Abstract
The protein levels of β-catenin are tightly regulated by the ubiquitin/proteasome system. We provide evidence that two distinct ubiquitin-dependent degradation pathways for β-catenin are active in the same Burkitt's lymphoma cells: Along with the classical glycogen-synthase kinase 3β-dependent destruction machinery, degradation of β-catenin through seven in absentia homolog 1 (Siah-1) ubiquitin ligase is functional in these cells. We show that inhibition of endogenous Siah-1 stabilizes and activates β-catenin in B cells. The principal Epstein–Barr virus oncoprotein, latent membrane protein 1, is involved in β-catenin up-regulation, and expression of latent membrane protein 1 in B lymphoma cells is associated with decreased Siah-1 RNA and protein levels. Thus, we demonstrate the significance of the endogenous Siah-1-dependent ubiquitin/proteasome pathway for β-catenin degradation in malignant human cells and its regulation by a viral oncogene.
Keywords: B lymphoma, ubiquitin/proteasomal degradation, Epstein–Barr virus, latent membrane protein 1
The protein β-catenin has critical functions in several normal and malignant intracellular processes. In cell-adhesion signaling, β-catenin binds the cytoplasmic domain of cadherin adhesion receptors along with α-catenin to transmit signals from cadherins to the underlying actin cytoskeleton (1). The cytoskeletal network of adherent cells is considered essential for normal tissue architecture and morphogenesis. When part of the transcriptional machinery in the nucleus, β-catenin can bind to Tcf/Lef factors and serve as a transcriptional coordinator for activation of target genes (2). The balance of β-catenin between these two functional complexes might be explained by distinct molecular forms of β-catenin that exist in cells (3), but the mechanism of this regulation still remains to be explored. In the meantime, it is well established that, in general, the protein levels of β-catenin are regulated by the ubiquitin-proteasome system (UPS) (2, 4).
Understanding of the mechanism of β-catenin regulation through the UPS came primarily from the study of canonical Wnt signaling in which β-catenin stabilization plays a central role (4). According to the most accepted model, β-catenin is efficiently targeted for ubiquitination and degradation by a multiprotein complex, including the serine/threonine kinase, glycogen-synthase kinase 3β (GSK3β), the scaffold proteins, Axin/conductin, and the adenomatosis polyposis coli tumor-suppressor protein (2, 4). Mutations in the components, such as Axin or adenomatosis polyposis coli, can be sufficient for functional activity of this destruction complex (2). Initial phosphorylation of β-catenin by casein kinase 1 precedes its further phosphorylation by GSK3β on the NH2 terminus of β-catenin (5). Phosphorylation of β-catenin by GSK3β is essential for the ubiquitination of β-catenin mediated by the SCFβ-TrCP ubiquitin ligase, in combination with the Skp1 and Cullin1 ubiquitin-conjugating complex (6). Initiation of classic Wnt signaling leads to inhibition of GSK3β-dependent phosphorylation and degradation of β-catenin and, as a result, activation of the β-catenin/Tcf transcriptional pathway (4). Because of the fact that inhibition of GSK3β is a key point in rescue of β-catenin from phosphorylation-dependent destruction, other cell signaling pathways affecting GSK3β kinase activity have been implicated in β-catenin signaling (7).
Also, a different pathway of β-catenin ubiquitination and proteasomal degradation has been uncovered. In this case, a distinct ubiquitin–ligase complex, Siah-1-SIP-Skp1-Ebi, promotes degradation of β-catenin through a mechanism independent of GSK3β-mediated phosphorylation (8, 9).
Siah (seven in absentia homolog) proteins are members of an evolutionarily highly conserved family of E3 ubiquitin ligases. They bind ubiquitin-conjugating enzymes via an N-terminal RING domain and target other proteins for ubiquitination and proteasomal degradation (10). Besides β-catenin, several important targets of the Siah-dependent ubiquitin/proteasome pathway have been identified (11, 12). Siah proteins are implicated in a variety of cellular processes, such as apoptosis and tumor suppression. Some studies have also suggested that human Siah-1 acts as a downstream effector of p53 (8, 9). However, the biological significance of Siah-dependent β-catenin ubiquitination and its regulation in normal and cancer cells is largely unknown.
The ability of Epstein–Barr virus (EBV) to transform normal resting B cells into indefinitely proliferating lymphoblastoid cell lines is a successful viral strategy for survival and the basis of several human malignancies. EBV immortalizes B lymphocytes through viral proteins that modify cellular gene expression at the level of transcription directly or indirectly by dysregulating cell signaling pathways. In type III EBV latency, exemplified by EBV lymphoproliferative disease in immunocompromised hosts, all nine viral latency products are expressed (13). Among them, latent membrane protein 1 (LMP1) is of particular interest as a primary EBV oncogene because it transforms rodent fibroblasts in vitro and induces lymphomas in transgenic mice (13). In addition, it can dysregulate cell signaling pathways and induce a variety of cellular genes that enhance cell survival and adhesive, invasive, and angiogenic potential (13, 14).
Human tumor viruses, including EBV, can activate the β-catenin signaling by different mechanisms (15–17). In this study, we show that two distinct pathways of β-catenin destruction through the ubiquitin-proteasome system coexist in the same lymphoid cells, and that the EBV oncoprotein LMP1 up-regulates β-catenin by increasing its stability through inhibition of Siah-1-mediated ubiquitination.
Materials and Methods
Plasmids. Wild-type pcLMP1 has been described in ref. 18. Tcf reporter plasmids, TOPFlash (optimal Tcf-binding site) and FOP-Flash (mutated Tcf-binding site) were obtained from Upstate Biotechnology. pHA-ubiquitin encodes a hemagglutinin (HA)-tagged ubiquitin was a gift from Y. Xiong (University of North Carolina). Both wild-type and mutant forms (S37A) of β-catenin-expressing plasmids were kindly provided by S.-G Hwang, (Kwangju Institute of Science and Technology, Gwangju, Korea). pSiahΔ1–75 plasmid, which expresses a Siah-1 dominant-negative mutant (Siah-1DN), has been described in ref. 9. The small interfering RNA (siRNA) duplexes were synthesized and purified by Qiagen (Cambridge, MA). Siah-1L target sequence was as follows: siSiah-1L, 5′-AACTCCTGCCTCCTTATGTATTT-3′. The nonsilencing siRNA (control siRNA) sequence was as follows: siCTR, 5′-AAGAGCCGTCAGACTGCTACA-3′.
Cell Culture and Transient Transfection. DG75 is an EBV-negative Burkitt's lymphoma (BL) cell line (19). Sav I and Sav III are genetically identical BL cell lines that differ in their EBV latency status (20). BL41-P3HR1 and BL41-B95-8 are cell lines prepared by infection of EBV-negative BL41 cells with the two different EBV strains, P3HR1 and B95-8 (21). Lymphoblastoid cell lines (LCL-23, -45, -67, and -89), generated by infecting B cells from anonymous healthy donors with B95-8 EBV strain, were provided by the Tissue Culture Facility of the Lineberger Cancer Center. All cells were maintained in RPMI 1640 medium plus 10% FBS. Cells were transfected by an electroporation method with the use of the Bio-Rad Gene Pulser at 210 V and 975 μF at the indicated concentrations of the LMP1-expressing plasmid. Vector DNA was added to equalize the total amount of DNA (10 μg) used in all transfections. After electroporation, cells were resuspended in 10 ml of complete medium and incubated for 48 h before harvesting.
Western Blot Analysis. Cells were lysed in lysis buffer [50 mM Hepes, pH 7.4/150 mM NaCl/10% glycerol/1 mM EDTA/1 mM sodium orthovanadate/100 mM NaF/1% Triton X-100/protease inhibitor mixture (Roche Diagnostics)]. Protein concentration was determined by the Bradford assay (Bio-Rad). Total cell proteins were resolved on SDS/PAGE, transferred to nitrocellulose membrane (Osmonics), blocked in 5% milk/Tris-buffered saline solution, and incubated at room temperature for 2 h with β-catenin (BD Transduction Laboratories), LMP1 (DAKO), and γ-tubulin (Sigma), Siah-1 (Transgenic) and Myc-tag (Cell Signaling Technology) antibodies. After washing with TBST for 10 min three times, the membrane was incubated with appropriate secondary antibody at room temperature for 1 h, washed three times with TBST as before, treated with SuperSignal (Pierce) detection reagents, and exposed to Kodak XAR-5 film.
Luciferase Reporter Assay. For dual-luciferase reporter assay, DG75 cells were transiently transfected with 3 μg of Tcf reporter plasmids, TOPFlash or FOPFlash, and the indicated amounts of the effector plasmid as described above. To control for transfection efficiency, a control reporter, pRL-TK (0.1 μg), which contains a herpes simplex virus thymidine kinase promoter driving a Renilla luciferase gene, was cotransfected. After 48 h, cells were lysed in passive lysis buffer, and luciferase activities were monitored in cell lysate with the use of Dual-Luciferase assay reagents (Promega) as described by the manufacturer. All reporter assay results presented are from two independent experiments prepared in triplicate.
Immunoprecipitation. DG75 cells were cotransfected with HA-tagged ubiquitin-encoding plasmid and increasing amounts of LMP1-expressing plasmid as described. Whole-cell lysates were precleared with A/G PLUS-Agarose beads for 30 min at 4°C and incubated with anti-HA tag antibody (Santa Cruz Biotechnology) for an additional hour and then with beads overnight at 4°C. After intensive washing and centrifugation, immune complexes were separated by SDS/PAGE and probed with β-catenin antibody by Western blotting.
RT-PCR. Total cellular RNA was extracted from cells with the use of TRIzol (GIBCO). DNase I-digested RNA (3 μg) was reverse-transcribed with the corresponding antisense primer. One-quarter of the reverse-transcribed RNA was amplified with Taq polymerase (95°C for 5 min; 30 cycles at 95°C for 1 min, 56°C for 1 min, 72°C for 30 sec, and 72°C for 5 min) by using sense primers 5′-GAC TGG CAC AAC TGC ATC CA-3′,5′-CTT CGA AAG AGA CCT TCT CT-3′, and 5′-ATG GGG AAG GTG AAG GTC GG-3′ and antisense primers 5′-AGC CAA GTT GCG AAT GGA TC-3′, 5′-ACA ATG CCT GTC CGT GCA AA-3′, and 5′-TGG AGG GAT CTC GCT TG-3′ for Siah-1, LMP1, and GAPDH, respectively.
The real-time RT-PCR reactions were performed in a volume of 20 μl containing oligonucleotide primers (5 μM of each), MgCl2 (5 mM), one-tenth of the cDNA (2 μl), and DNA Master SYBR green (Roche Molecular Biochemicals) containing Taq DNA polymerase, reaction buffer, dNTP, and the double-stranded DNA-specific fluorescent dye SYBR Green I. Amplification was carried out in two steps: preincubation at 95°C for 10 min and 45 cycles with denaturation at 95°C for 15 sec, annealing at 56°C for 7 sec, and extension at 72°C for 9 sec. Acquisition of the fluorescent signal from the samples was carried out at the end of the elongation step.
Results
First, we compared endogenous levels of LMP1 and β-catenin in several B lymphocytic lines to find whether there was any correlation between LMP1 and β-catenin expression. We used several Burkitt's lymphoma cell lines, including BL41-P3HR1 and BL41-B95, infected by the prototype EBV B95–8 strain or the defective P3HR1 virus, respectively. BL41-P3HR1 cells express very low levels of LMP1, whereas BL41-B95 cells express high levels. When these lines were compared with the parental line by Western blotting, only BL41-B95-8 cells had a high level of β-catenin (Fig. 1a, lanes 1–3). These results are consistent with the previous report by Everly et al. (22), in which the authors observed an increase in the level of total β-catenin protein in EBV-infected Burkitt's lymphomas and no difference in mRNA levels. Additional evidence came from comparison of LMP1 and β-catenin levels in four different type III latently EBV-infected lymphoblastoid cell lines (LCLs) that expressed different levels of LMP1. A 40% decrease in LMP1 levels correlates with 6.5-fold lower β-catenin levels in LCLs (Fig. 1a, lanes 4–7). These observations suggest that LMP1 as a product of type III latency up-regulates β-catenin in B cells.
Fig. 1.
LMP1 up-regulates β-catenin in B-cell lines. (a) Correlation of β-catenin and LMP1 protein levels in EBV-positive and -negative B cell lines. Total cell lysates were probed with β-catenin and LMP1 antibodies. Each band was quantified with the use of bioproril bio 1d image analysis software (Vilber Lourmat, Marne-la-Vallée, France). (b) LMP1 expression increases β-catenin levels in EBV-negative B lymphoma cells. Increasing amounts of LMP1-expressing vector (pcLMP1) were transfected into DG75 for 48 h, and protein levels of β-catenin and LMP1 were detected with indicated antibodies. (c) Increase of β-catenin/Tcf transcriptional activity by LMP1. pcLMP1 (1 μg) was cotransfected with either wild-type (TOPFlash) or mutant (FOPFlash) reporter plasmids into DG75 cells. The total amount of DNA was kept at 10 μg. After 48 h of transfection, luciferase activity was determined. The data represent two independent experiments prepared in triplicate.
To investigate whether LMP1 is sufficient for the up-regulation of β-catenin, an LMP1-expressing plasmid was introduced into an EBV-negative B cell line, DG75. The exogenously introduced LMP1 increased the level of β-catenin in a dose-dependent manner (Fig. 1b), confirming the direct role of LMP1 in the accumulation of β-catenin in B cells. To determine whether the increased β-catenin is transcriptionally active, we transfected cells with reporter plasmids containing wild-type (TOPFlash) and mutant (FOPFlash) binding sites for Tcf (23). Fig. 1c shows that LMP1 specifically induces Tcf reporter activity. These results indicate that at least some of the LMP1-induced β-catenin is transcriptionally active.
The level of β-catenin is normally regulated by UPS. As shown in Fig. 2a, lane 1, the β-catenin level in DG75 cells is maintained at a low level, but in the presence of the proteasome inhibitor MG101, it accumulates dramatically (Fig. 2a, lane 3), indicating that β-catenin in these B lymphoma cells is constitutively recycled by proteasome machinery. LMP1 partly stabilizes β-catenin (Fig. 2a, lane 2), but in the presence of MG101, there is additional accumulation of the protein (Fig. 2a, lane 4). Ubiquitination is usually a necessary step that precedes proteasomal degradation (24). To examine whether LMP1 affects β-catenin on the level of ubiquitination, we introduced HA-tagged ubiquitin along with LMP1 into DG75 cells and immunoprecipitated ubiquitin-complexed products. As shown in Fig. 2b, the amount of ubiquitin-complexed β-catenin decreased significantly in the presence of LMP1, which indicates that LMP1 reduces β-catenin degradation at the level of ubiquitination or upstream of it.
Fig. 2.
LMP1 reduces ubiquitination of β-catenin in B lymphoma cells. (a) DG75 cells were either mock-treated (lanes 1 and 2) or treated with 10 μM MG101 (lanes 3 and 4) in the presence (lanes 2 and 4) or absence (lanes 1 and 3) of LMP1 for 4 h, and the level of β-catenin was determined. (b) Increasing amounts of pLMP1 (0, 1, and 3 μg, respectively) were transfected along with pHA-ubiquitin (3 μg) into DG75 cells for 48 h. The total amount of DNA was kept at 10 μg. Ubiquitin-conjugated products were immunoprecipitated with HA antibody and pulled down with A/G PLUS-Agarose. The level of ubiquitin-attached β-catenin was detected by Western blotting with β-catenin antibody. As a loading control, the amount of IgG in the precipitates and the level of tubulin in the total lysates are shown. Each band was quantified as described in Fig. 1a. (b Right) The correlation between total and ubiquitinated β-catenin.
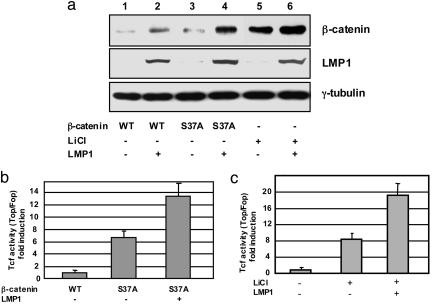
As part of the ligase complex, β-TrCP1 recognizes β-catenin as a substrate for ubiquitination only when it is phosphorylated by GSK3β at both Ser-33 and Ser-37 residues (25). Indeed, use of nonphosphorylable mutants of β-catenin and the pharmacological inhibitor of GSK3β, LiCl, has demonstrated activation of β-catenin through inhibition of ubiquitin-dependent degradation (26). We therefore introduced a nonphosphorylable form of β-catenin, S37A, which has Ala instead of Ser at position 37, into DG75 cells. The results indicate that the β-catenin S37A mutant is more stable and transcriptionally active than wild type β-catenin (Fig. 3 a, lanes 1 and 3, and b, columns 1 and 2). Similar results were obtained in the presence of LiCl; inhibition of GSK3β activity increases β-catenin protein levels (Fig. 3a, lanes 1 and 5) and Tcf reporter activity (Fig. 3c, columns 1 and 2). These results are consistent with our previous observation that the GSK3β/Axin/adenomatosis polyposis coli destruction complex of β-catenin is intact and functionally active in EBV-infected B lymphoma cells (20). Taken together, these data lead to the conclusion that GSK3β-dependent degradation machinery for β-catenin is active in B lymphoma cell lines.
Fig. 3.
LMP1 is involved in GSK3β-independent activation of β-catenin. (a) The level of β-catenin in DG75 cells after transfection with either wild-type (WT) or mutant form (S37A) of β-catenin (1 μg) for 48 h in the presence or absence of LMP1 (1 μg) was compared. For lanes 5 and 6, cells were treated with 20 mM LiCl for 24 h. (b) Tcf/Lef transcriptional activity in DG75 cells transfected with either WT or mutant form of β-catenin in the presence or absence of LMP1. The data indicate the average fold of TOPFlash/FOPFlash from two independent experiments prepared in triplicate. (c) LMP1 (0, 1, and 3 μg) was cotransfected with luciferase reporter into DG75 cells in the presence or absence of 20 mM LiCl, and luciferase assay was performed as described in Fig. 3b. In each experiment, the total amount of DNA was kept at 10 μg.
Unexpectedly, expression of LMP1 further increased stabilization of the nonphosphorylable form of β-catenin (Fig. 3a, compare lanes 3 and 4) and the transcriptional activity of β-catenin/Tcf complex (Fig. 3b, column 3). Moreover, LMP1 amplified protein accumulation and transcriptional activation of β-catenin induced by inhibition of GSK3β with LiCl (Fig. 3 a, lane 6, and c, column 3). These results do not rule out that LMP1 can inhibit GSK3β-dependent β-catenin destruction machinery but suggest that LMP1 is also involved in inhibition of another degradation pathway for β-catenin in these cells.
In contrast to GSK3β-dependent ubiquitin/proteasomal degradation of β-catenin through SCFβ-TrCP ubiquitin ligase complex, ubiquitination by Siah-1SIP-Skp1-Ebi does not requires β-catenin phosphorylation at its N terminus (8, 9). Because LMP1 appears to inhibit degradation of β-catenin at least, in part, through a mechanism other than GSK3β-dependent ubiquitination, we next explored whether Siah-1-dependent destruction is involved in the regulation of β-catenin by LMP1. First, we examined Siah-1 and β-catenin levels after introducing LMP1 into DG75 cells and found that with increasing amounts of LMP1, the protein levels of Siah-1 decreased, which directly correlates with β-catenin stabilization (Fig. 4a, lanes 1–3). To provide biological significance for this finding, we investigated whether Siah-1 expression correlates with the latency status of EBV-infected B-cells. We compared Siah-1, β-catenin, and LMP1 endogenous levels in the genetically identical BL cell lines Sav I and Sav III that differ in their EBV latency status: type I and type III, respectively (Fig. 4a, lanes 4 and 5), and observed a similar effect: The protein level of Siah-1 is higher in type I latency than in type III. To examine whether the ubiquitin ligase activity of Siah-1 is important for β-catenin degradation in B cells, we introduced a Siah-1 inactive mutant, Siah-1DN (9), into DG75 cells. The functionally inert Siah-1 mutant significantly increased transcriptional activity of β-catenin (Fig. 4b, compare columns 1 and 3) and protein levels (Fig. 4c, compare lanes 1 and 3), indicating that Siah-1 ubiquitinating activity plays a certain role in down-regulation of β-catenin in these cells. Moreover, the presence of LMP1 did not alter β-catenin activation by Siah-1DN (Fig. 4 b and c). In addition, we used Siah-1 siRNA to inhibit the expression of endogenous Siah-1. Introduction of Siah-1 siRNA into DG75 cells inhibited Siah-1 expression in a dose-dependent manner and, as a result, the expression level of β-catenin was increased (Fig. 4d). Consistent with results of the Siah-1 dominant-negative experiment, the stabilizing effect of LMP1 on β-catenin was abolished when Siah-1 expression was inhibited by the siRNA.
Fig. 4.
Inhibition of Siah-1 ubiquitin ligase up-regulates β-catenin in B lymphoma cells. (a) LMP1-dependent reduction of Siah-1 expression. DG75 cells were transfected with 0, 1, and 3 μg of pcLMP1 (lanes 1–3). After 48 h of transfection, the protein levels of LMP1, β-catenin and Siah-1 were determined with the indicated antibodies. In addition, genetically identical BL cell lines, Sav I and III, were used (lanes 4 and 5). (b) Functional inhibition of Siah-1 ubiquitin-ligase increasesβ-catenin/Tcf transcriptional activity. One microgram of Myc-tagged Siah-1 dominant-negative mutant-expressing plasmid (pSiah-1DN) was transfected into DG75 in the presence or absence of pcLMP1 (1 μg). The data indicate the average fold of TOPFlash/FOPFlash from two independent experiments prepared in triplicate. (c) Functional inhibition of Siah-1 ubiquitin-ligase increases β-catenin protein stabilization. One microgram of pSiah-1DN was transfected into DG75 cells in the presence or absence of pcLMP1 (1 μg). In each experiment, the total amount of DNA was kept at 10 μg. The expression of Siah-1DN was detected with an anti-c-Myc antibody. (d) Inhibition of endogenous Siah-1 expression stabilizes β-catenin. Siah-1 siRNA was transfected at the indicated concentration in the absence (lanes 1, 3, and 5) or presence (lanes 2, 4, and 6) of pcLMP1 (1 μg). The final concentration of siRNA in each transfection was made up to 50 nM by supplementation with a control siRNA. Protein levels of β-catenin, Siah-1, and LMP1 were detected with indicated antibodies.
There are several possible mechanisms whereby LMP1 can regulate Siah-1 expression. LMP1 interrupts signaling pathways on different levels, including at the level of transcription. To investigate whether LMP1 regulates Siah-1 transcriptionally, we performed semiquantitative and real-time RT-PCR assays with Siah-1 mRNA from DG75 cells in the presence and absence of LMP1. As shown in Fig. 5 exogenous expression of LMP1 reduces endogenous levels of Siah-1 mRNA in a dose-dependent manner. These experiments indicate that LMP1 signaling can regulate Siah-1 gene expression.
Fig. 5.
LMP1 inhibits Siah-1 expression on the transcriptional level. Total RNA purified from LMP1-transfected DG75 cells was subjected to RT-PCR (Left). The level of Siah-1 transcripts was quantified by real-time RT-PCR (Right). Siah-1 expression in each LMP1-transfected sample is shown in term of relative qualification compared with expression of GAPDH, which has been ascribed an arbitrary value of 1. (Error bars = SD, n = 2 in each group.)
Taken together, our results demonstrate that both GSK3β- and Siah-1-dependent pathways of β-catenin ubiquitin/proteasomal degradation are active in human B lymphoma cells, and the viral oncogene LMP1 participates in stabilization of β-catenin in these cells through inhibition of Siah-1-mediated destruction.
Discussion
The UPS is one of the most powerful mechanisms that controls cellular processes (27). For many major participants in cell signaling, including β-catenin, the lifetime of the target protein depends on coherence of all components of UPS. Dysregulation of the mechanism at any level disrupts the functional balance in cells and accounts for many diseases (28).
The regulation of β-catenin is an example of a cell strategy in which an expressed protein is a constitutive target for destruction by the proteasome machinery. In most adherent cells, a certain amount of β-catenin is necessary for normal function of cell-cell adherent junctions, and as a part of the cytoskeletal complex, β-catenin is considered to be invisible to the proteasome machinery (1). In contrast to adherent cells, studies with B lymphoma cells that grow in suspension reveal low levels of total β-catenin (refs. 15, 20, and 22; this work); even overexpressed wild-type β-catenin is barely detectable (Fig. 3a). At the same time, the amount of β-catenin increases dramatically in the presence of a proteasome inhibitor (ref. 20 and Fig. 2a). These observations indicate that the proteasome machinery in these human cancer cells (specifically, BLs) is extremely effective for the destruction of β-catenin.
As with other tumor viruses, EBV up-regulates β-catenin signaling in different cell systems through distinct signaling pathways (16). In epithelial cells, the EBV product LMP2A activates β-catenin through inhibition of the PI3/Akt pathway (29, 30). The EBV type III latency state also stabilizes and actives β-catenin in B lymphocyte lines (20). Here, we show that the EBV oncogene, LMP1, inhibits ubiquitin-dependent proteasomal degradation of β-catenin in B cells (Fig. 2) and that at least part of the stabilized β-catenin can activate the Tcf promoter (Fig. 3). A recent study by Everly et al. (22) indicates that stabilized β-catenin in EBV-infected B cells is preferably localized in cytoplasm and not in the nucleus. Indeed, detection of β-catenin in the nucleus indicates its possible involvement in transcription, but the amount of the protein in the nucleus does not necessarily reflect the level of transcriptional activity (3). In this case, the use of a Tcf reporter can show the differences in β-catenin transcriptional activity. In type III latently EBV-infected B cells, we detected stabilized β-catenin in both nucleus and cytoplasm (C. Mayer and J.S., unpublished data). Observations on β-catenin cytoplasmic localization raise important questions about other possible functions of stabilized β-catenin in EBV-infected B cells. The presence of transcriptionally active β-catenin does not eliminate the ability of another pool of β-catenin to take part in cell adhesion signaling, but the role of β-catenin as part of adhesion signaling in lymphoid cells requires further investigation.
The step of ubiquitination is critical for the normal elimination process through UPS (24). Regulation of β-catenin by phosphorylation-dependent elimination through UPS and the role of GSK3β in this process have been well documented (2, 5, 6). In perfect conformity with previous studies, our results indicate that GSK3β-dependent destruction machinery is active in B lymphoma cells: As expected, both nonphosphorylable mutant β-catenin and the GSK3β pharmacological inhibitor, LiCl, successfully stabilize and activate β-catenin (Fig. 3). At the same time, the observation that LMP1 is able to enhance β-catenin stabilization and transcriptional activity further when GSK3β is inhibited (Fig. 3) indicates that, besides the GSK3β-dependent pathway, other destruction machinery for β-catenin is functional in these cells. Evidence of this possibility for β-catenin has been reported by different research groups in distinct model systems (31–36).
Our results implicate the Siah-1 ubiquitin ligase as a part of the β-catenin ubiquitin-proteasome destruction machinery active in B lymphoma cells. Experiments with enzymatically inert mutant indicate that ubiquitinating activity of Siah-1 is required for reduction of β-catenin protein levels (Fig. 4c) and transcriptional activity (Fig. 4b). Furthermore, the inhibition of Siah-1 expression by siRNA has a stabilizing effect on β-catenin levels (Fig. 4d). Our results, along with recent studies of inactivating mutations of the Siah-1 gene in gastric cancer (32), support the role of the endogenous Siah-1 pathway in β-catenin destruction.
Our data demonstrate that the EBV oncogene LMP1 up-regulates β-catenin in B-cells, and we suggest that LMP1 stabilizes β-catenin by inhibiting the Siah-1 ligase. Our results show that expression of LMP1 reduces Siah-1 RNA levels (Fig. 5), which indicates that LMP1 might reduce transcription of Siah1 gene. The expression of Siah-1 has been attributed to regulation by p53 (8, 9); aberrant accumulation of β-catenin in tumors is associated with inactivation of the p53 gene in hepatocellular carcinoma (37). Stabilization of β-catenin is not a hallmark of BLs (20, 22). On the other hand, differences observed in β-catenin levels in B lymphoma cell lines (15, 22) might be explained, for example, by variations in p53 transcriptional activity due to p53 mutations that are frequently detected in BLs (13). There is also a possibility that LMP1 might affect the expression level of Siah-1 through the inhibition of activity of other transcriptional factors. The promoter region of Siah-1 was recently identified (38), but its regulation is still unexplored.
Finally, Siah-1 itself is a target for ubiquitination and proteasomal degradation (10), so it is possible that LMP1 may regulate Siah1 not only at the RNA level, but at the level of protein stability as well. In type III EBV latency, β-catenin is stabilized through regulation of the ubiquitin system, specifically through deubiquitination (20). Possibly both ubiquitination and deubiquitination then might be involved in β-catenin regulation not only directly, but also indirectly, in upstream steps.
In general, the existence of more than one pathway for ubiquitin-dependent degradation of β-catenin in the same cells may seem redundant but simultaneously provides an important step in understanding how cells can regulate β-catenin levels. It is tempting to speculate that different ubiquitin-proteasome machineries might be, for example, cell compartment-specific. In any event, destruction of β-catenin in BLs sets an example through which the regulation of the same target protein lifetime by UPS can be effectively controlled by distinct molecular mechanisms. The implication of Siah-1 in β-catenin proteasomal degradation in B lymphoma cells raises many important questions about the regulation of Siah-1-dependent ubiquitin ligase complex in these cells in general and by an oncogenic virus specifically. It should be noted that besides β-catenin, there are other known targets for Siah-1-dependent ubiquitination (11, 12). Therefore, the ultimate biological significance of our observation that an EBV oncogene regulates the Siah-1 ubiquitin ligase remains to be established.
This study demonstrates the coexistence of two ubiquitin-proteasome systems of β-catenin degradation (namely, GSK3β-dependent and Siah-1-dependent) in the same human cancer cells, and it shows that the product of a tumor virus can dysregulate the latter system on the level of ubiquitin ligase, thus pointing to a biological significance for the endogenous Siah-1-dependent mechanism (Fig. 6). Participation of LMP1 in β-catenin regulation, however, does not necessarily mean that LMP1 is the major determinant of β-catenin levels in EBV-transformed B cells. From the virus perspective, it is likely that different EBV products in type III latency can synergistically cooperate in stabilization and activation of β-catenin and operate through distinct signaling pathways (15, 17).
Fig. 6.
Working model of ubiquitin-dependent proteasome pathways for β-catenin in B lymphoma cells. Ubiquitin/proteasome machinery for β-catenin destruction is highly effective in BL cells. At least two ubiquitin-dependent proteasome pathways for β-catenin degradation are active in these cells. One pathway is the classical degradation of β-catenin through the SCFβ-TrCP ubiquitin ligase complex that depends on phosphorylation of β-catenin by GSK3β kinase. Another pathway is through ubiquitination of β-catenin by the Siah-1SIP-Skp1-Ebi complex. The EBV oncogene LMP1 can up-regulate β-catenin in B lymphoma cells by inhibition of the latter: Siah-1-dependent ubiquitination.
Acknowledgments
We thank M. Peifer and E. Gershburg for valuable discussions. This work was supported by National Cancer Institute Grant CA 19014 and by Korean Research Foundation Grant KRF-2003-013-C00064.
Author contributions: K.L.J. and J.S. designed research; K.L.J. and S.Y.S. performed research; K.L.J., J.S., and J.S.P. analyzed data; and K.L.J., J.S., and J.S.P. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: BL, Burkitt's lymphoma; EBV, Epstein–Barr virus; GSK3β, glycogen-synthase kinase 3β; HA, hemagglutinin; Siah, seven in absentia homolog; UPS, ubiquitin-proteasome system.
References
- 1.Gottardi, C. J. & Gumbiner, B. M. (2001) Curr. Biol. 11, R792–R794. [DOI] [PubMed] [Google Scholar]
- 2.Peifer, M. & Polakis, P. (2000) Science 287, 1606–1609. [DOI] [PubMed] [Google Scholar]
- 3.Gottardi, C. J. & Gumbiner, B. M. (2004) J. Cell Biol. 167, 339–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nelson, W. J. & Nusse, R. (2004) Science 303, 1483–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Polakis, P. (2002) Curr. Biol. 12, R499–R501. [DOI] [PubMed] [Google Scholar]
- 6.Kikuchi, A. (2000) Biochem. Biophys. Res. Commun. 268, 243–248. [DOI] [PubMed] [Google Scholar]
- 7.Meijer, L., Flajolet, M. & Greengard, P. (2004) Trends Pharmacol. Sci. 25, 471–480. [DOI] [PubMed] [Google Scholar]
- 8.Liu, J., Stevens, J., Rote, C. A., Yost, H. J., Hu, Y., Neufeld, K. L., White, R. L. & Matsunami, N. (2001) Mol. Cell 7, 927–936. [DOI] [PubMed] [Google Scholar]
- 9.Matsuzawa, S. I. & Reed, J. C. (2001) Mol. Cell 7, 915–926. [DOI] [PubMed] [Google Scholar]
- 10.Hu, G. & Fearon, E. R. (1999) Mol. Cell. Biol. 19, 724–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.House, C. M., Frew, I. J., Huang, H.-L., Wiche, G., Traficante, N., Nice, E., Catimel, B. & Bowtell, D. D. L. (2003) Proc. Natl. Acad. Sci. USA 100, 3101–3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fanelli, M., Fantozzi, A., De Luca, P., Caprodossi, S., Matsuzawa, S., Lazar, M. A., Pelicci, P. G. & Minucci, S. (2004) J. Biol. Chem. 279, 5374–5379. [DOI] [PubMed] [Google Scholar]
- 13.Rickinson, A. & Kieff, E. (2001) in Virology, eds. Fields, B. N., Knipe, D. M. & Howley, P. M. (Lippincott-Raven, Philadelphia).
- 14.Yoshizaki, T., Wakisaka, N. & Pagano, J. (2005) in Epstein–Barr Virus: Infection, Pathogenesis, and Control, ed. Robertson, E. (Horizon, Norfolk, U.K.).
- 15.Shackelford, J. & Pagano, J. S. (2004) Mol. Cell. Biol. 24, 5089–5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pagano, J. S., Blaser, M., Buendia, M. A., Damania, B., Khalili, K., Raab-Traub, N. & Roizman, B. (2004) Semin. Cancer Biol. 14, 453–471. [DOI] [PubMed] [Google Scholar]
- 17.Shackelford, J. & Pagano, J. S. (2005) in The Ubiquitin-Proteasome System, eds. Mayer, J., Layfield, R. (Portland, London), Vol. 41.
- 18.Zhang, L., Wu, L., Hong, K. & Pagano, J. S. (2001) J. Virol. 75, 12393–12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ben-Bassat, H., Goldblum, N., Mitrani, S., Goldblum, T., Yoffey, J. M., Cohen, M. M., Bentwich, Z., Ramot, B., Klein, E. & Klein, G. (1977) Int. J. Cancer 19, 27–33. [DOI] [PubMed] [Google Scholar]
- 20.Shackelford, J., Maier, C. & Pagano, J. S. (2003) Proc. Natl. Acad. Sci. USA 100, 15572–15576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calender, A., Billaud, M., Aubry, J.-P., Banchereau, J., Vuillaume, M. & Lenoir, G. M. (1987) Proc. Natl. Acad. Sci. USA 84, 8060–8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Everly, D. N., Jr., Kusano, S. & Raab-Traub, N. (2004) J. Virol. 78, 11648–11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van de Wetering, M., Oosterwegel, M., Dooijes, D. & Clevers, H. (1991) EMBO J. 10, 123–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ciechanover, A. & Ben-Saadon, R. (2004) Trends Cell Biol. 14, 103–106. [DOI] [PubMed] [Google Scholar]
- 25.Polakis, P. (2000) Genes Dev. 14, 1837–1851. [PubMed] [Google Scholar]
- 26.van Noort, M., Meeldijk, J., van der Zee, R., Destree, O. & Clevers, H. (2002) J. Biol. Chem. 277, 17901–17905. [DOI] [PubMed] [Google Scholar]
- 27.Ciechanover, A. & Schwartz, A. L. (2004) Biochim. Biophys. Acta 1695, 3–17. [DOI] [PubMed] [Google Scholar]
- 28.Ciechanover, A. (2003) Biochem. Soc. Trans. 31, 474–481. [DOI] [PubMed] [Google Scholar]
- 29.Morrison, J. A., Klingelhutz, A. J. & Raab-Traub, N. (2003) J. Virol. 77, 12276–12284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morrison, J. A. & Raab-Traub, N. (2005) J. Virol. 79, 2375–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharma, C., Pradeep, A., Wong, L., Rana, A. & Rana, B. (2004) J. Biol. Chem. 279, 35583–35594. [DOI] [PubMed] [Google Scholar]
- 32.Kim, C. J., Cho, Y. G., Park, C. H., Jeong, S. W., Nam, S. W., Kim, S. Y., Lee, S. H., Yoo, N. J., Lee, J. Y. & Park, W. S. (2004) Oncogene 23, 8591–8596. [DOI] [PubMed] [Google Scholar]
- 33.Topol, L., Jiang, X., Choi, H., Garrett-Beal, L., Carolan, P. J. & Yang, Y. (2003) J. Cell Biol. 162, 899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu, Z. & Hunter, T. (2004) Cell Cycle 3, 571–573. [PubMed] [Google Scholar]
- 35.Xiao, J. H., Ghosn, C., Hinchman, C., Forbes, C., Wang, J., Snider, N., Cordrey, A., Zhao, Y. & Chandraratna, R. A. (2003) J. Biol. Chem. 278, 29954–29962. [DOI] [PubMed] [Google Scholar]
- 36.Bonvini, P., Hwang, S. G., El-Gamil, M., Robbins, P., Kim, J. S., Trepel, J. & Neckers, L. (2000) Biochim. Biophys. Acta 1495, 308–318. [DOI] [PubMed] [Google Scholar]
- 37.Cagatay, T. & Ozturk, M. (2002) Oncogene 21, 7971–7980. [DOI] [PubMed] [Google Scholar]
- 38.Fiucci, G., Beaucourt, S., Duflaut, D., Lespagnol, A., Stumptner-Cuvelette, P., Geant, A., Buchwalter, G., Tuynder, M., Susini, L., Lassalle, J. M., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 3510–3515. [DOI] [PMC free article] [PubMed] [Google Scholar]