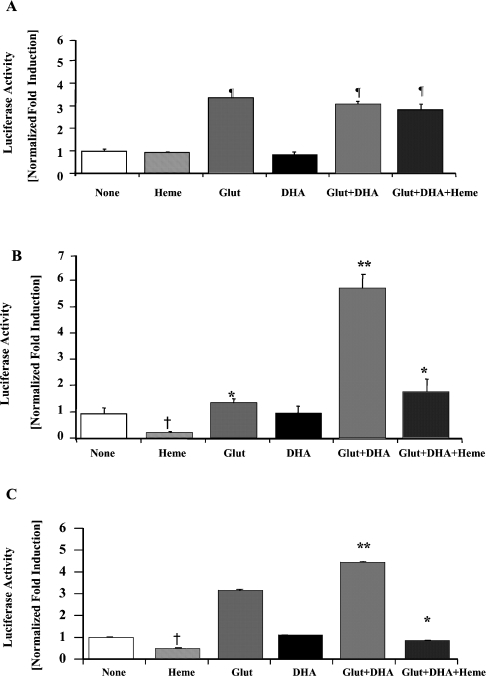
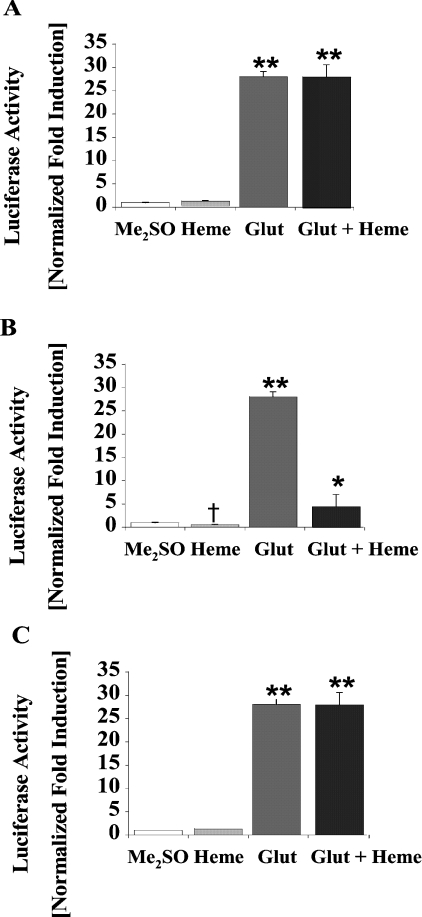
To determine if the effect of DHA on pGcALAS9.1-Luc was mediated through its ability to decrease haem levels in the cells, we assessed the induction of this construct by Glut and DHA in the presence of exogenous haem (Figure 4). Haem (20 μM, 16 h) treatment was found to repress significantly both the basal and Glut- and DHA-induced luciferase activities of this construct by 75 and 70% respectively (Figure 4B). In contrast, there was no effect of 20 μM haem either on the basal or Glut- and DHA-induced reporter gene expression of pGcALAS3.5-Luc (Figure 4A). These results are consistent with the −9.1 to −3.5 kb region of the cALAS-1 promoter containing one or more cis-acting elements that mediate haem repression of ALAS-1 and with the notion that DHA acts indirectly upon the ALAS-1 promoter by reducing cellular haem levels. The haem-repressive region was further localized to between −6.3 and −3.5 kb with the construct pGcALAS6.3-Luc (Figures 1 and 4C). Haem (20 μM) significantly repressed the basal and Glut- and DHA-induced luciferase activities of this construct to 53 and 20% of control respectively (Figure 4C).
Figure 4. Effects of Glut, DHA and haem in LMH cells transiently transfected with pGcALAS3.5-Luc (A), pGcALAS9.1-Luc (B) or pGcALAS6.3-Luc (C).
Transfected cells were harvested 16 h after chemical treatment, and lysates were used to measure luciferase, β-gal and protein levels. Concentrations of the chemicals were as follows: Glut, 50 μM; DHA, 250 μM; and haem, 20 μM. Results are means±S.E.M. for triplicate determinations, and representative of five independent experiments. Luciferase activities are expressed as fold induction compared with vehicle-treated control, normalized to β-gal and protein levels. *Differs from Glut+DHA, P<0.001. **Differs from others, P<0.001. ¶Differs from Me2SO, haem or DHA alone, P<0.01. In (A), the three groups treated with Glut did not differ significantly from one another. †Significant decrease from cells treated with Me2SO, P<0.001.
The haem-responsive region can repress drug induction from the upstream DRES
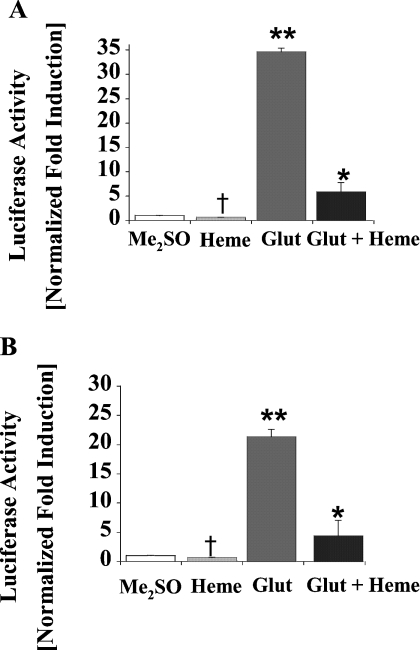
Previously, two separate DRESs approx. 13 kb upstream of the transcription start site were shown to play a key role in drug-mediated activation of the cALAS-1 gene [27]. In order to determine if the haem-responsive region identified in the present study could also regulate the activity of the drug-responsive enhancer sequences, constructs containing these DRESs and either the entire haem-responsive region or the 5′ (−6.3 to −4.5 kb) and the 3′ (−4.5 to −3.5 kb) portions were tested for drug induction in the presence of exogenous haem (Figures 5 and 6). As expected, Glut caused a marked increase in reporter gene activity from pGL3DRES-Luc (Figure 5A), but not from pGL3-Prom-Luc that lacked the DRES (results not shown). Haem alone did not affect the luciferase activity from either construct (Figure 5A, and results not shown). In contrast, haem treatment of cells transfected with constructs containing either the entire haem-responsive region (−6.3 to −3.5 kb) or the proximal (−6.3 to −4.5 kb) or distal (−4.5 to −3.5 kb) portions caused a significant decrease in both basal and Glut-mediated reporter gene activity compared with cells exposed to vehicle alone (Figures 5B, 6A and 6B).
Figure 5. Effects of Glut and haem on LMH cells transiently transfected with pGL3DRESLuc (A), pGcALASDRES6.3-3.5-Luc (B) or pGcALASDRES3.5-6.3-Luc (C).
Transfected cells were treated, harvested and assayed as described in the Materials and methods section and the legend to Figure 4. Results are means±S.E.M. for triplicate determinations and representative of five independent experiments. **Glut differs significantly from vehicle (Me2SO) alone or haem alone, P<0.001; *Glut + Haem differs significantly from Glut alone, P<0.001. †Significant decrease from cells treated with Me2SO, P<0.01.
Figure 6. Effects of Glut, DHA and haem on LMH cells transiently transfected with pGcDRES 6.3-4.5-Luc (A) or pGL3 DRES 4.5-3.5-Luc (B).
Transfected cells were treated, harvested and assayed as described in the Materials and methods section and the legend to Figure 4. Results are means±S.E.M. for triplicate determinations and representative of five independent experiments. †Differs from vehicle alone (Me2SO) or Glut + Haem, P<0.01. **Differs from all other treatment groups, P<0.0001. *Differs from Glut alone, P<0.001.
Interestingly, when the haem-responsive region was placed in the reverse orientation in pGcALASDRES3.5-6.3-Luc (Figure 1), no reduction in basal or Glut-mediated reporter gene activity in haem-treated cells was detected (Figure 5C). These results suggest that the ability of haem to down-regulate the cALAS-1 promoter is both sequence-specific and dependent on the orientation of the haem-repressive region. The two constructs, pGcALASDRES6.3-4.5-Luc and pGcALASDRES4.5-3.5-Luc, containing non-overlapping portions of the 2.8 kb region were also haem-responsive, with both constructs producing a degree of haem-dependent repression of reporter activity similar to that seen for the entire haem-responsive region in pGcALAS-DRES6.3-3.5-Luc (Figure 6). Therefore the cALAS-1 promoter contains at least two haem-responsive regions that can function independently. The fact that reporter gene activities in the plasmids pGcALAS3.5-Luc, pGL3DRES-Luc and pGcALAS-DRES3.5-6.3-Luc (reverse orientation of haem-responsive region) were unaffected by haem (Figures 4A, 5A and 5C) indicates that haem, at the concentration used (20 μM), was not toxic to LMH cells, a finding consistent with other results [27,28]. Additional experiments demonstrated that haem, but not iron or protoporphyrin alone, was required to repress the basal and Glut-mediated induction of the ALAS-1 promoter mediated by the −6.3 to −3.5 kb region (results not shown).
DISCUSSION
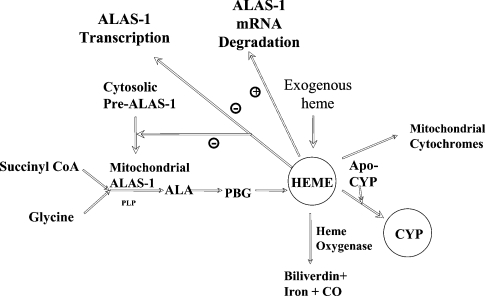
Haem has been proposed to regulate ALAS-1 negatively by a variety of mechanisms, including repression of transcription, increased degradation of mRNA and inhibition of mitochondrial translocation, as described below and as summarized in Figure 7. The observed effects of haem on hepatic ALAS-1 led to the formulation of a concept referred to as the ‘regulatory’, or ‘unassigned’ haem pool [2,4,42,43]. According to this hypothesis, hepatocytes contain a small, but crucial, haem pool that functions as a barometer of the cell's haem requirements. Haem is envisioned to exchange between this pool and the various haemoproteins of the cells (Figure 7).
Figure 7. Effects of the regulatory haem pool on hepatic haem metabolism.
Haem exerts regulatory effects on both ALAS-1 and HO-1 to control both its synthesis and degradation. ALAS-1 catalyses the formation of ALA from glycine and succinyl-CoA. Haem is synthesized from ALA through a series of intermediary steps catalysed by the enzymes of the haem biosynthetic pathway. Several steps in the production of functional mature ALAS-1 protein are down-regulated by exogenous or endogenous haem, as indicated by the minus signs, or by the plus sign indicating increased degradation of ALAS-1 mRNA. Haem is also incorporated into apo-haemoproteins [such as CYP (cytochrome P450) proteins and mitochondrial cytochromes] as the prosthetic group to form functional holo-haemoproteins.
Although it has been known for many years that ALAS-1 is the rate-controlling enzyme for hepatic haem biosynthesis and that levels of its mRNA and activity can be increased severalfold by porphyrogenic chemicals [2], the molecular mechanisms that regulate the expression of the ALAS-1 housekeeping gene are poorly understood. The half-life, t1/2, of the mRNA of hepatic ALAS-1 is short (1–5 h) [11,12]. Because of this, transcriptional repression by haem could be an effective mechanism for regulation of ALAS-1 [8–10]. However, to date the evidence for haem repression of cALAS-1 gene transcription has been controversial [8,10,12]. Early hypotheses [3], according to which drugs were believed to induce ALAS-1 transcription indirectly by lowering haem levels [10], have since been proved to be incorrect with the identification of two drug-responsive regions at approx. −13 kb of the cALAS-1 promoter [35,36]. However, these findings do not rule out the possibility that additional haem-responsive elements modulate the basal and drug-induced transcription of ALAS-1. Indirect evidence for an effect of this haem pool on drug-mediated induction of ALAS-1 was obtained from studies in mice with targeted disruptions of the gene for porphobilinogen deaminase. Such mice have a deficiency in haem synthesis that resembles the defect in the human disease acute intermittent porphyria, and augmented drug-mediated induction of ALAS-1 mRNA [34] suggesting that normal cellular haem levels can influence the degree of drug induction.
We were the first to demonstrate that LMH cells, a chicken hepatoma cell line, simply cultured on 0.1% gelatin, retain inducibility of ALAS-1 and HO-1 mRNA and activity in response to various treatments [27,28]. These cells have proven to be a useful, relevant model for study of porphyrin and haem metabolism and for testing porphyrogenicity of drugs and chemicals [27,29,44]. In the present study, the transcriptional regulation of ALAS-1 by drugs, haem and DHA was assessed using promoter-reporter constructs. Specifically, the cis-acting elements in the 5′-flanking region of ALAS-1 that mediate haem-dependent regulation of ALAS-1 gene transcription were studied with the long-term goal of demonstrating clearly whether there is an effect of haem to down-regulate ALAS-1 gene transcription and, if so, to identify the haem-repressive element(s) that mediate this effect. Our major findings are: (i) a second drug-responsive region is located between −3.5 and 0.3 kb, since Glut induced up to a 3-fold increase in reporter activity from constructs containing at least 3.5 kb of promoter sequence; (ii) haem significantly reduced both basal and Glut-induced reporter gene activity, in a dose-dependent manner, when constructs contained sequences between −6.3 and −3.5 kb, and this region could be further divided into two independent haem-responsive regions; and (iii) these haem-responsive regions could also down-regulate the expression of constructs containing the far-upstream DRES. This paper demonstrates for the first time that haem can repress markedly the transcription of the cALAS-1 gene. These results are consistent with earlier findings in adult rat liver where haem or its precursor ALA was able to repress transcription as measured by nuclear run-on assays [3,8,32]. Previous studies that did not find any effect of haem on ALAS-1 transcription in avian hepatocytes were performed in primary cultures of chick embryo liver cells, and did not involve reporter constructs. It is possible that the effects of haem on the half-life of ALAS-1 mRNA [12] together with the low level of ALAS-1 transcription in these cells may have masked the down-regulation of ALAS-1 transcription by haem. Alternatively, the contrasting results of the earlier studies and the present study may reflect differences between the regulation of ALAS-1 transcription in embryonic and adult-derived hepatocytes such as LMH cells used in this study. The concentrations of the haem required for repression of ALAS-1 gene transcription (10–20 μM; Figure 3) are approximately an order of magnitude greater than those sufficient to block mitochondrial uptake of the enzyme [15,17] or to increase its mRNA breakdown [12,13]. Thus haem effects may be manifold and redundant and depend upon the degree and duration of excess intracellular haem. LMH cells may provide a useful model for study of the integration of these haem-dependent down-regulatory effects.
Expression of several other genes is known to be regulated by haem, including HO-1, the rate-controlling enzyme of haem breakdown [2,38,45,46], and globin genes in developing erythrocytes [47], raising the possibility that these genes share common haem-regulatory elements. However, we have been unable to find any significant sequence similarity between the −6.3 to −3.5 kb region of cALAS-1 and the 5′-flanking regions of these genes. This may reflect the fact that haem up-regulates HO-1 and globin genes, whereas it down-regulates hepatic ALAS-1 and may therefore use different transcription factors. It is still unclear whether the 5′-flanking regions of ALAS-1 genes from other species contain candidate haem-responsive elements. To date, there are no functional data available delineating where putative haem-regulated elements may lie in the ALAS-1 promoter of other species. To determine if there are any sequences or transcription factors common to the avian haem-responsive region and the ALAS-1 promoter of other species, we have compared the avian haem-responsive region sequence with the entire 5′-flanking regions of the human and murine genes [35,36] using Genomatrix DiAllign TF software program (http://www.genomatix.de/index.html). In a stretch of highly conserved sequences, two short sequences present within the avian haem-responsive region (−6.3 to −3.5 kb) were also found in human and murine ALAS-1 5′-flanking regions. The first, at avian −4590 to −4565 bp, corresponds to −1863 to −1839 bp of the human and −2393 to −2369 bp of the mouse. The second, at avian −3494 to −3473 bp, corresponds to −1107 to −1086 bp of the human and −2088 to −2066 bp of the mouse (tsp=+1). Interestingly, these two common regions separate into the two independent haem-responsive regions (−6.3 to −4.5 kb) and (−4.5 to −3.5 kb) of cALAS-1 identified in the present study (Figure 6). Whether any of these is functional or responsive to haem remains to be seen.
In summary, our studies in LMH cells show that haem represses both basal and drug-mediated induction of ALAS-1 by one or more cis-acting elements in the −6.3 to −3.5 kb 5′-flanking region. The ability of this region to regulate the known drug-responsive regions identified in the ALAS-1 promoter suggests that this region plays an important role in modifying the transcriptional response of the cell to drugs. Fine mapping of this region, identification of key DNA-binding proteins and further delineation of haem, protein and DNA interactions are ongoing in our laboratory.
Acknowledgments
We thank Dr R. Lambrecht and Dr Y. Shan of the Departments of Pharmacology and Medicine respectively, University of Connecticut Health Center, for helpful discussion, and J. Clark and S. Donohue for expert assistance in the preparation of this paper. This work was supported by the U.S. Public Health Service grants RO1-DK38825 and MO1-RR06192, and contracts N01-DK92326 and U01-DK-065193. The opinions expressed herein are those of the authors and do not necessarily reflect the views of the U.S. Public Health Service or the University of Connecticut Health Center.
References
- 1.Ades I. Z. Heme production in animal tissues: the regulation of biogenesis of delta-aminolevulinate synthase. Int. J. Biochem. 1990;22:565–578. doi: 10.1016/0020-711x(90)90032-x. [DOI] [PubMed] [Google Scholar]
- 2.Bonkovsky H. L. Porphyrin and heme metabolism and the porphyrias. In: Zakim D., Boyer T., editors. Hepatology: A Textbook of Liver Disease. Philadelphia, PA: W.B. Saunders; 1990. pp. 378–424. [Google Scholar]
- 3.May B. K., Dogra S. C., Sadlon T. J., Bhasker C. R., Cox T. C., Bottomley S. S. Molecular regulation of heme biosynthesis in higher vertebrates. Prog. Nucleic Acid Res. Mol. Biol. 1995;51:1–51. doi: 10.1016/s0079-6603(08)60875-2. [DOI] [PubMed] [Google Scholar]
- 4.Kappas A., Sassa S., Galbraith R. A., Nordmann Y. The porphyrias. In: Scriver C. R., Beaudet A. L., Sly W. S., Valle D., editors. The Metabolic Basis of Inherited Disease. New York: McGraw-Hill; 1995. pp. 2103–2160. [Google Scholar]
- 5.Riddle R. D., Yamamoto M., Engel J. D. Expression of delta-aminolevulinate synthase in avian cells: separate genes encode erythroid-specific and nonspecific isozymes. Proc. Natl. Acad. Sci. U.S.A. 1989;86:792–796. doi: 10.1073/pnas.86.3.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bishop D. F., Henderson A. S., Astrin K. H. Human delta-aminolevulinate synthase: assignment of the housekeeping gene to 3p21 and the erythroid-specific gene to the X chromosome. Genomics. 1990;7:207–214. doi: 10.1016/0888-7543(90)90542-3. [DOI] [PubMed] [Google Scholar]
- 7.Sassa S., Nagai T. The role of heme in gene expression. Int. J. Hematol. 1996;63:167–178. doi: 10.1016/0925-5710(96)00449-5. [DOI] [PubMed] [Google Scholar]
- 8.Srivastava G., Borthwick I. A., Maguire D. J., Elferink C. J., Bawden M. J., Mercer J. F., May B. K. Regulation of 5-aminolevulinate synthase mRNA in different rat tissues. J. Biol. Chem. 1988;263:5202–5209. [PubMed] [Google Scholar]
- 9.Yamamoto M., Kure S., Engel J. D., Hiraga K. Structure, turnover, and heme-mediated suppression of the level of mRNA encoding rat liver delta-aminolevulinate synthase. J. Biol. Chem. 1988;263:15973–15979. [PubMed] [Google Scholar]
- 10.Srivastava G., Hansen A. J., Bawden M. J., May B. K. Hemin administration to rats reduces levels of hepatic mRNAs for phenobarbitone-inducible enzymes. Mol. Pharmacol. 1990;38:486–493. [PubMed] [Google Scholar]
- 11.Drew P. D., Ades I. Z. Regulation of the stability of chicken embryo liver delta-aminolevulinate synthase mRNA by hemin. Biochem. Biophys. Res. Commun. 1989;162:102–107. doi: 10.1016/0006-291x(89)91968-2. [DOI] [PubMed] [Google Scholar]
- 12.Hamilton J. W., Bement W. J., Sinclair P. R., Sinclair J. F., Alcedo J. A., Wetterhahn K. E. Heme regulates hepatic 5-aminolevulinate synthase mRNA expression by decreasing mRNA half-life and not by altering its rate of transcription. Arch. Biochem. Biophys. 1991;289:387–392. doi: 10.1016/0003-9861(91)90428-l. [DOI] [PubMed] [Google Scholar]
- 13.Cable E. E., Miller T. G., Isom H. C. Regulation of heme metabolism in rat hepatocytes and hepatocyte cell lines: delta-aminolevulinic acid synthase and heme oxygenase are regulated by different heme-dependent mechanisms. Arch. Biochem. Biophys. 2000;384:280–295. doi: 10.1006/abbi.2000.2117. [DOI] [PubMed] [Google Scholar]
- 14.Ohashi A., Sinohara H. Incorporation of cytosol delta-aminolevulinate synthase of rat liver into the mitochondria in vitro. Biochem. Biophys. Res. Commun. 1978;84:76–82. doi: 10.1016/0006-291x(78)90265-6. [DOI] [PubMed] [Google Scholar]
- 15.Hayashi N., Watanabe N., Kikuchi G. Inhibition by hemin of in vitro translocation of chicken liver delta-aminolevulinate synthase into mitochondria. Biochem. Biophys. Res. Commun. 1983;115:700–706. doi: 10.1016/s0006-291x(83)80201-0. [DOI] [PubMed] [Google Scholar]
- 16.Srivastava G., Borthwick I. A., Brooker J. D., Wallace J. C., May B. K., Elliott W. H. Hemin inhibits transfer of pre-delta-aminolevulinate synthase into chick embryo liver mitochondria. Biochem. Biophys. Res. Commun. 1983;117:344–349. doi: 10.1016/0006-291x(83)91582-6. [DOI] [PubMed] [Google Scholar]
- 17.Lathrop J. T., Timko M. P. Regulation by heme of mitochondrial protein transport through a conserved amino acid motif. Science. 1993;259:522–525. doi: 10.1126/science.8424176. [DOI] [PubMed] [Google Scholar]
- 18.Wolfson S. J., Bartczak A., Bloomer J. R. Effect of endogenous heme generation on delta-aminolevulinic acid synthase activity in rat liver mitochondria. J. Biol. Chem. 1979;254:3543–3546. [PubMed] [Google Scholar]
- 19.Bonkowsky H. L., Tschudy D. P., Collins A., Doherty J. M. Control of δ-aminolevulinic acid synthetase and tyrosine aminotransferase in tumors and livers of tumor-bearing rats. J. Natl. Cancer Inst. 1973;50:1215–1225. doi: 10.1093/jnci/50.5.1215. [DOI] [PubMed] [Google Scholar]
- 20.Iwasa F., Sassa S., Kappas A. δ-Aminolaevulinate synthase in human HepG2 hepatoma cells. Repression by haemin and induction by chemicals. Biochem. J. 1989;262:807–813. doi: 10.1042/bj2620807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bissell D. M., Bearn A. G. Liver cell culture: current status and prospects for the study of inherited diseases. Birth Defects Orig. Artic. Ser. 1980;16:283–291. [PubMed] [Google Scholar]
- 22.Decad G. M., Hsieh D. P., Byard J. L. Maintenance of cytochrome P-450 and metabolism of aflatoxin B1 in primary hepatocyte cultures. Biochem. Biophys. Res. Commun. 1977;78:279–287. doi: 10.1016/0006-291x(77)91251-7. [DOI] [PubMed] [Google Scholar]
- 23.Schuetz E. G., Wrighton S. A., Safe S. H., Guzelian P. S. Regulation of cytochrome P-450p by phenobarbital and phenobarbital-like inducers in adult rat hepatocytes in primary monolayer culture and in vivo. Biochemistry. 1986;25:1124–1133. doi: 10.1021/bi00353a027. [DOI] [PubMed] [Google Scholar]
- 24.Schuetz E. G., Schuetz J. D., May B., Guzelian P. S. Regulation of cytochrome P-450b/e and P-450p gene expression by growth hormone in adult rat hepatocytes cultured on a reconstituted basement membrane. J. Biol. Chem. 1990;265:1188–1192. [PubMed] [Google Scholar]
- 25.Ben Ze'ev A., Robinson G. S., Bucher N. L., Farmer S. R. Cell-cell and cell-matrix interactions differentially regulate the expression of hepatic and cytoskeletal genes in primary cultures of rat hepatocytes. Proc. Natl. Acad. Sci. U.S.A. 1988;85:2161–2165. doi: 10.1073/pnas.85.7.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evarts R. P., Marsden E., Thorgeirsson S. S. Regulation of heme metabolism and cytochrome P-450 levels in primary culture of rat hepatocytes in a defined medium. Biochem. Pharmacol. 1984;33:565–569. doi: 10.1016/0006-2952(84)90308-3. [DOI] [PubMed] [Google Scholar]
- 27.Kolluri S., Elbirt K. K., Bonkovsky H. L. Heme biosynthesis in a chicken hepatoma cell line (LMH): comparison with primary chick embryo liver cells (CELC) Biochim. Biophys. Acta. 1999;1472:658–667. doi: 10.1016/s0304-4165(99)00159-2. [DOI] [PubMed] [Google Scholar]
- 28.Gabis K. K., Gildemeister O. S., Pepe J. A., Lambrecht R. W., Bonkovsky H. L. Induction of heme oxygenase-1 in LMH cells. Comparison of LMH cells to primary cultures of chick embryo liver cells. Biochim. Biophys. Acta. 1996;1290:113–120. doi: 10.1016/0304-4165(96)00009-8. [DOI] [PubMed] [Google Scholar]
- 29.Russo S. M., Pepe J. A., Cable E. E., Lambrecht R. W., Bonkovsky H. L. Repression of ALA synthase by heme and zinc-mesoporphyrin in a chick embryo liver cell culture model of acute porphyria. Eur. J. Clin. Invest. 1994;24:406–415. doi: 10.1111/j.1365-2362.1994.tb02184.x. [DOI] [PubMed] [Google Scholar]
- 30.Handschin C., Podvinec M., Meyer U. A. CXR, a chicken xenobiotic-sensing orphan nuclear receptor, is related to both mammalian pregnane X receptor (PXR) and constitutive androstane receptor (CAR) Proc. Natl. Acad. Sci. U.S.A. 2000;97:10769–10774. doi: 10.1073/pnas.97.20.10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamilton J. W., Bement W. J., Sinclair P. R., Sinclair J. F., Wetterhahn K. E. Expression of 5-aminolaevulinate synthase and cytochrome P-450 mRNAs in chicken embryo hepatocytes in vivo and in culture. Effect of porphyrinogenic drugs and haem. Biochem. J. 1988;255:267–275. [PMC free article] [PubMed] [Google Scholar]
- 32.Dogra S. C., May B. K. Phenobarbital-induced activation of CYP2H1 and 5-aminolevulinate synthase genes in chick embryo hepatocytes is blocked by an inhibitor of protein phosphorylation. Arch. Biochem. Biophys. 1996;327:271–278. doi: 10.1006/abbi.1996.0121. [DOI] [PubMed] [Google Scholar]
- 33.Louis C. A., Wood S. G., Walton H. S., Sinclair P. R., Sinclair J. F. Mechanism of the synergistic induction of CYP2H by isopentanol plus ethanol: comparison to glutethimide and relation to induction of 5-aminolevulinate synthase. Arch. Biochem. Biophys. 1998;360:239–247. doi: 10.1006/abbi.1998.0956. [DOI] [PubMed] [Google Scholar]
- 34.Jover R., Hoffmann F., Scheffler-Koch V., Lindberg R. L. Limited heme synthesis in porphobilinogen deaminase-deficient mice impairs transcriptional activation of specific cytochrome P450 genes by phenobarbital. Eur. J. Biochem. 2000;267:7128–7137. doi: 10.1046/j.1432-1327.2000.01815.x. [DOI] [PubMed] [Google Scholar]
- 35.Fraser D. J., Podvinec M., Kaufmann M. R., Meyer U. A. Drugs mediate the transcriptional activation of the 5-aminolevulinic acid synthase (ALAS1) gene via the chicken xenobiotic-sensing nuclear receptor (CXR) J. Biol. Chem. 2002;277:34717–34726. doi: 10.1074/jbc.M204699200. [DOI] [PubMed] [Google Scholar]
- 36.Fraser D. J., Zumsteg A., Meyer U. A. Nuclear receptors constitutive androstane receptor and pregnane X receptor activate a drug-responsive enhancer of the murine 5-aminolevulinic acid synthase gene. J. Biol. Chem. 2003;278:39392–39401. doi: 10.1074/jbc.M306148200. [DOI] [PubMed] [Google Scholar]
- 37.Kawaguchi T., Nomura K., Hirayama Y., Kitagawa T. Establishment and characterization of a chicken hepatocellular carcinoma cell line, LMH. Cancer Res. 1987;47:4460–4464. [PubMed] [Google Scholar]
- 38.Lincoln B. C., Healey J. F., Bonkovsky H. L. Regulation of hepatic haem metabolism. Disparate mechanisms of induction of haem oxygenase by drugs and metals. Biochem. J. 1988;250:189–196. doi: 10.1042/bj2500189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maguire D. J., Day A. R., Borthwick I. A., Srivastava G., Wigley P. L., May B. K., Elliott W. H. Nucleotide sequence of the chicken 5-aminolevulinate synthase gene. Nucleic Acids Res. 1986;14:1379–1391. doi: 10.1093/nar/14.3.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook J., Fritsch E. F., Maniatis T. Plainview, NY: Cold Spring Harbor Laboratory Press; 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 41.Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 42.Sinclair P. R., Granick S. Heme control on the synthesis of delta-aminolevulinic acid synthetase in cultured chick embryo liver cells. Ann. N.Y. Acad. Sci. 1975;244:509–520. doi: 10.1111/j.1749-6632.1975.tb41551.x. [DOI] [PubMed] [Google Scholar]
- 43.Granick S., Sinclair P., Sassa S., Grieninger G. Effects by heme, insulin, and serum albumin on heme and protein synthesis in chick embryo liver cells cultured in a chemically defined medium, and a spectrofluorometric assay for porphyrin composition. J. Biol. Chem. 1975;250:9215–9225. [PubMed] [Google Scholar]
- 44.Handschin C., Meyer U. A. A conserved nuclear receptor consensus sequence (DR-4) mediates transcriptional activation of the chicken CYP2H1 gene by phenobarbital in a hepatoma cell line. J. Biol. Chem. 2000;275:13362–13369. doi: 10.1074/jbc.275.18.13362. [DOI] [PubMed] [Google Scholar]
- 45.Shan Y., Pepe J., Lu T. H., Elbirt K. K., Lambrecht R. W., Bonkovsky H. L. Induction of the heme oxygenase-1 gene by metalloporphyrins. Arch. Biochem. Biophys. 2000;380:219–227. doi: 10.1006/abbi.2000.1921. [DOI] [PubMed] [Google Scholar]
- 46.Shan Y., Pepe J., Lambrecht R. W., Bonkovsky H. L. Mapping of the chick heme oxygenase-1 proximal promoter for responsiveness to metalloporphyrins. Arch. Biochem. Biophys. 2002;399:159–166. doi: 10.1006/abbi.2001.2742. [DOI] [PubMed] [Google Scholar]
- 47.Tahara T., Sun J., Igarashi K., Taketani S. Heme-dependent up-regulation of the alpha-globin gene expression by transcriptional repressor Bach1 in erythroid cells. Biochem. Biophys. Res. Commun. 2004;324:77–85. doi: 10.1016/j.bbrc.2004.09.022. [DOI] [PubMed] [Google Scholar]