Abstract
Cyp26A1 encodes an RA (retinoic acid)-catabolizing CYP (cytochrome P450) protein that plays a critical role in regulating RA distribution in vivo. Cyp26A1 expression is inducible by RA, and the locus has previously been shown to contain a RARE (RA response element), R1, within the minimal promoter [Loudig, Babichuk, White, Abu-Abed, Mueller and Petkovich (2000) Mol. Endocrinol. 14, 1483–1497]. In the present study, we report the identification of a second functional RARE (R2) located 2.0 kb upstream of the Cyp26A1 transcriptional start site. Constructs containing murine sequences encompassing both R1 and R2 showed that these elements work together to generate higher transcriptional activity upon treatment with RA than those containing R1 alone. Inclusion of R2 also dramatically enhanced the sensitivity of reporter constructs to RA, as even treatment with 10−8 M RA resulted in a 5-fold induction of reporter activity. Mutational analysis identified R2 as the functional element responsible for the increased RA inducibility of promoter constructs. The element was shown to bind RARγ (RA receptor γ)/RXRα (retinoid X receptor α) heterodimers in vitro, and inclusion of nuclear receptors in transfections boosted the transcriptional response. A construct containing both R1 and R2 was used to generate a stable luciferase reporter cell line that can be used as a tool to identify factors regulating Cyp26A1 expression. The analysis of R1 and R2 has led to the proposal that the two elements work synergistically to provide a maximal response to RA and that R2 is an upstream enhancer.
Keywords: Cyp26A1, embryonic development, nuclear receptor, retinoic acid, retinoic acid response element (RARE), transcriptional regulation
Abbreviations: APL, acute promyelocytic leukaemia; CYP, cytochrome P450; EMSA, electrophoretic mobility-shift assay; HDAC, histone deacetylase; RA, retinoic acid; RALDH, retinaldehyde dehydrogenase; RAR, RA receptor; RARE, RA response element; RXR, retinoid X receptor; TSA, trichostatin A; wt, wild-type
INTRODUCTION
Retinoic acid (RA), the active form of vitamin A, is essential for establishing the balance between cell proliferation, differentiation and apoptosis during critical stages of tissue morphogenesis. Thus the spatial and temporal dispersal of RA must be tightly controlled. This is accomplished by co-ordinated expression of RA-synthesizing enzymes (RALDH, retinaldehyde dehydrogenase) and RA-catabolizing enzymes (Cyp26) in various tissues undergoing patterning changes. Factors controlling the expression of these enzymes underlie the earliest stages of tissue differentiation.
RA is synthesized by a family of RALDHs (RALDH1, RALDH2, RALDH3 and RALDH4) that irreversibly oxidize retinaldehyde to RA [1–4]. Although each gene exhibits a unique embryonic expression pattern, RALDH2 is the most widely expressed with transcripts detected in undifferentiated somites, limb buds and various organs [5,6]. The catabolism of RA is mediated by a family of CYP (cytochrome P450) enzymes, Cyp26A1, Cyp26B1 and Cyp26C1, that act to oxidize RA to more polar metabolites including 4-oxo-RA, 4-OH-RA and 18-OH-RA, which are believed to be inactive in vivo [7–9]. During mid-gestation, Cyp26A1 is expressed in the tailbud neuroepithelium and cervical mesenchyme, Cyp26B1 is present in the hindbrain, branchial arches and limb bud, and Cyp26C1 is transiently expressed in the hindbrain [10,11]. When the expression of Cyp26A1, Cyp26B1 and RALDH2 are compared in E9.5 (embryonic day 9.5) embryos, RALDH2 is located in the trunk of the embryo, flanked by Cyp26B1 anteriorly and Cyp26A1 posteriorly [5,10]. Therefore the complementary expression of these enzymes is responsible for the establishment of locally restricted RA levels in the embryo. Generation of transgenic mice lacking either Cyp26A1 or RALDH2 results in embryonic lethal phenotypes, establishing the importance of regulating RA distribution for normal embryogenesis [12–14].
RA induces gene expression through binding to the nuclear receptors RAR (RA receptor) and RXR (retinoid X receptor) [15,16]. all-trans-RA acts as a ligand for RAR, while the isomer 9-cis-RA can be bound by both RAR and RXR. For each receptor, there are three subtypes (α, β and γ) and several isoforms, which differ in their tissue distribution [17,18]. RAR and RXR bind to each other forming functional heterodimers. According to a current model of transcriptional activation, in the absence of ligand, RAR/RXR heterodimers are bound to DNA, and they recruit co-repressors with HDAC (histone deacetylase) activity, resulting in chromatin condensation and gene silencing [19]. Upon ligand binding, RAR and RXR undergo conformational changes that favour the dissociation of co-repressors and the recruitment of other proteins with histone acetylase activity, which opens up the chromatin, making it accessible to transcriptional machinery to initiate transcription.
The binding site for RAR/RXR heterodimers is a specific DNA sequence known as a RARE (RA response element). RAREs consist of a direct repeat of a core hexameric sequence, PuG(G/T)-TCA, separated by 1, 2 or 5 base-pairs (DR1, DR2 and DR5) [15]. More than 500 genes with diverse functions are regulated by RA, and RAREs have been localized in many of these genes including RARβ, CRBPI, CRABPII and members of the Hox and HNF gene families [20].
Although certain tissues express Cyp26A1 in an apparent RA-independent manner so as to form a barrier to RA, Cyp26A1 has been shown to be inducible by RA both in vivo and in cell lines, indicating that regulation of RA catabolism may include a positive feedback loop [21,22]. Given the sensitivity of embryonic tissues to RA, it is believed that this induction plays a critical role in protecting some regions of the embryo from fluctuations in maternal RA that could potentially result in teratogenesis [23].
Clinically, the RA-mediated induction of Cyp26A1 may be significant as retinoids can be used as pharmacological agents. Cyp26A1 is inducible in skin where retinoids have been used extensively to treat conditions such as acne, psoriasis and ich-thyosis [24]. It is conceivable that prolonged treatment of skin can induce Cyp26A1-mediated metabolism, thereby limiting effective therapeutic doses of retinoids. Also, RA is often used for the treatment of APL (acute promyelocytic leukaemia), and a high proportion of patients initially undergo remission. However, there is a very high chance of relapse as patients begin to exhibit RA resistance [25]. Studies in APL cell lines have shown that Cyp26A1 mRNA is inducible by RA; thus Cyp26A1 induction may be responsible for acquired retinoid resistance [26].
We are interested in determining transcription factor interactions with the Cyp26A1 promoter leading to its expression and inducibility. The proximal region of the Cyp26A1 promoter has been studied and observed to contain a RARE (R1), as well as a G-rich element that is able to bind Sp1/Sp3 [27]. The present study has uncovered a conserved second RARE (R2) occurring 2 kb upstream of the transcription start site, which appears to function synergistically with the R1 element to provide maximal induction of Cyp26A1 in response to RA.
MATERIALS AND METHODS
DNA constructs for transient transfections
All DNA constructs for transient transfections were cloned into the luciferase reporter vector, pGL3-Basic (Promega, Madison, WI, U.S.A.). The cloning of the constructs Cyp26A1-163wt (where wt stands for wild-type), Cyp26A1-238wt and Cyp26A1-238-3′TAATmut has been described previously [27]. Cyp26A1-238 RARE-flipped is a construct in which the RARE was inverted by amplifying a 159 bp fragment using the PCR primers 5′-CAGGGGCCCGATCCGCAATTAAAGAAGTTCACCCAAAGTTCAAATTTGTCT and 5′-GAAAGCTT- GGCACGCTTCAGCCTCCCGCG. The product was then digested with ApaI and HindIII and used to replace the fragment removed from Cyp26A1-238wt digested with the same enzymes. The Cyp26A1-1.4 kb construct was generated by PCR using the primers 5′-CCAAAGCTTTGTGGCGGGAGGACATGAGAAGCC and 5′-GACCATGGCACGCTTCAGCCTCCCGCG. The resulting product was digested with HindIII and NcoI and ligated into the appropriate sites of pGL3-Basic. Cyp26A1-1.4 kb RAREmut was made by taking the fragment amplified using the primers 5′-CAGGGGCCCGATCCGCAATTAAAGATGAACTCCCAATGAACTAATTTGTCT and 5′-AGCTCCATGGGGCACGCTTCAGCCTCCCGCG, digesting it with ApaI and NcoI and using it to replace the wt ApaI/NcoI fragment in Cyp26A1-1.4 kb. The primers 5′-GATCGAATTCCCAACGGTTGGGCAAGGG and 5′-GATCGAATTCGGCACGCTTCAGCCTCCCGCG were used to amplify 2.6 kb of the promoter region. The PCR product was digested with HindIII, and the 1198 bp upstream piece was cloned into the unique HindIII site of Cyp26A1-1.4 wt and Cyp26A1-1.4 kb RAREmut to generate the Cyp26A1-2.6 kb wt and Cyp26A1-2.6 kb RARE1mut constructs. The same fragment was inverted and cloned into Cyp26A1-1.4 wt to create Cyp26A1-2.6 kb-1198rev. The Cyp26A1-Δ1.4 kb construct was generated by inserting the 1198 bp fragment containing the R2 element into pGL3-Basic. Finally, Cyp26A1-2.6 kb RARE2mut construct was made by amplifying a 301 bp fragment containing R2 using the primers 5′-CCGGCCTGCAGGGGTTTCAGGCGGGTTTGGCTAAAGGATTGGG and 5′-GCCGGGATGCCCATGGGTTGTA- TTTCGGGG. The fragment was digested with PstI and NcoI, and used to replace the wt sequence in the Cyp26A1-2.6 kb construct. For all PCRs, BAC clones containing the mCyp26A1 locus were used as the template [27].
Cell culture and transfections
P19 (mouse teratocarcinoma) cells and MCF-7 (human breast adenocarcinoma) cells were cultured in MEM (minimal essential medium; pH 7.2) and DMEM (Dulbecco's modified Eagle's medium; pH 7.2) respectively. All media were supplemented with 10% (v/v) fetal calf serum, 0.5% penicillin–streptomycin, 0.1% gentamicin and 0.1% fungizone, and cells were grown at 37 °C with 5% CO2.
All transient transfections were performed using FuGENE transfection reagent (Roche Diagnostics, Laval, QC, Canada) using a 1:3 ratio of DNA and FuGENE. Cells were seeded in plates 24 h before transfection: for P19 cells, 1×105 cells/well in a 24-well plate, and for MCF-7 cells, 1×105 cells/well in a 12-well plate. P19 cells were transfected with 825 ng of DNA, while 250 ng was used for MCF-7 cells. Cells were treated, 24 h post-transfection, with either DMSO or all-trans-RA resuspended in DMSO. After treatment, plates were wrapped in aluminium foil and incubated for 24 h. Then, media were aspirated and cells were washed twice with 1×PBS. The PBS was aspirated and 100 μl of passive lysis buffer (Promega) was added and left for 15 min. Lysed cells were transferred to a 96-well assay plate, and readings were taken using an EG&G Berthold MicroLumat Plus LB 96V luminometer. All transfections were performed in triplicate, and experiments were repeated three times.
Generation and maintenance of the stable cell line
To stably transfect P19 cells with the Cyp26A1-2.6 kb wt construct, the fragment was fused to a firefly luciferase gene and cloned into pcDNA 3.1 (Invitrogen Life Technologies, Carlsbad, CA, U.S.A.). The construct was transfected into P19 cells in 24-well plates with 1×105 cells/well using FuGENE. Following a 24 h incubation, cells were trypsinized and transferred to a 100 mm plate. After a further 24 h, cells were selected using 0.4 mg/ml Geneticin (G418; Invitrogen Life Technologies) and selection was maintained for 2 weeks after which monoclonal cells were selected, cultured separately and screened for luciferase activity.
EMSAs (electrophoretic mobility-shift assays)
EMSAs were performed as described previously [27]. For in vitro transcription and translation of RARα and RXRγ, plasmids containing each gene were linearized with XhoI, precipitated and resuspended in nuclease-free water. Transcription and translation were carried out using the TNT T7/T3-coupled reticulocyte lysate system kit as per the manufacturer's instructions (Promega).
Northern-blot analysis
P19 and MCF-7 cells were seeded separately in T175 flasks. When cells had grown to approx. 70% confluence, media were removed and replaced with media supplemented with either DMSO or DMSO/10 μM RA. After 24 h, media were aspirated and 7 ml of TRIzol® (Invitrogen Life Technologies) was added directly to the cells. Total RNA was then extracted as per the manufacturer's instructions. Northern blotting was performed using a standard capillary transfer with 10×SSC (1×SSC is 0.15 M NaCl/0.015 M sodium citrate) as the transfer buffer, and hybridization was performed using Quickhyb (Stratagene, La Jolla, CA, U.S.A.), as explained previously [22]. For Cyp26A1, a full-length cDNA probe was used, and for 36B4, a 564 bp PstI fragment was used.
Southern-blot analysis
Genomic DNA extraction, enzymatic digestion and Southern blotting were performed as explained previously [13]. The probe for hybridization was a HindIII fragment spanning the distal 1.2 kb of the Cyp26A1-2.6 kb wt construct.
RESULTS
A second putative RARE upstream of Cyp26A1 is embedded in a region of the promoter highly conserved between mouse and human
Alignments of human and mouse sequence upstream of the Cyp26A1 transcriptional start site revealed two homologous domains with 94 and 82% similarity between the two species (Figure 1A). By comparison, the similarity of the entire coding sequence of Cyp26A1 between human and mouse is 89%. The first upstream domain exhibiting 94% similarity spans from −12 to −223 (counting the transcriptional start site as 0) has been previously characterized and shown to contain a RARE (labelled as R1), a TATA box and a binding site for SP1/SP3 [27]. The second region, which we describe in the present study, was located between −1745 and −2005 of the murine gene. Within this region there is a perfectly conserved 42 bp stretch that contains a DR5 motif reminiscent of a classical RARE (Figure 1B, labelled as R2). In between the two shaded areas (Figure 1A), there are no regions of significant (>80%) similarity between the mouse and human sequences.
Figure 1. Conserved regions between the human and mouse Cyp26A1 upstream sequences.
(A) The mouse Cyp26A1 gene has two putative RAREs, each embedded in a region that is highly similar to the human locus. The RAREs are denoted as R1 and R2 and represented by black bars. Conserved regions between mouse and human are shown as light grey boxes, and the percentage similarity is labelled below. Numbers below the schematic are bp relative to the transcriptional start site (0). (B) The sequence surrounding the R2 element is highly conserved (82%) between mouse and human. R2 is located within a 42 bp region shaded in grey that is perfectly conserved. The hexameric repeats of the RARE are boxed.
Inclusion of additional upstream sequences enhances RA inducibility of Cyp26A1 in vitro
As shown by Northern-blot analysis (Figure 2A), both P19 (mouse teratocarcinoma) and MCF-7 (human breast adenocarcinoma) cells exhibit induction of endogenous Cyp26A1 transcripts after 24 h of treatment with 10 μM RA, in agreement with previous reports [28,29]. Blots were stripped and reprobed with the ubiquitously expressed 36B4 to show even loading of samples (Figure 2A). This indicates that both cell lines are appropriate models for studying the RA-mediated induction of Cyp26A1.
Figure 2. The 2.6 kb fragment of the Cyp26A1 promoter is highly inducible in P19 cells.
(A) Endogenous Cyp26A1 is inducible by treatment with 10−6 M RA for 24 h in both P19 and MCF-7 cell lines as shown by Northern-blot analysis. The same blot was probed with 36B4 to control for even RNA loading. (B) Transient transfections with reporter constructs show that the 2.6 kb promoter fragment exhibits the highest RA inducibility. Cells were treated with either DMSO or DMSO/10−6 M RA, and co-transfected with RARγ and RXRα.
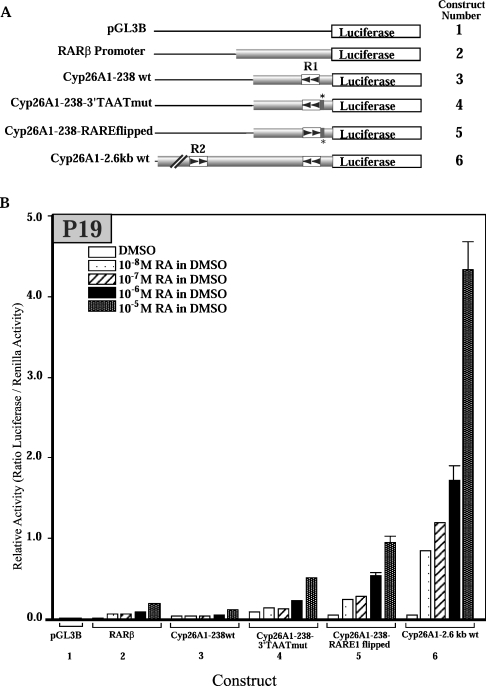
It has been previously shown that a Cyp26A1 promoter-reporter gene construct containing 238 bp of sequence upstream of the Cyp26A1 transcriptional start site is inducible by RA [27]. To determine if elements further upstream were involved in RA-mediated induction of Cyp26A1, a number of constructs were generated containing between 163 bp and 2.6 kb of the putative mouse Cyp26A1 promoter fused to a luciferase reporter gene (Figure 2B). These constructs were transiently transfected into P19 cells, and cells were treated with 10 μM RA for 24 h to determine whether there was any increase in luciferase activity.
Constructs containing only the R1 element exhibited mild induction upon RA treatment, with the highest increase being a 2.5-fold induction by the Cyp26A1-238 wt construct (Figure 2B). In the construct Cyp26A1-2.6 kb wt, containing 2.6 kb of upstream sequence, including both the R1 and R2 elements, there was a 4-fold induction of reporter activity with RA treatment, and the magnitude of activity was higher than that of any of the other constructs. When RARγ and RXRα were co-transfected into cells, there were slight effects on reporter activity generated by constructs containing only the R1 element. The highest fold induction was 4-fold induction for the Cyp26A1-238 wt construct. However, in the construct containing both R1 and R2, co-transfection with nuclear receptors resulted in a 10-fold induction of reporter activity.
Both the R1 and R2 elements are required for optimal RA induction of Cyp26A1 in vitro
To determine whether (i) the R2 element is responsible for increased inducibility of the 2.6 kb wt construct and (ii) the R1 and R2 elements synergistically regulate Cyp26A1 expression, a series of constructs were generated in which one or both of the RAREs were mutated (Figure 3A). Transient transfections in both P19 and MCF-7 cell lines showed that the R2 element is necessary for maximal reporter induction. Constructs containing R2 (Figures 3B and 3C, lanes 4–6 and 9) showed the highest fold increase of reporter activity upon RA treatment. In the absence of R1 (lane 5), constructs containing R2 still showed significant reporter induction, but the fold increase was lower than when both elements were present (lanes 4 and 6). Conversely, in constructs containing only R1 (lane 2), there was a very low induction of reporter activity. When the R2 site was mutated to abolish the RARE, there was a loss of RA-mediated induction (lane 8), indicating that the R2 was responsible for the induction observed in the 2.6 kb wt construct. When the proximal part of the construct is deleted, leaving only the R2 (lane 10), there was no reporter activity, indicating that the R2 element alone is incapable of initiating transcription.
Figure 3. Both R1 and R2 elements are required for optimal Cyp26A1 inducibility by RA in vitro.
(A) Constructs used for transient transfections to test the effect of R1 and R2 on reporter expression. Black boxes indicate where an element has been mutated, and numbers below the constructs refer to the number of base pairs upstream of the transcriptional start site. (B) Transient transfection of reporter constructs into P19 cells, treated with either DMSO or DMSO/10−6 M RA. Constructs containing R2 (lanes 4–6 and 9) show a higher fold induction, and the greatest induction is observed when both elements are present (lanes 4, 6 and 9). (C) Similar results were obtained when transfections were performed in MCF-7 cells.
R1 and R2 elements are able to bind RARγ/RXRα heterodimers in vitro
Two double-stranded oligonucleotides (Figure 4A) corresponding to R1 or R2 were radiolabelled and used as probes for EMSAs. The RAREs in each oligonucleotide have been marked by the square bracket and the direct repeats are shaded (Figure 4A).
Figure 4. The R2 element binds RAR/RXR in vitro.
(A) Double-stranded oligonucleotides for R1 and R2 that were used for bandshift experiments. In each sequence, the DR5 RARE is shaded. (B) Bandshift experiments with probes for R1 and R2 show formation of a complex (arrowhead) only in the presence of both RARγ and RXRα.
To determine if the murine R1 and R2 elements were able to bind RAR or RXR, oligonucleotides were radiolabelled and added to reticulocyte lysates containing in vitro translated RARγ and/or RXRα. As shown in Figure 4(B), RARγ or RXRα alone were unable to bind the R1 element. However, a specific complex (Figure 4B, arrowhead) was formed when both receptors were added, in agreement with previously published results [27]. Similarly, incubation of the R2 element with RARγ and RXRα resulted in the appearance of a novel complex indicative of RAR/RXR heterodimer formation and binding to the DR5 motif (Figure 4B, arrowhead).
The R2 element is sensitive to low concentrations of RA
To investigate the responsiveness of Cyp26A1 promoter elements to different concentrations of RA, transient transfections were performed in P19 cells. Several constructs (Figure 5A) were transfected to compare the sensitivity of a construct containing R2 with other previously characterized RA-inducible constructs.
Figure 5. The R2 element is sensitive to low doses of RA.
(A) Constructs used for transient transfections to test the sensitivity of different promoter elements to RA. (B) Transient transfections were performed in P19 cells and treated with DMSO or DMSO/RA (concentrations as indicated). P19 cells transfected with the 2.6 kb wt fragment containing R1 and R2 show the greatest fold induction at all RA concentrations tested.
The RARβ promoter has been shown to be inducible by RA [30], and in the present study displays greater RA-responsiveness than the Cyp26A1-238 wt construct when treated with 10−8 to 10−5 M RA (Figure 5B). It has been previously shown that mutating a 5′-TAAT motif overlapping the Cyp26A1 promoter R1 RARE enhances the RA inducibility of this construct [27]. Two constructs containing mutations of this motif were included (Cyp26A1-238wt-3′TAATmut and Cyp26A1-238wt-RARE1 flipped), and as expected both constructs were more responsive than either the 238 wt fragment or the RARβ promoter. However, the 2.6 kb wt construct was much more responsive than any of the other tested constructs (Figure 5B). When treated with a dose of 10−8 M RA, there was a 5-fold induction in reporter activity compared with DMSO-treated cells. Treatment with 10−5 M RA resulted in a greater than 50-fold induction.
Characterization of a stable cell line containing the 2.6 kb reporter construct
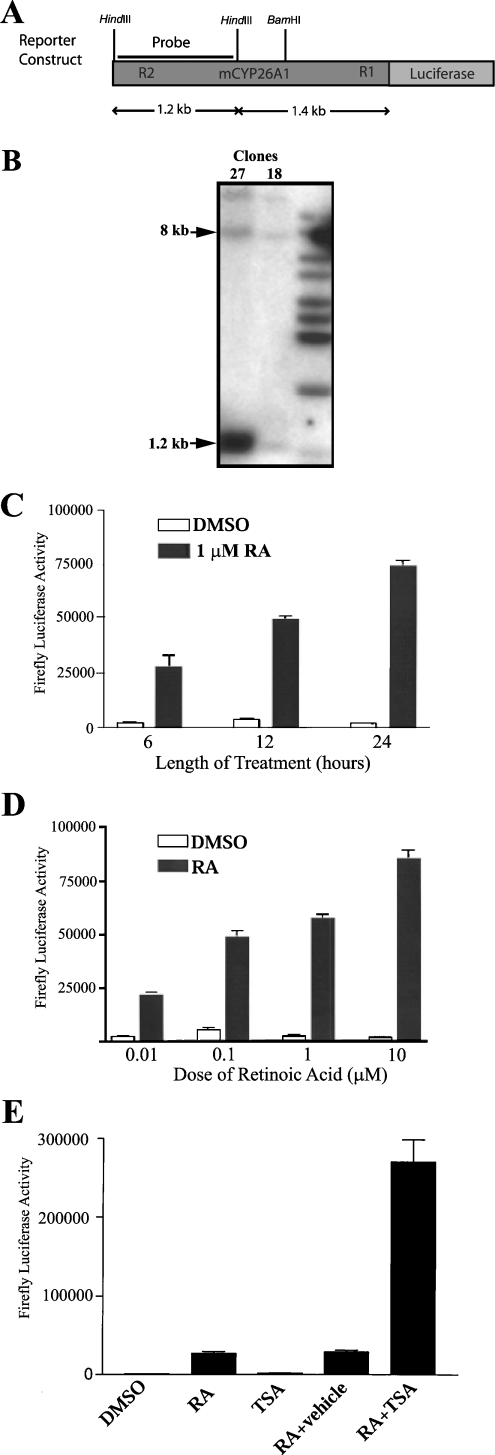
To study the regulation of this gene in stably transfected cells, a stable cell line was established containing the R1 and R2 elements upstream of a reporter gene. The 2.6 kb of sequence upstream of the mouse Cyp26A1 transcriptional start site was fused to a luciferase reporter gene and cloned into the vector pcDNA 3.1, which contains a neomycin resistance marker (Figure 6A). P19 cells were transfected with the construct and then grown in media supplemented with Geneticin for 2 weeks to allow for selection of clones in which the construct had stably integrated into genomic DNA. Isolated clones (27 in total) were picked, cultured separately and tested for luciferase activity after treatment with RA. Four clones showed high luciferase activity after RA treatment (results not shown), and these clones were analysed by Southern blotting to confirm stable integration of the construct. HindIII-digested genomic DNA was blotted and probed with a 1.2 kb fragment corresponding to the distal region of the construct. With this strategy, an 8 kb band would represent endogenous Cyp26A1 alleles, while the integrated construct would yield a 1.2 kb band. When tested, clone 27 (Figure 6B) clearly showed both bands, indicating that the construct had been stably integrated, and this clone was chosen for further analysis.
Figure 6. Characterization of a stable cell line containing the R2-Cyp26A1 reporter construct.
(A) Design of the construct used for generation of the stable cell line; 2.6 kb of the mouse Cyp26A1 promoter was fused to a luciferase reporter gene. (B) Southern blot for two clones (27 and 18) showing integration of the construct into stably transfected P19 cells. A HindIII genomic digest was performed for the blot, and the probe used is indicated in (A). Bands at 8 kb (endogenous locus) and 1.2 kb (construct) are indicated by arrowheads. (C) Induction of clone 27 reporter activity over 24 h treatment with either DMSO or DMSO/1 μM RA. (D) Response of clone 27 to treatment for 24 h with increasing doses of RA. (E) Treatment of the stable cell line with 100 ng/ml of the HDAC inhibitor TSA either alone or with 1 μM RA. TSA was dissolved in ethanol, and was the vehicle that was used along with RA to treat cells in the fourth sample set.
To measure the transcriptional activation of the Cyp26A1 promoter construct over time, luciferase activity was measured 6, 12 and 24 h after treatment with 1 μM RA (Figure 6C). After 6 h, reporter activity was induced 8-fold, and by 24 h there was a 24-fold induction of luciferase activity. This agrees with previous results that have shown that, in vitro, Cyp26-mediated hydroxylase activity increases between 8 and 24 h after RA treatment [31].
The sensitivity of the stable cell line to different doses of RA was tested by treating cells for 24 h (Figure 6D). Treatment for 24 h with 0.01 μM RA caused a 10-fold induction, and fold induction increased with concentration of RA, reaching a 48-fold induction with treatment of 10 μM RA. These results are similar to the dose response observed when the construct was transiently transfected (Figure 5B), and confirm the sensitivity of the integrated construct to low concentrations of RA.
Finally, to examine whether chromatin remodelling had an effect on transcriptional activity of the integrated construct, the cell line was treated with the HDAC inhibitor TSA (trichostatin A). Treatment with RA alone results in an induction of luciferase activity in the stable cell line (Figure 6E). However, this induction was increased a further 7-fold when cells were treated with both RA and 100 ng/ml of TSA, reflecting synergism between TSA and RA as has been reported in [32]. Cells treated with only TSA showed no increased reporter activity, and cells treated with RA and ethanol (vehicle for TSA) responded identically to cells treated with RA alone.
DISCUSSION
Cyp26A1 is an RA-metabolizing enzyme, the expression of which is inducible by RA in tissues and cell lines from mouse, human, chick, zebrafish and frog [7,21,28,33,34]. Considering the potent effects of RA as a morphogen, Cyp26A1 induction by its substrate probably plays a critical physiological role in regulating and equilibrating RA distribution both in the embryo and differentiating tissues in the adult. It has been previously reported that there is a RARE (R1) in the proximal promoter of Cyp26A1 which is responsible for low-level transcriptional induction by RA [27]. In the present study, we have identified a second, upstream RARE (R2) that acts synergistically with R1 to enhance the transcriptional response of the Cyp26A1 promoter to RA.
The R2 element is located 2.5 kb upstream of the murine Cyp26A1 transcriptional start site, and is found within a 42 bp region that is perfectly conserved between human and mouse. Further analysis of the two genomes showed that the R1 and R2 elements are embedded within highly conserved regions and that there is no significant similarity in the intervening sequence. This indicates that both elements may play significant functional roles in the regulation of Cyp26A1 expression.
The R2 element is an example of a classical DR5 RARE. EMSA experiments show that the R2 element forms a specific complex with RARγ and RXRα. Additionally, when RARγ and RXRα were co-transfected along with a construct containing both R1 and R2 elements, induction by RA increased from 4-fold to 10-fold. RXRα is ubiquitously expressed in the murine embryo, while RARγ isoforms are expressed in the tailbud and precartilaginous condensations [17,18], which are regions where Cyp26A1 is up-regulated following administration of RA [21] (G. A. MacLean, unpublished work). Coupled with the fact that Cyp26A1 induction is lost in RARγ/RXRα −/− cell lines [22], these findings support the possibility that the RARγ/RXRα heterodimer is responsible for Cyp26A1 induction in vivo.
Through deletion and point mutation analysis, it was shown that inclusion of the R2 element enhances Cyp26A1 induction by RA. The R2 element was able to enhance induction regardless of orientation. It was also unable to support transcription in the absence of R1 and its surrounding sequences. Therefore we propose that the Cyp26A1 gene contains a single promoter that includes R1 and that R2 is an upstream enhancer element that is necessary for complete RA inducibility, but is not by itself sufficient for transcription. The genes for murine Cyp26A1 and Cyp26C1 are separated by 4.7 kb, with R2 located between the loci (A. Tahayato and M. Petkovich, unpublished work). The proximity of R2 with Cyp26C1 suggests that the activity of this enhancer may affect the expression of Cyp26C1. Preliminary studies indicate that Cyp26C1 is indeed inducible by RA albeit to a much lesser magnitude than that for Cyp26A1 [9]. Also, there are several tissues where Cyp26A1 and Cyp26C1 expression overlaps, suggesting that certain elements of transcriptional regulation may be shared [11].
RA has been observed to affect the expression of more than 530 genes involved in many diverse processes [20]. RARβ has been well characterized as inducible by RA, and is often used in experiments as a positive control for RA inducibility. We have shown that a reporter construct containing both R1 and R2 elements can be induced >50-fold when treated with 10−5 M RA in P19 cells. By comparison, under the same conditions, RARβ is induced 4-fold. The Cyp26A1 construct even when treated with 10−8 M RA shows a 5-fold induction. Considering the function of Cyp26A1, it is logical that the locus would be highly sensitive to RA to respond to minute fluctuations in retinoid levels. The high sensitivity of our construct to RA in vitro indicates that we have isolated the elements responsible for RA induction in vivo.
A stable cell line containing the 2.6 kb-Cyp26A1 construct was generated to study the role of chromatin remodelling in regulation of Cyp26A1 expression. In the absence of ligand, RAR/RXR heterodimers bind to RAREs and recruit co-repressors with HDAC activity. Unliganded RAR/RXR is required for proper expression of several genes including those involved in chondrocyte maturation [35], and head formation in frog [36]. There is evidence from vitamin A-deficient quail that Cyp26A1 expression is maintained in anterior tissues in the absence of RA, raising the possibility that unliganded RAR/RXR is involved in expression [37]. However, treatment of the 2.6 kb-Cyp26A1 cell line with the HDAC inhibitor TSA alone had no effect on reporter expression, indicating that R1 and R2 are elements required for the induction of Cyp26A1, but not basal expression. Given that other non-RAREs in the Cyp26A1 promoter are also highly conserved, it is likely that other gene regulation pathways are important for basal Cyp26A1 expression.
We note that transcriptional synergism between vitamin D-responsive elements has also been observed in the 25-hydroxy-vitamin D3 24-hydroxylase (Cyp24) promoter [38]. Since Cyp24 plays a catabolic role in vitamin D signalling analogous to that of Cyp26 in RA signalling, our findings suggest that co-operativity between distinct nuclear response elements is a conserved mechanism for generating high-level expression of catabolizing enzymes in this type of autoregulatory feedback loop.
The highly sensitive Cyp26A1-promoter reporter line we have described above may be a useful tool as an indicator cell line to detect low levels of RA. This stable line may also be useful for studying the signalling pathways that co-ordinate to induce or repress RA-mediated transcriptional regulation. Elucidation of factors involved in controlling Cyp26 regulation will be necessary to understand the initiation of pattern formation in vivo.
Acknowledgments
Operating grants from the Canadian Institute of Health Research and the National Cancer Institute of Canada supported this research. G.A.M. is supported by an R.S. McLaughlin Fellowship and a studentship from the National Science and Engineering Council. We thank D. Cameron, T. Pennimpede and K. Stewart (Cancer Research Institute, Queen's University) for critical comments on this paper.
References
- 1.Penzes P., Wang X., Sperkova Z., Napoli J. L. Cloning of a rat cDNA encoding retinal dehydrogenase isozyme type I and its expression in E. coli. Gene. 1997;191:167–172. doi: 10.1016/s0378-1119(97)00054-1. [DOI] [PubMed] [Google Scholar]
- 2.Zhao D., McCaffery P., Ivins K. J., Neve R. L., Hogan P., Chin W. W., Drager U. C. Molecular identification of a major retinoic-acid-synthesizing enzyme, a retinaldehyde-specific dehydrogenase. Eur. J. Biochem. 1996;240:15–22. doi: 10.1111/j.1432-1033.1996.0015h.x. [DOI] [PubMed] [Google Scholar]
- 3.Lin M., Zhang M., Abraham M., Smith S. M., Napoli J. L. Mouse retinal dehydrogenase 4 (RALDH4), molecular cloning, cellular expression, and activity in 9-cis-retinoic acid biosynthesis in intact cells. J. Biol. Chem. 2003;278:9856–9861. doi: 10.1074/jbc.M211417200. [DOI] [PubMed] [Google Scholar]
- 4.Mic F. A., Molotkov A., Fan X., Cuenca A. E., Duester G. RALDH3, a retinaldehyde dehydrogenase that generates retinoic acid, is expressed in the ventral retina, otic vesicle and olfactory pit during mouse development. Mech. Dev. 2000;97:227–230. doi: 10.1016/s0925-4773(00)00434-2. [DOI] [PubMed] [Google Scholar]
- 5.Niederreither K., McCaffery P., Drager U. C., Chambon P., Dolle P. Restricted expression and retinoic acid-induced downregulation of the retinaldehyde dehydrogenase type 2 (RALDH-2) gene during mouse development. Mech. Dev. 1997;62:67–78. doi: 10.1016/s0925-4773(96)00653-3. [DOI] [PubMed] [Google Scholar]
- 6.Niederreither K., Fraulob V., Garnier J. M., Chambon P., Dolle P. Differential expression of retinoic acid-synthesizing (RALDH) enzymes during fetal development and organ differentiation in the mouse. Mech. Dev. 2002;110:165–171. doi: 10.1016/s0925-4773(01)00561-5. [DOI] [PubMed] [Google Scholar]
- 7.White J. A., Guo Y. D., Baetz K., Beckett-Jones B., Bonasoro J., Hsu K. E., Dilworth F. J., Jones G., Petkovich M. Identification of the retinoic acid-inducible all-trans-retinoic acid 4-hydroxylase. J. Biol. Chem. 1996;271:29922–29927. doi: 10.1074/jbc.271.47.29922. [DOI] [PubMed] [Google Scholar]
- 8.White J. A., Ramshaw H., Taimi M., Stangle W., Zhang A., Everingham S., Creighton S., Tam S. P., Jones G., Petkovich M. Identification of the human cytochrome P450, P450RAI-2, which is predominantly expressed in the adult cerebellum and is responsible for all-trans-retinoic acid metabolism. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6403–6408. doi: 10.1073/pnas.120161397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taimi M., Helvig C., Wisniewski J., Ramshaw H., White J., Amad M., Korczak B., Petkovich M. A novel human cytochrome P450, CYP26C1, involved in metabolism of 9-cis and all-trans isomers of retinoic acid. J. Biol. Chem. 2004;279:77–85. doi: 10.1074/jbc.M308337200. [DOI] [PubMed] [Google Scholar]
- 10.MacLean G., Abu-Abed S., Dollé P., Tahayato A., Chambon P., Petkovich M. Cloning of a novel retinoic acid-metabolizing cytochrome P450, Cyp26B1, and comparative expression analysis with Cyp26A1 during early murine development. Mech. Dev. 2001;107:195–201. doi: 10.1016/s0925-4773(01)00463-4. [DOI] [PubMed] [Google Scholar]
- 11.Tahayato A., Dolle P., Petkovich M. Cyp26C1 encodes a novel retinoic acid-metabolizing enzyme expressed in the hindbrain, inner ear, first branchial arch and tooth buds during murine development. Gene Expr. Patterns. 2003;3:449–454. doi: 10.1016/s1567-133x(03)00066-8. [DOI] [PubMed] [Google Scholar]
- 12.Niederreither K., Subbarayan V., Dolle P., Chambon P. Embryonic retinoic acid synthesis is essential for early mouse post-implantation development. Nat. Genet. 1999;21:444–448. doi: 10.1038/7788. [DOI] [PubMed] [Google Scholar]
- 13.Abu-Abed S., Dolle P., Metzger D., Beckett B., Chambon P., Petkovich M. The retinoic acid-metabolizing enzyme, CYP26A1, is essential for normal hindbrain patterning, vertebral identity, and development of posterior structures. Genes Dev. 2001;15:226–240. doi: 10.1101/gad.855001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sakai Y., Meno C., Fujii H., Nishino J., Shiratori H., Saijoh Y., Rossant J., Hamada H. The retinoic acid-inactivating enzyme CYP26 is essential for establishing an uneven distribution of retinoic acid along the anterio-posterior axis within the mouse embryo. Genes Dev. 2001;15:213–225. doi: 10.1101/gad.851501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chambon P. A decade of molecular biology of retinoic acid receptors. FASEB J. 1996;10:940–954. [PubMed] [Google Scholar]
- 16.Wei L. N. Retinoid receptors and their coregulators. Annu. Rev. Pharmacol. Toxicol. 2003;43:47–72. doi: 10.1146/annurev.pharmtox.43.100901.140301. [DOI] [PubMed] [Google Scholar]
- 17.Dolle P., Fraulob V., Kastner P., Chambon P. Developmental expression of murine retinoid X receptor (RXR) genes. Mech. Dev. 1994;45:91–104. doi: 10.1016/0925-4773(94)90023-x. [DOI] [PubMed] [Google Scholar]
- 18.Mollard R., Viville S., Ward S. J., Decimo D., Chambon P., Dolle P. Tissue-specific expression of retinoic acid receptor isoform transcripts in the mouse embryo. Mech. Dev. 2000;94:223–232. doi: 10.1016/s0925-4773(00)00303-8. [DOI] [PubMed] [Google Scholar]
- 19.Dilworth F. J., Chambon P. Nuclear receptors coordinate the activities of chromatin remodeling complexes and coactivators to facilitate initiation of transcription. Oncogene. 2001;20:3047–3054. doi: 10.1038/sj.onc.1204329. [DOI] [PubMed] [Google Scholar]
- 20.Balmer J. E., Blomhoff R. Gene expression regulation by retinoic acid. J. Lipid Res. 2002;43:1773–1808. doi: 10.1194/jlr.r100015-jlr200. [DOI] [PubMed] [Google Scholar]
- 21.Iulianella A., Beckett B., Petkovich M., Lohnes D. A molecular basis for retinoic acid-induced axial truncation. Dev. Biol. 1999;205:33–48. doi: 10.1006/dbio.1998.9110. [DOI] [PubMed] [Google Scholar]
- 22.Abu-Abed S. S., Beckett B. R., Chiba H., Chithalen J. V., Jones G., Metzger D., Chambon P., Petkovich M. Mouse P450RAI (CYP26) expression and retinoic acid-inducible retinoic acid metabolism in F9 cells are regulated by retinoic acid receptor gamma and retinoid X receptor alpha. J. Biol. Chem. 1998;273:2409–2415. doi: 10.1074/jbc.273.4.2409. [DOI] [PubMed] [Google Scholar]
- 23.McCaffery P. J., Adams J., Maden M., Rosa-Molinar E. Too much of a good thing: retinoic acid as an endogenous regulator of neural differentiation and exogenous teratogen. Eur. J. Neurosci. 2003;18:457–472. doi: 10.1046/j.1460-9568.2003.02765.x. [DOI] [PubMed] [Google Scholar]
- 24.Winterfield L., Cather J., Menter A. Changing paradigms in dermatology: nuclear hormone receptors. Clin. Dermatol. 2003;21:447–454. doi: 10.1016/j.clindermatol.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 25.Gallagher R. E. Retinoic acid resistance in acute promyelocytic leukemia. Leukemia. 2002;16:1940–1958. doi: 10.1038/sj.leu.2402719. [DOI] [PubMed] [Google Scholar]
- 26.Ozpolat B., Mehta K., Tari A. M., Lopez-Berestein G. all-trans-Retinoic acid-induced expression and regulation of retinoic acid 4-hydroxylase (CYP26) in human promyelocytic leukemia. Am. J. Hematol. 2002;70:39–47. doi: 10.1002/ajh.10099. [DOI] [PubMed] [Google Scholar]
- 27.Loudig O., Babichuk C., White J., Abu-Abed S., Mueller C., Petkovich M. Cytochrome P450RAI(CYP26) promoter: a distinct composite retinoic acid response element underlies the complex regulation of retinoic acid metabolism. Mol. Endocrinol. 2000;14:1483–1497. doi: 10.1210/mend.14.9.0518. [DOI] [PubMed] [Google Scholar]
- 28.White J. A., Beckett-Jones B., Guo Y. D., Dilworth F. J., Bonasoro J., Jones G., Petkovich M. cDNA cloning of human retinoic acid-metabolizing enzyme (hP450RAI) identifies a novel family of cytochromes P450. J. Biol. Chem. 1997;272:18538–18541. doi: 10.1074/jbc.272.30.18538. [DOI] [PubMed] [Google Scholar]
- 29.Fujii H., Sato T., Kaneko S., Gotoh O., Fujii-Kuriyama Y., Osawa K., Kato S., Hamada H. Metabolic inactivation of retinoic acid by a novel P450 differentially expressed in developing mouse embryos. EMBO J. 1997;16:4163–4173. doi: 10.1093/emboj/16.14.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de The H., Marchio A., Tiollais P., Dejean A. Differential expression and ligand regulation of the retinoic acid receptor alpha and beta genes. EMBO J. 1989;8:429–433. doi: 10.1002/j.1460-2075.1989.tb03394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marikar Y., Wang Z., Duell E. A., Petkovich M., Voorhees J. J., Fisher G. J. Retinoic acid receptors regulate expression of retinoic acid 4-hydroxylase that specifically inactivates all-trans retinoic acid in human keratinocyte HaCaT cells. J. Invest. Dermatol. 1998;111:434–439. doi: 10.1046/j.1523-1747.1998.00297.x. [DOI] [PubMed] [Google Scholar]
- 32.Minucci S., Horn V., Bhattacharyya N., Russanova V., Ogryzko V. V., Gabriele L., Howard B. H., Ozato K. A histone deacetylase inhibitor potentiates retinoid receptor action in embryonal carcinoma cells. Proc. Natl. Acad. Sci. U.S.A. 1997;94:11295–11300. doi: 10.1073/pnas.94.21.11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hollemann T., Chen Y., Grunz H., Pieler T. Regionalized metabolic activity establishes boundaries of retinoic acid signalling. EMBO J. 1998;17:7361–7372. doi: 10.1093/emboj/17.24.7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swindell E. C., Thaller C., Sockanathan S., Petkovich M., Jessell T. M., Eichele G. Complementary domains of retinoic acid production and degradation in the early chick embryo. Dev. Biol. 1999;216:282–296. doi: 10.1006/dbio.1999.9487. [DOI] [PubMed] [Google Scholar]
- 35.Weston A. D., Chandraratna R. A., Torchia J., Underhill T. M. Requirement for RAR-mediated gene repression in skeletal progenitor differentiation. J. Cell Biol. 2002;158:39–51. doi: 10.1083/jcb.200112029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koide T., Downes M., Chandraratna R. A., Blumberg B., Umesono K. Active repression of RAR signaling is required for head formation. Genes Dev. 2001;15:2111–2121. doi: 10.1101/gad.908801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reijntjes S., Gale E., Maden M. Generating gradients of retinoic acid in the chick embryo: Cyp26C1 expression and a comparative analysis of the Cyp26 enzymes. Dev. Dyn. 2004;230:509–517. doi: 10.1002/dvdy.20025. [DOI] [PubMed] [Google Scholar]
- 38.Kerry D. M., Dwivedi P. P., Hahn C. N., Morris H. A., Omdahl J. L., May B. K. Transcriptional synergism between vitamin D-responsive elements in the rat 25-hydroxyvitamin D3 24-hydroxylase (CYP24) promoter. J. Biol. Chem. 1996;271:29715–29721. doi: 10.1074/jbc.271.47.29715. [DOI] [PubMed] [Google Scholar]