Abstract
Vitamin A signals play critical roles during embryonic development. In particular, heart morphogenesis depends on vitamin A signals mediated by the retinoid X receptor α (RXRα), as the systemic mutation of this receptor results in thinning of the myocardium and embryonic lethality. However, the molecular and cellular mechanisms controlled by RXRα signaling in this process are unclear, because a myocardium-restricted RXRα mutation does not perturb heart morphogenesis. Here, we analyze a series of tissue-restricted mutations of the RXRα gene in the cardiac neural crest, endothelial, and epicardial lineages, and we show that RXRα signaling in the epicardium is required for proper cardiac morphogenesis. Moreover, we detect an additional phenotype of defective coronary arteriogenesis associated with RXRα deficiency and identify a retinoid-dependent Wnt signaling pathway that cooperates in epicardial epithelial-to-mesenchymal transformation.
Keywords: coronary vessels, epicardium, retinoids, wnt, FGF
Retinoids exert their functions by binding to the nuclear receptors, retinoid A receptor and retinoid X receptor (RXR), both of which are members of the superfamily of ligand-inducible transcriptional regulators (1, 2). The ligand-activated receptors differentially regulate gene expression through specific hormone responsive DNA elements, where the receptors can bind as heterodimers or homodimers (3). Knockout mice carrying single or combined mutations of retinoid receptors (4) demonstrated that the only single mutation in its homozygous state that confers a cardiac phenotype and embryonic lethality is that of RXRα. Midgestation lethality in the RXRα mutant is due to heart failure (5) that results, in part, from thinning of the myocardial compact zone (6–8). The specific partner for RXRα responsible for the cardiac defect is currently unknown. Despite the major cardiac muscle defect of the systemic RXRα mutants, ventricular-specific RXRα mutant mice are viable and indistinguishable from their wild-type littermates (9), and in chimeric embryos made with embryonic stem cells lacking RXRα, cardiomyocytes deficient in RXRα developed normally and contributed to the ventricular chamber wall in a normal manner (10). Together, these data demonstrated that thinning of the myocardium is secondary to arrest of RXRα signaling in neighboring tissues.
Recent reports point to a putative role of the epicardium in the transduction of retinoid signaling. First, studies using retinoic acid response elements in transgenic mice indicated that epicardial cells have the potential to respond to retinoids (11). Second, some mutations that affect the epicardium (12–14) result in a thin myocardial wall, and finally, combined epicardial/placental expression of a dominant-negative retinoid A receptor in transgenic mice induces prominent cardiac defects (15). However, the absence of epicardial-restricted promoters to target Cre expression made it impossible to discriminate the role of the epicardium from the function of placental tissues in transducing retinoid signaling and direct proper cardiac morphogenesis. This is an important consideration, because mutation of other nuclear receptors expressed in the placenta have severe implications in cardiac development (16).
Here, we analyze a series of tissue-restricted mutations of the RXRα gene in the cardiac neural crest, endothelial, and epicardial lineages, for which we have generated transgenic mice driving Cre expression selectively into each of these derivatives. We show that RXRα signaling in the epicardium is critical for proper morphogenesis of the heart. Moreover, we detect an additional phenotype of defective coronary arteriogenesis associated with RXRα deficiency in the epicardium, and identify a retinoid-dependent Wnt signaling pathway that cooperates in epicardial epithelial-to-mesenchymal transformation (EMT).
Methods
Generation of Epicardial-Restricted RXRα Mutants. Transgenic mice expressing Cre-recombinase in an epicardial-restricted manner (G5 line) were generated by the use of a construct in which a 4.8-kb NotI/KpnI fragment of the GATA-5 promoter (17) was ligated upstream of the cDNA encoding phage P1 Cre-recombinase. ROSA26-indicator mice were obtained from The Jackson Laboratory.
Epicardial-restricted RXRα mutant mice were generated upon crossing G5 with a previously described mouse line that carries a RXRα floxed allele (RXRαfl/fl) (9).
Generation of Neural Crest-Restricted and Endothelial-Restricted RXRα Mutants. Neural crest-restricted RXRα mutant mice were generated by cross breeding RXRαfl/fl with Wnt-1-Cre transgenic mice (18). Double transgenic mice were characterized by PCR using the following primers: Wnt1P1, 5′-GAG CAC CCT GGA TGT GAA GT-3′ and Wnt1P2, 5′-ATT CTC CCA CCG TCA GTA CG-3′; RXRαfl: RXRp1, 5′-ACC AAG CAC ATC TGT GCT ATC T-3′ and RXRp3, 5′-ATG AAA CTG CAA GTG GCC TTG A-3′.
Endothelial-restricted RXRα mutants were generated by crossing the RXRαfl/fl line with a tie2-cre transgenic line (19). For breeding, RXRαfl/fl females were crossed with males heterozygous for the floxed RXRα allele (RXRαfl/+) and heterozygous for the Cre transgene. Primers for genotyping were: Tie2 cre: Tie2creP1, 5′-CCC TGT GCT CAG ACA GAA ATG AGA-3′; Tie2creP2, 5′-CGC ATA ACC AGT GAA ACA GCA TTG C-3′.
Histology, Immunohistochemistry, and β-Galactosidase Staining. Histological analysis was performed according to standard protocols for paraffin embedding. For immunohistochemistry, embryos were cryopreserved in 30% sucrose and embedded in OCT. Cryosections were cut at a thickness of 15 μm, unless described otherwise. Antibodies used were as follows: 1:100 α-smooth muscle actin (Sigma, clone 1A4), 1:50 β-catenin (Affinity Research Products), 1:50 FGF2 (Santa Cruz Biotechnology, sc-79), 1:50 α4 integrin (Chemicon), 1:50 TGF-β2 (Santa Cruz Biotechnology, sc-90). β-Galactosidase staining was performed on either whole-mount or frozen sections, as indicated for each panel in the figure legends.
In Situ Hybridization. Tissues were fixed overnight in 4% paraformaldehyde and embedded in paraffin. Sections were cut at a thickness of 10 μm, and in situ hybridization was performed with [35S]UTP-labeled probes, using standard procedures (20). Whole-mount in situ hybridization was performed essentially as described (21).
Collagen Gel Assay. Mouse epicardial cell monolayers were grown on three-dimensional gels containing 1% collagen type I (Vitrogen 100; Collagen Aesthetics, Palo Alto, CA), as described (22). Hearts from embryonic day 12.5 (E12.5) mouse embryos were dissected and ventricular chambers were placed epicardial side down on the collagen gel. After 48 h, the explanted hearts were removed and the gels were incubated for 60 h in DMEM containing 10% FCS and supplemented with penicillin/streptomycin and glutamine. Transformation to mesenchymal cells was defined as the invasion of cells into the gel, as described (22). The number of transformed mesenchymal cells per explant was counted by using an inverted microscope equipped with phase contrast or Hoffman modulation optics.
Chicken Embryo Manipulations. Chick embryos were obtained from macintyre Poultry (San Diego). Eggs were incubated at 38°C and staged according to Hamburger and Hamilton (23). Recombinant adenoviruses encoding full-length chicken Wnt9b (Ad-Wnt9b), a constitutively active form of Xenopus β-catenin (24) containing the internal Armadillo repeats (Ad-actβcat) and egfp (Ad-egfp) were produced and purified as previously described (25). Recombinant FGF2 (R & D Systems) was loaded into heparin acrylic beads (Sigma) for 1 h. Concentrated adenoviruses or FGF2 beads were microinjected or grafted, respectively, into the proepicardium of Hamburger–Hamilton stage 17 embryos. Embryos were harvested after 7 days and processed for immunostaining or in situ hybridization.
Results
Removal of RXRα Expression in Endothelial or Cardiac Neural Crest Lineages Does Not Affect Cardiac Morphogenesis. To map the source of inductive RXRα-dependent signaling in the heart, we have generated a collection of RXRα mutants restricted to individual cardiac cell derivatives including endothelial, neural crest, and epicardial cells, using a Cre-recombination strategy with cell-type-specific promoters (26) and an RXRα floxed allele (9). We used a Tie2-Cre transgenic mouse line (19) to delete RXRα in endothelial lineages. Adult endothelial-restricted RXRα mutants appeared at the expected Mendelian frequency (Table 1), and showed no overt cardiac malformations (Fig. 5, which is published as supporting information on the PNAS web site). Similarly, the mutation of RXRα in neural crest derivatives achieved by using Wnt-1-Cre transgenic mice (18) produced viable offspring at the expected Mendelian ratios (Table 1) with cardiac morphogenesis indistinguishable from that of control animals (Fig. 5). Moreover, the combined deletion of RXRα in endothelial- and neural crest-derived structures did not affect animal survival nor induced visible cardiac phenotypes (Table 1 and Fig. 5). These results indicate that RXRα signaling in endothelial and/or neural crest derivatives is dispensable for cardiac morphogenesis.
Table 1. Survival of mice after cell lineage-specific gene targeting of the RXRα floxed allele.
| Genotype
|
Survival, no. observed/no. expected | ||||||
|---|---|---|---|---|---|---|---|
| Strain | n | nCre+ | Age, days | Breeding | cre | RXRα | |
| tie2-cre | 56 | 39 | P20 | Tie2cre/+ RXRαf/+ × RXRαf/f | tie2 | RXRα f/+ | 22/14 |
| tie2 | RXRα f/f | 17/14 | |||||
| wnt1-cre | 97 | 66 | P20 | W1cre/+ RXRαf/+ × W1cre/+ RXRαf/+ | wnt1 | RXRα +/+ | 20/18 |
| wnt1 | RXRα f/+ | 27/36 | |||||
| wnt1 | RXRα f/+ | 19/18 | |||||
| tie2-cre mlc2V-cre | 57 | 46 | P20 | Tie2cre/+ RXRαf/f × M2vcre/+ RXRαf/f | tie2 | RXRα f/f | 13/14 |
| tie2 mlc2v | RXRα f/f | 10/14 | |||||
| mlc2v | RXRα f/f | 23/14 | |||||
| tie2-cre wnt1-cre | 21 | 15 | P20 | Tie2cre/+ RXRαf/f × W1cre/+ RXRαf/f | tie2 | RXRα f/f | 3/5 |
| tie2 wnt1 | RXRα f/f | 8/5 | |||||
| wnt1 | RXRα f/f | 4/5 | |||||
| g5-cre | 100 | 51 | E13 | G5cre/+ RXRαf/+ × RXRαf/f | g5 | RXRα f/+ | 26/25 |
| g5 | RXRα f/f | 25/25 | |||||
| g5-cre | 64 | 31 | E16 | g5 | RXRα f/+ | 20/16 | |
| g5 | RXRα f/f | 11/16 | |||||
| g5-cre | 43 | 18 | P20 | g5 | RXRα f/+ | 14/11 | |
| g5 | RXRα f/f | 4/11 | |||||
Animals hemizygous for either tie2-cre (endothelial-specific), wnt-1-Cre (neural crest-specific), a combination of tie2-cre and mlc2v-cre (ventricular myocyte-specific), a combination of tie2-cre and wnt-1-cre, or the epicardial-specific G5-cre were crossed with the RXRα floxed allele mouse to obtain each of the above cre animals that were also hemizygous for RXRα-flox allele. These animals were then bred to the homozygous RXRα-flox to obtain offspring with lineage-specific removal of RXRα. Survival rates and respective genotypes were scored. Genotypes corresponding to Cre- animals are not indicated. n, the total number of animals resulting from each breeding scheme; nCre+, the number of Cre positive animals, Pn, postnatal day n.
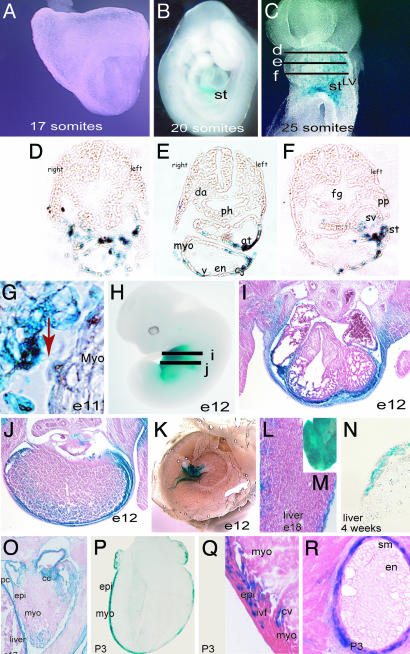
Generation of Epicardial-Restricted Cre Transgenic Line. To direct epicardial-restricted expression of Cre, we generated a transgenic mouse line containing a 4.8-kb fragment of the chicken GATA-5 promoter and Cre-recombinase (G5-Cre). As indicated by interbreeding with a ROSA26 reporter line (27), Cre-activity was preferentially restricted to epicardial derivatives during development, although it was also detected in the body wall, pericardium, and the outer envelop of some internal organs. The onset of Cre activity from the G5-Cre line occurs at the 20-somite stage (around E9.25) and it is restricted to the proepicardium (Fig. 1 A and B). At the 25-somite stage, delamination of LacZ-positive cells from the septum transversum can be observed. These cells migrate rostrally and reach the left portion of the undivided common atrium and left ventricle. Between E9.5 (25 somites) and E10 (32 somites), LacZ-positive cells envelop the caudal surface of the common ventricular chamber myocardium (Fig. 1 C–F). No expression is detected in hindgut, foregut, or the aorta. At E12, LacZ-positive cells are detected in the epicardium as well as in the body wall and in the hepatic capsule (Fig. 1 H–J). LacZ-positive cells can also be observed in the outer epithelium of the liver (Fig. 1 L–O) and kidneys. In postnatal hearts (postnatal day 3), LacZ activity is detected in the epicardium and subepicardium as well as in a subset of intermyocardial fibroblasts and smooth muscle cells of the coronary arteries. We did not observe Cre activity in the coronary endothelium (Fig. 1 P–R).
Fig. 1.
Specificity of Cre expression in the epicardium of G5-Cre transgenic mice. (A and B) The onset of Cre activity from the G5-Cre line maps at the 20-somite stage (around E9.25) and is restricted to the distal portion of the septum transversum, as shown by intercross with the ROSA indicator mouse. (C) At the 25-somite stage, delamination of LacZ-positive cells from the septum transversum can be observed in whole-mount LacZ staining. These cells migrate rostrally and reach the left portion of the common atrium and left ventricle. Between day E9.5 (25 somites) and E10 (32 somites), LacZ-positive cells envelop the caudal surface of the common left ventricular myocardium. No expression is detected in hindgut, foregut, or the aorta. (D–F) Transverse cryosections of a separate 25-somite embryo where performed at the approximate embryonic regions indicated in C and subsequently stained for β-galactosidase activity. LacZ-positive cells (Cre activity) in the septum transversum, sinus venosus, and over the myocardium in the caudal aspects of the heart are shown. (G) Delamination of epicardial cells to the outer surface of the myocardium and cytoplasmic expansions from the septum transversum to the myocardium (red arrow). (H–J) Whole-mount staining and histological sections of an E12 embryo showing Cre activity in the epicardium, pericardium, and body wall. (J) Staining also was detected in the hepatic capsule as well as some background staining within the liver. (K) Cre activity was not detected in the placenta. (L and M) Whole-mount and histological section of an E18 liver showing LacZ staining in the connective tissue covering the liver (not in the hepatocytes) (N) Frozen section of a 4-week-old liver, showing absence of staining in the hepatocytes. (O) Frozen section of an E17 embryonic heart illustrating LacZ staining in the epicardium, diaphragm, pericardium, and cardiac cushion. (P–R) Frozen sections stained for β-galactosidase activity. (P) In postnatal hearts (postnatal day 3), LacZ activity is detected in the epicardium and subepicardial layers and in a subset of intermyocardial fibroblasts (Q) and in the smooth muscle of the coronary arteries (R). Note the absence of Cre activity in the coronary endothelium (R). st, septum transversum; LV, left ventricle; pa, pharyngeal arches; v, ventricle; epi, epicardium; myo, myocardium; ivf, interventricular fibroblasts; cv, coronary vessel; sm, smooth muscle; en, endocardium; cc, cardiac cushion; pc, pericardium. Note that, to avoid penetration artefacts, older samples were LacZ-stained on cryosectioned tissues in N–R.
Cardiovascular Defects in Epicardial-Restricted RXRα Mutant Mice. Intercrossing of G5-Cre mice hemizygous for the RXRα-floxed allele with the homozygous RXRα floxed line (G5Cre;RXRαf/+ × RXRαf/f) resulted in postnatal recovery of only 36.4% of the expected Cre+;RXRαf/f offspring (four observed, 11 expected, n = 43), herein referred to as epi-RXRα mutant, demonstrating that the absence of epicardial RXRα impairs embryonic survival (Table 1). The surviving epi-RXRα mutants reach adulthood and reproduce normally; however, they have a reduced lifespan, and none of them survived >11 months of age.
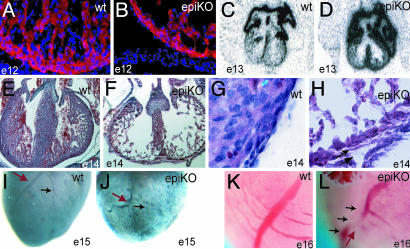
From G5cre/+;RXRαf/+ × RXRαf/f intercrosses, epi-RXRα mutant mice were scored at the expected Mendelian rate at E13 (25 observed, 25 expected, n = 51 Cre+) Because we include only the Cre-positive (nCre+) offspring in our analysis, we expect half of the Cre+ offspring to also be RXRαf/f (25 observed, 25 expected, n = 51 Cre+). However, this ratio decreased to less than the expected 50% by E16 (11 observed, 16 expected, n = 31 Cre+). Morphological analyses of the heart of epi-RXRα mutant embryos showed thinning in the myocardial wall (3.3 ± 1.1 versus 6.4 ± 1.4 cell layers deep in control siblings, n = 10 and 14, respectively; Fig. 2). These abnormalities were accompanied by detachment of the epicardium and thinning of the subepicardial component (Fig. 2 G and H). Detachment of the epicardium has been recently characterized as part of the phenotype in the systemic RXRα mutant (28). The lack of compactation was a consequence of the decreased number of myocytes in the ventricular compact layer, as indicated by staining with sarcomeric α-actinin (Fig. 2 A and B). The expression of atrial myosin light chain-2 (mlc2a), which is normally down-regulated in the ventricular compartment of E13 control embryos (5, 29, 30), persisted in epi-RXRα mutants (Fig. 2 C and D).
Fig. 2.
Histologic and morphologic analysis of epicardial mutant RXRα embryos. (A and B) A thin myocardium is detected as early as E12 and corresponds to decreased number of myocytes, as indicated by staining with the muscle marker sarcomeric α-actinin (red, α-actinin; blue, DAPI nuclear staining). (C and D) Epicardial RXRα mutation is sufficient to impair the normal down-regulation of mlc2a in E13 ventricles, and reflects deficient specification in the mutant myocardium. (E and F) Epi-RXRα mutant embryos at E14 display a lack of myocardial compaction, as indicated by hematoxylin–eosin staining. (G and H) E14 epi-RXRα mutant ventricles also exhibit epicardial detachment and thickened subepicardial space, suggesting deficient formation and/or activation of the epicardial epithelium (arrows). (I–L) Epi-RXRα mutant ventricles at E15 present altered coronary arteriogenesis, with tortuous vessels (J, red arrow) and defective branching (J, black arrow) as shown by whole-mount immunohistochemistry using a Pecam-1 antibody. (K and L) Photographs of freshly dissected E16 hearts.
We also detected a phenotype not previously characterized in the systemic knockout of RXRα. Epi-RXRα mutant embryos displayed a defect in coronary arteriogenesis as evidenced by the abnormal arterial branching over the surface of the heart (Fig. 2 I–L). Formation of the coronaries from epicardial precursors involves mesenchymal transformation to invade the subepicardial space or myocardial interstitium, whereas other precursors remain in the epicardium (22, 31). Therefore, this finding is consistent with the development of an abnormal epicardium. Furthermore, we also observed evident alterations in the coronaries of homozygous mutant mice that survived to adulthood (Fig. 6, which is published as supporting information on the PNAS web site), which may be causative of their reduced lifespan.
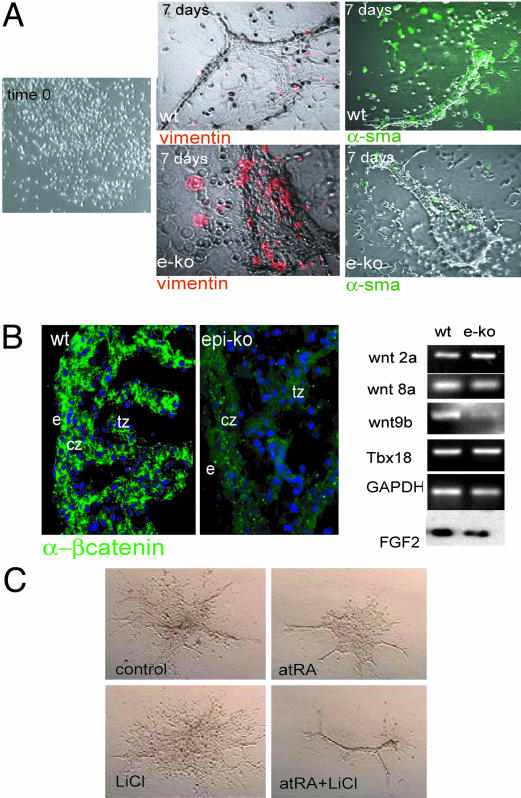
A Wnt-FGF Pathway Is Downstream of Epicardial RXRα. To analyze the role of RXRα in epicardial signaling, we cultured epicardial explants from E12–12.5 wild-type or epi-RXRα mutant embryos under conditions that promote EMT in vitro (Fig. 3). In these assays, wild-type epicardial explants form vessel-like structures that are positive for α-smooth muscle actin and express low levels of the mesenchymal transitional marker vimentin. In contrast, epicardial explants from epi-RXRα mutant embryos failed to form vessel-like structures and maintained a vimentin-positive phenotype (Fig. 3A), suggesting that RXRα signaling restricts the mesenchymal lineage.
Fig. 3.
Altered EMT and defective maturation of myocardial progenitors in epicardial RXRα mutant embryos. (A) Epithelial monolayers cultured in collagen matrix were generated from E12–E12.5 mouse ventricles as described in Methods. After a period of 7 days in culture, the initial epicardial epithelium retracts and forms vessel-like structures under high serum conditions. Explants from wild-type embryos are largely negative for the transition marker vimentin (red signal; 15 vimentin-positive cells per 120 total cells) and positive for α-smooth muscle actin (green signal). Epicardial RXRα mutant explants fail to down-regulate vimentin (red signal, 117 of 122 total cells) and are deficient in α-smooth muscle expression (green). (B) Epicardial-mutant embryos display decreased β-catenin stability, concomitant with a specific decrease in wnt9b expression, as shown by RT-PCR on isolated hearts. No changes in the other Wnt genes, Wnt 8a, Wnt 8b, and Wnt 2a, were detected at the mRNA level. No changes in the epicardial marker Tbx18 were detected. GAPDH was used as control. FGF2 protein content is also decreased in mutant embryos as detected by Western blot in isolated hearts. (C) In serum-free conditions, simultaneous treatment of wild-type epicardial monolayers with all-trans retinoic acid (atRA) and the β-catenin activator lithium chloride (LiCl) induced differentiation of explanted monolayers to a vascular-like phenotype.
To determine the downstream regulatory genes responsible for the phenotype in epi-RXRα mutants, we analyzed the expression of genes known to be involved in myocardial proliferation, epicardial activation, or EMT. These genes include erythropoietin and its receptor (32), Fgf2 (33), Fog-2 (14), α-4 integrin (12), Tbx18 (34), and β-catenin (35). Among these, we detected a down-regulation of FGF2 and β-catenin levels in the epicardium of epi-RXRα mutants (Fig. 3B and Fig. 7, which is published as supporting information on the PNAS web site). The decreased levels of β-catenin were not the result of a reduced expression of the β-catenin mRNA (data not shown), indicating a decreased stability of this protein in epi-RXRα mutants. These results are consistent with a decreased activity of Wnt signaling through the canonical pathway. Importantly, the expression of Wnt9b, which encodes a canonical Wnt ligand, was dramatically down-regulated in the epicardium of epi-RXRα mutant embryos (Figs. 3B and 7). The effect of RXRα deficiency appeared specific for Wnt9b, because the levels of other Wnt ligands expressed in neighboring tissues, such as Wnt8a, Wnt8b, and Wnt2a, were not altered in epi-RXRα mutants (Figs. 3C and 7). These results indicate that the epicardial expression of Wnt9b depends on RXRα signaling, and suggest that Wnt9b is, at least in part, responsible for the regulation of β-catenin stability during epicardial development. Consistent with this notion, simultaneous treatment of wild-type epicardial monolayers with all trans retinoic acid and the β-catenin activator LiCl promoted the differentiation of epicardial explants to a vascular phenotype (Fig. 3C).
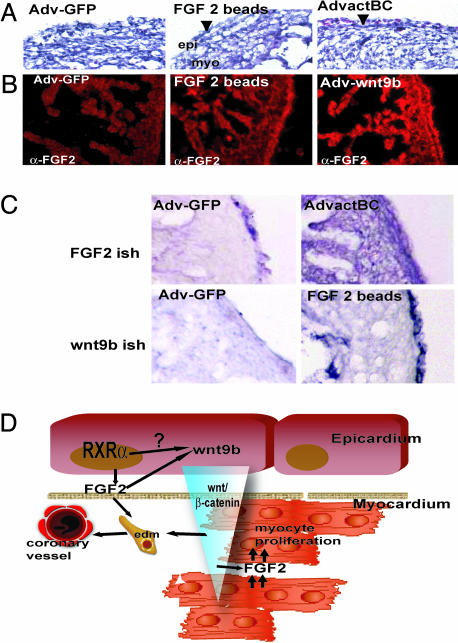
FGF2-dependent signaling is central to myocyte proliferation in the developing heart. Fgf2 and FgfR1 mRNA levels are reduced upon microsurgical inhibition of epicardial formation (36), suggesting that epicardium-derived signals regulate the levels of FGF2 in the myocardium and control myocyte proliferation (36). FGF2 plays an additional role as a modulator of epicardial EMT, as blocking antisera directed against FGF2 inhibited conditioned medium-stimulated EMT in vitro (37). The coregulation of Wnt9b and Fgf2 expression in the epicardial RXRα mutation prompted us to investigate whether Wnt and FGF signaling function in a common pathway in the crosstalk between the epicardium and myocardium. We addressed this possibility by epicardial-restricted gain-of-function experiments in chick embryos. For this purpose, we grafted beads soaked in FGF2 or injected adenovirus overexpressing either constitutively active β-catenin (Ad-actβcat) or Wnt9b (Ad-Wnt9b) in the proepicardial organ of Hamburger–Hamilton stage 17 chick embryos. Implantation of FGF2-soaked beads in the proepicardium resulted in hearts that displayed a two-layered nonvascular epicardium (Fig. 4A) in which FGF2 immunoreactivity was increased only in the epicardium (Fig. 4B). Importantly, the epicardial expression of Wnt9b was up-regulated in response to local FGF2 treatment (Fig. 4C). In turn, targeted overexpression of activated β-catenin or Wnt9b in the epicardium resulted in increased FGF2 immunoreactivity and Fgf2 expression in both epicardium and myocardium (Fig. 4 B and C). Furthermore, epicardial overexpression of Wnt9b or activated β-catenin resulted in increased vasculogenesis as indicated by the presence of subepicardial blood cells (Fig. 4A, arrowheads).
Fig. 4.
A retinoid–FGF–wnt pathway for epicardial activation and regulation of myocardial growth. (A) Histological analysis of chicken embryos overexpressing control GFP adenovirus (Adv-GFP), constitutively active β-catenin (AdvactBC), Wnt9b (Adv-wnt9b), or FGF2 beads. Observe the increased vascularization as demonstrated by the presence of blood cells in the subepicardial zone (arrowhead) upon β-catenin treatment and after grafting of FGF2-soaked beads. (B) Increased FGF2 expression is restricted to the epicardium upon FGF2 treatment. This finding is in contrast to induction of myocardial FGF2 expression after Wnt9b treatment. (C) Schematic showing the predicted relationship between wnt/β-catenin/FGF2 activation. Overexpression of active β-catenin induces FGF2 mRNA accumulation in the myocardium. Reciprocally, overexpression of epicardial FGF2 results in the accumulation of wnt9b mRNA in the epicardium. (D) Schematic of a proposed molecular mechanism for RXRα regulation of coronary formation and myocardial growth. RXRα activates the expression of epicardial FGF2. Epicardial FGF2 stimulates EMT of epithelial epicardial cells and induces epicardial wnt9b expression. Wnt9b then stabilizes myocardial β-catenin, inducing myocardial synthesis of FGF2, which in turn stimulates myocardial proliferation.
Discussion
Systemic mutation of RXRα in the mouse results in a spectrum of defects in which the most prominent alteration is the thinning of the myocardial wall, suggesting that RXRα expression regulates the proliferation and differentiation of cardiomyocytes. However, mutation of RXRα in ventricular myocytes does not affect the thickness of the myocardium or the viability of the embryos (9). Further support for a nonmyocyte requirement of RXRα comes from the analysis of chimeric embryos, in which RXRα-null myocytes populate the heart and contribute to chamber development to the same extent as wild-type cells (10). Therefore, the thickening of the myocardial wall is a myocardium response to retinoid-dependent signals originating in adjacent cell lineages. Here, we have generated mutations of the RXRα gene in the different cardiac cell lineages, including the neural crest, the endothelium, a combination of both neural crest and endothelium, and the epicardium. Of all these lineage-specific mutations, only the removal of epicardial RXRα expression demonstrated a cardiovascular phenotype.
Recent work has suggested a role for epicardial derivatives in heart development. Mutations in several epicardial genes, including α-4 integrin/VCAM (12, 38, 39), Wilm's tumor (13, 40) and erythropoietin (32) lead to defects in cardiac morphogenesis. Others have established the existence of a cross-talk between myocardium and epicardium through Friend-of-gata2 (Fog2) function. Thus, mutations in the GATA4 coactivator Fog2 resulted in a thin ventricular myocardium, common atrioventricular canal, and coronary defects. These defects were rescued by myocardial reexpression of Fog-2 (14). Interestingly, we found no differences in Fog-2 expression between wild-type and RXRα mutant embryos (not shown). Similarly, the expression of α-4 integrin, Wilm's tumor and erythropoietin was unchanged in epi-RXRα mutant embryos (data not shown). These results indicate the existence of an alternative regulatory pathway for epicardial signaling in the heart that depends on RXRα but is independent of Fog-2/GATA4 signaling.
Importantly, the effect of epicardial RXRα mutation is not as severe as observed in the systemic mutant. The myocardium of epicardial RXRα mutant mice is thin and fails compactation, but is never observed as a single-layered tissue. Therefore, the question arises as to whether this milder phenotype of epi-RXRα mutant mice is due to incomplete recombination of the floxed allele or the requirement of RXRα expression in a combination of lineages in the heart, or reflects the function of RXRα in extracardiac tissues. Direct determination of the rate of recombination in the epicardium is technically challenging, because Cre expression generates a truncated RXRα form that cannot be distinguished from the wild-type RXRα by immunocytochemistry (41). Even though our histochemical analyses using RO-SA26 indicator mice demonstrate Cre activity in most epicardial cells, we cannot formally rule out the possibility that background levels of RXRα could account for the milder phenotype of the epicardial RXRα mutants.
A significant portion of epi-RXRα mutant mice survive to adulthood, indicating the existence of compensatory signals, probably outside the epicardium, that regulate retinoid-mediated myocyte growth. Other defects observed in the systemic mutant (5–7) are also absent in epi-RXRα mutant mice, most significantly those related to the formation of the cardiac cushion mesenchyme and outflow tract. The alterations in cardiac cushion mesenchyme in the systemic RXRα mutants correlate with an increased myocardial expression of TGF-β2, which results in enhanced apoptosis in both the sinistroventralconal cushion and dextrodorsalconal cushion, and malformations in both the outflow tract and the aortic sac (8). The absence of TGF-β2 up-regulation in the epicardial RXRα mutants (data not shown) may be related to the normal mosphogenesis of the cardiac cushion mesenchyme and outflow tract in these animals. Taken together, our data demonstrate that epicardial RXRα is required for normal cardiac morphogenesis, and suggest that RXRα-expressing cells in addition to the epicardium are required to achieve the full spectrum of defects observed in the null RXRα mutant mice. In the outflow tract in particular, it is likely that RXRα is required, not only in the epicardium, but also in at least one of the other cell lineages including myocardial, endocardial, neural crest, and mesenchymal cells, to prevent the full spectrum of defects induced in the systemic mutation. Further studies are needed to examine the effects of removing epicardial RXRα in combination with these other lineages to test combinatorial effects of the RXRα mutation.
Formation of the coronary vessels from epicardial precursors is a process that occurs, in part, from some epicardial cells undergoing mesenchymal transformation and invading the subepicardial space or myocardial interstitium (22, 31). The anatomical defects in the coronary vessels of epi-RXRα mutants, together with the retention of a mesenchymal phenotype in epicardial explants of epi-RXRα mutants, indicate that RXRα-dependent signaling restricts the extent of mesenchymal transformation of epicardial cells. Recent studies underscore the importance of restraining EMT in coronary formation. For instance, inhibition of α4-integrin stimulates the maintenance of the mesenchymal phenotype and impairs coronary formation (31). Although the coronary defect of epi-RXRa mutants appears to be independent of α4-integrin expression, it is possible that RXRα and α4-integrin share some downstream signaling factors.
Previous evidence from experiments using conditioned media suggested the existence of epicardial factors that induce myocyte proliferation (15). Our data show that at least two different families of secreted factors are downstream of epicardial RXRα signaling. We have identified Wnt-9b and FGF-2 as genes responsive to the loss of RXRα in the epicardium. These data support a recent finding suggesting that Wnt genes may regulate myocyte proliferation (42).
Our findings provide a mechanistic insight into the roles of retinoic acid signaling during ventricular chamber development (Fig. 4D). We demonstrate a pleiotropic function for epicardial RXRα signaling through an FGF2/Wnt9b loop that controls the maturation of epicardial-derived mesenchymal cells and the secretion of mitotic factors. We propose a working model where down-regulation of this loop, as in epi-RXRα mutants, would result in impaired epicardial EMT and associated defects in vasculogenesis, as well as in a loss of FGF-mediated myocyte proliferation (36). These events would then lead to a thinning of the myocardium. FGFs other than FGF2 may also act on the myocardium as effectors of Wnt-β-catenin signaling because recent evidence has uncovered a mitogenic function of epicardial FGF9, which is also responsive to retinoid signaling (43). Future experiments using epicardial-specific loss-of-function of these factors are needed to test this hypothesis.
Supplementary Material
Acknowledgments
We thank Tiffany Neal and Jacob Andrade for excellent technical assistance. E.M. is a recipient of National Institutes of Health Training Grant, P.K. is a Ramon y Cajal Awardee, and M.Z. is a recipient of an Award from the Spanish Ministerio de Educacion y Ciencia. A.R. is partially supported by a postdoctoral fellowship from Fundación Inbiomed (San Sebastián, Spain). This work has been supported by grants from Fundación Inbiomed (to J.C.I.B.), The G. Harold and Leila Mathers Charitable Fund (to J.C.I.B.), and the National Institutes of Health (to J.C.I.B., S.W.K., K.R.C., and P.R.-L.).
Author contributions: P.R.-L. designed research; E.M., M.Z., A.R., Y.K., J.W., X.Z., S.W.K., and P.R.-L. performed research; J.B. and P.R.-L. contributed new reagents/analytic tools; E.M., M.Z., A.R., P.K., J.C.I.B., K.R.C., and P.R.-L. analyzed data; and P.R.-L. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: RXR, retinoid X receptor; EMT, epithelial-to-mesenchymal transformation; En, embryonic day n.
References
- 1.Evans, R. M. (1988) Science 240, 889–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Green, S. & Chambon, P. (1988) Trends Genet. 4, 309–314. [DOI] [PubMed] [Google Scholar]
- 3.Umesono, K., Murakami, K. K., Thompson, C. C. & Evans, R. M. (1991) Cell 65, 1255–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mark, M., Ghyselinck, N. B., Wendling, O., Dupe, V., Mascrez, B., Kastner, P. & Chambon, P. (1999) Proc. Nutr. Soc. 58, 609–613. [DOI] [PubMed] [Google Scholar]
- 5.Dyson, E., Sucov, H. M., Kubalak, S. W., Schmid-Schonbein, G. W., DeLano, F. A., Evans, R. M., Ross, J., Jr., & Chien, K. R. (1995) Proc. Natl. Acad. Sci. USA 92, 7386–7390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sucov, H. M., Dyson, E., Gumeringer, C. L., Price, J., Chien, K. R. & Evans, R. M. (1994) Genes Dev. 8, 1007–1018. [DOI] [PubMed] [Google Scholar]
- 7.Gruber, P. J., Kubalak, S. W., Pexieder, T., Sucov, H. M., Evans, R. M. & Chien, K. R. (1996) J. Clin. Invest. 98, 1332–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kubalak, S. W., Hutson, D. R., Scott, K. K. & Shannon, R. A. (2002) Development (Cambridge, U.K.) 129, 733–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, J., Kubalak, S. W. & Chien, K. R. (1998) Development (Cambridge, U.K.) 125, 1943–1949. [DOI] [PubMed] [Google Scholar]
- 10.Tran, C. M. & Sucov, H. M. (1998) Development (Cambridge, U.K.) 125, 1951–1956. [DOI] [PubMed] [Google Scholar]
- 11.Xavier-Neto, J., Neville, C. M., Shapiro, M. D., Houghton, L., Wang, G. F., Nikovits, W., Stockdale, F. E. & Rosenthal, N. (1999) Development (Cambridge, U.K.) 126, 2677–2687. [DOI] [PubMed] [Google Scholar]
- 12.Yang, J. T., Rayburn, H. & Hynes, R. O. (1995) Development (Cambridge, U.K.) 121, 549–560. [DOI] [PubMed] [Google Scholar]
- 13.Moore, A. W., Schedl, A., McInnes, L., Doyle, M., Hecksher-Sorensen, J. & Hastie, N. D. (1998) Mech. Dev. 79, 169–184. [DOI] [PubMed] [Google Scholar]
- 14.Tevosian, S. G., Deconinck, A. E., Tanaka, M., Schinke, M., Litovsky, S. H., Izumo, S., Fujiwara, Y. & Orkin, S. H. (2000) Cell 101, 729–739. [DOI] [PubMed] [Google Scholar]
- 15.Chen, T. H., Chang, T.-C., Kang, J.-O., Choudhary, B., Makta, T., Tran, C. M., Burch, J. B., Eid, H. & Sucov, H. M. (2002) Dev. Biol. 250, 198–207. [DOI] [PubMed] [Google Scholar]
- 16.Barak, Y., Nelson, M. C., Ong, E. S., Jones, Y. Z., Ruiz-Lozano, P., Chien, K. R., Koder, A. & Evans, R. M. (1999) Mol Cell 4, 585–595. [DOI] [PubMed] [Google Scholar]
- 17.MacNeill, C., French, R., Evans, T., Wessels, A. & Burch, J. B. (2000) Dev. Biol. 217, 62–76. [DOI] [PubMed] [Google Scholar]
- 18.Danielian, P. S., Muccino, D., Rowitch, D. H., Michael, S. K. & McMahon, A. P. (1998) Curr. Biol. 8, 1323–1326. [DOI] [PubMed] [Google Scholar]
- 19.Kisanuki, Y. Y., Hammer, R. E., Miyazaki, J., Williams, S. C., Richardson, J. A. & Yanagisawa, M. (2001) Dev. Biol. 230, 230–242. [DOI] [PubMed] [Google Scholar]
- 20.Ruiz-Lozano, P., Smith, S. M., Perkins, G., Kubalak, S. W., Boss, G. R., Sucov, H. M., Evans, R. M. & Chien, K. R. (1998) Development (Cambridge, U.K.) 125, 533–544. [DOI] [PubMed] [Google Scholar]
- 21.Wilkinson, D. G. (1992) in In Situ Hybridization, ed. Wilkinson, D. G. (Oxford Univ. Press, Oxford), pp. 75–83.
- 22.Dettman, R. W., Denetclaw, W., Jr., Ordahl, C. P. & Bristow, J. (1998) Dev. Biol. 193, 169–181. [DOI] [PubMed] [Google Scholar]
- 23.Hamburger, V. & Hamilton, H. L. (1951) J. Morphol. 88, 49–92. [PubMed] [Google Scholar]
- 24.Funayama, N., Fagotto, F., McCrea, P. & Gumbiner, B. M. (1995) J. Cell Biol. 128, 959–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyake, S., Makimura, M., Kanegae, Y., Harada, S., Sato, Y., Takamori, K., Tokuda, C. & Saito, I. (1996) Proc. Natl. Acad. Sci. USA 93, 1320–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu, H., Marth, J. D., Orban, P. C., Mossmann, H. & Rajewsky, K. (1994) Science 265, 103–106. [DOI] [PubMed] [Google Scholar]
- 27.Soriano, P. (1999) Nat. Genet. 21, 70–71. [DOI] [PubMed] [Google Scholar]
- 28.Jenkins, S. J., Hutson, D. R. & Kubalak, S. W. (2005) Dev. Dyn. 233, 1091–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kubalak, S. W., Miller-Hance, W. C., O'Brien, T. X., Dyson, E. & Chien, K. R. (1994) J. Biol. Chem. 269, 16961–16970. [PubMed] [Google Scholar]
- 30.Kastner, P., Mark, M., Ghyselinck, N., Krezel, W., Dupe, V., Grondona, J. M. & Chambon, P. (1997) Development (Cambridge, U.K.) 124, 313–326. [DOI] [PubMed] [Google Scholar]
- 31.Dettman, R. W., Pae, S. H., Morabito, C. & Bristow, J. (2003) Dev. Biol. 257, 315–328. [DOI] [PubMed] [Google Scholar]
- 32.Wu, H., Lee, S. H., Gao, J., Liu, X. & Iruela-Arispe, M. L. (1999) Development (Cambridge, U.K.) 126, 3597–3605. [DOI] [PubMed] [Google Scholar]
- 33.Morabito, C. J., Kattan, J. & Bristow, J. (2002) Curr. Opin. Cardiol. 17, 235–241. [DOI] [PubMed] [Google Scholar]
- 34.Haenig, B. & Kispert, A. (2004) Dev. Genes Evol. 214, 407–411. [DOI] [PubMed] [Google Scholar]
- 35.Liebner, S., Cattelino, A., Gallini, R., Rudini, N., Iurlaro, M., Piccolo, S. & Dejana, E. (2004) J. Cell Biol. 166, 359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pennisi, D. J., Ballard, V. L. & Mikawa, T. (2003) Dev. Dyn. 228, 161–172. [DOI] [PubMed] [Google Scholar]
- 37.Morabito, C. J., Dettman, R. W., Kattan, J., Collier, J. M. & Bristow, J. (2001) Dev. Biol. 234, 204–215. [DOI] [PubMed] [Google Scholar]
- 38.Kwee, L., Baldwin, H. S., Shen, H. M., Stewart, C. L., Buck, C., Buck, C. A. & Labow, M. A. (1995) Development (Cambridge, U.K.) 121, 489–503. [DOI] [PubMed] [Google Scholar]
- 39.Sengbusch, J. K., He, W., Pinco, K. A. & Yang, J. T. (2002) J. Cell Biol. 157, 873–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moore, A. W., McInnes, L., Kreidberg, J., Hastie, N. D. & Schedl, A. (1999) Development (Cambridge, U.K.) 126, 1845–1857. [DOI] [PubMed] [Google Scholar]
- 41.Kastner, P., Grondona, J. M., Mark, M., Gansmuller, A., LeMeur, M., Decimo, D., Vonesch, J. L., Dolle, P. & Chambon, P. (1994) Cell 78, 987–1003. [DOI] [PubMed] [Google Scholar]
- 42.Nakamura, T., Sano, M., Songyang, Z. & Schneider, M. D. (2003) Proc. Natl. Acad. Sci. USA 100, 5834–5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lavine, K. J., Yu, K., White, A. C., Zhang, X., Smith, C., Partanen, J. & Ornitz, D. M. (2005) Dev. Cell 8, 85–95. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.