Abstract
Many bacterial activities, including expression of virulence factors, horizontal genetic transfer, and production of antibiotics, are controlled by intercellular signaling using small molecules. To date, understanding of the molecular mechanisms of peptide-mediated cell-cell signaling has been limited by a dearth of published information about the molecular structures of the signaling components. Here, we present the molecular structure of PrgX, a DNA- and peptide-binding protein that regulates expression of the conjugative transfer genes of the Enterococcus faecalis plasmid pCF10 in response to an intercellular peptide pheromone signal. Comparison of the structures of PrgX and the PrgX/pheromone complex suggests that pheromone binding destabilizes PrgX tetramers, opening a 70-bp pCF10 DNA loop required for conjugation repression.
Keywords: DNA binding, Gram-positive bacteria, inducible conjugation, x-ray crystallography, transcription factor
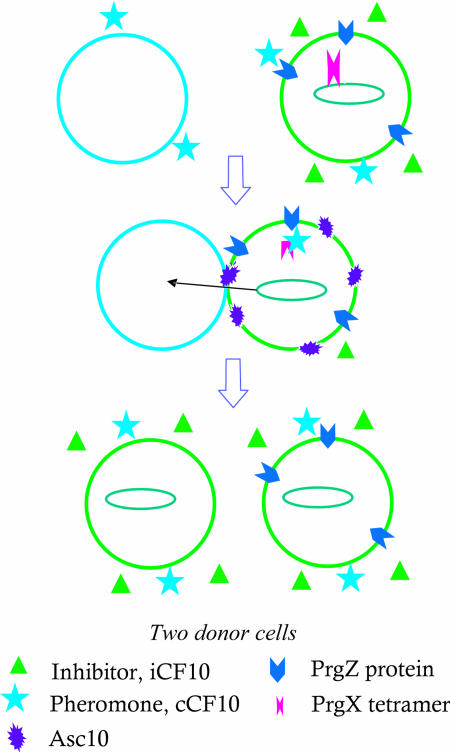
Multicellular behavior controlled by small intercellular signal molecules is an essential component of the biology of bacteria (1, 2). Although acyl-homoserine lactones generally serve as the intercellular signals in Gram-negative bacteria, oligopeptides most frequently carry out this function in Gram-positive bacteria (3). Despite the large number of peptide-mediated signaling systems identified and analyzed, there have been no published reports directly demonstrating the mechanism by which the interaction of a peptide signal molecule with its receptor triggers the biological response at the molecular level. An early bacterial cell-cell communication system in which a specific peptide signal was identified was the sex pheromone system of Enterococcus faecalis (4). The E. faecalis genome encodes the production of a variety of hydrophobic octa- and heptapeptides, each of which is capable of stimulating expression of conjugative transfer functions encoded by a cognate plasmid (3). The pheromone-responsive plasmids play an important role in dissemination of antibiotic resistance, and recently this signaling system has also been shown to play an essential role in expression of virulence in these opportunistic pathogens (5). As illustrated in Fig. 1, the signaling pathway entails production of peptide pheromone by plasmid-free cells (potential conjugative recipients) and import of the pheromone into the plasmid-containing conjugative donor cell, where the pheromone's interaction with a cytoplasmic receptor triggers a mating response that ultimately leads to transfer of the plasmid.
Fig. 1.
Model for pheromone-inducible conjugative transfer of pCF10. Recipient cell (left) produces enough pheromone (cCF10) to overcome the donor cell inhibitor (iCF10) production, leading to internalization of cCF10, expression of Asc10, aggregation of cells, and transfer of plasmid.
E. faecalis cells carrying pCF10 up-regulate expression of conjugation genes in response to the presence of cCF10 (amino acid sequence Leu-Val-Thr-Leu-Val-Phe-Val) produced by plasmid-free recipient cells (6). Several recent studies (7-10) have indicated that a critical molecular event in the initiation of the pheromone response in donor cells is the binding of internalized cCF10 to PrgX, a 33-kDa pCF10-encoded protein that negatively regulates transcription of the pCF10 prgQ conjugation operon in a pheromone-sensitive fashion (10, 11). Genetic and sequence analysis of PrgX suggests that it may have three functional domains: an N-terminal DNA-binding domain, a central dimerization domain, and a potential C-terminal pheromone-binding domain (9, 12). There is no extended region of high sequence similarity between PrgX and any other known protein, including the TraA proteins (13, 14) believed to be the functional equivalents of PrgX in other enterococcal pheromone plasmids. PrgX binds to two sites in pCF10 in a cooperative fashion (9). Genetic and biochemical studies (8, 12) suggest that PrgX oligomerization is essential for its function and that pheromone may affect the PrgX oligomerization state. These results have been incorporated into a DNA-looping model in which both binding sites are occupied in the repressed state, with the DNA-binding interactions at each site being strengthened by protein-protein interactions between the two sites (12). Pheromone binding to PrgX was predicted to disrupt the interactions between the proteins bound to the two sites, ultimately reducing PrgX occupancy of the prgQ operator. Although the direct and specific binding of purified PrgX to pCF10 DNA has been demonstrated, the behavior of the purified protein in solution has made it difficult to demonstrate a direct interaction between PrgX and cCF10. Previous genetic and biochemical data did not directly reveal the stoichiometry of binding to the two DNA-binding sites, nor did the data provide a strong indication of how pheromone binding to PrgX would cause conformational changes that reduce the oligomerization state and lead to induction of the pheromone response. We have addressed several key features of this model by determining the structures of PrgX and PrgX/cCF10.
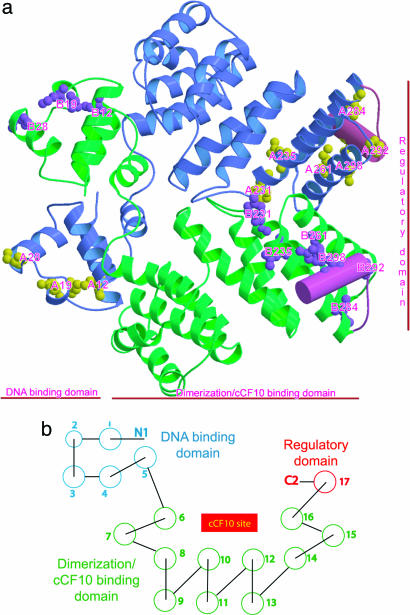
We have determined five crystal structures of PrgX: wild type (WT), two crystal forms of a Y153C mutant expressing a dominant negative phenotype (8), and two complexes of cCF10 with WT PrgX or with a C-terminal deletion mutant [ΔCT = (P291L, P292K, Δ294-317)], also expressing a dominant negative phenotype. The x-ray crystal structures reveal that PrgX is nearly all helical, containing a total of 17 α-helices (Fig. 2) connected by loops of various lengths. The three functional domains of PrgX suggested by genetic studies can be discerned in the crystal structure: an N-terminal DNA-binding domain, a large central dimerization (and pheromone-binding) domain, and a C-terminal regulatory domain. The oligomeric state of PrgX in crystals is a tetramer formed from two dimers, which can form the foundation of a DNA loop held together by the binding of one of the dimers to each DNA-binding site. The structures of the PrgX/cCF10 complexes provide a clear indication of the mechanism by which pheromone binding leads to a decrease in the PrgX oligomerization state in vivo. The structures also provide explanations for the phenotypic effects of several previously characterized point mutations affecting oligomerization, DNA binding, and other functions ascribed to this protein.
Fig. 2.
Topology of PrgX dimer. (a) Ribbon diagram of a PrgX homodimer showing the domain swapping of the N termini. Sites of PrgX mutations are shown with yellow and magenta balls. 12, 19, and 28 (in the N-terminal domains) are dominant negative mutants (B.K.K. and G.M.D., unpublished data) that lose the ability to bind DNA, most likely because they directly contact DNA. Mutations of residues 231, 235, and 261 (in the dimerization domains) affect PrgX dimerization (8, 10). Mutations of residues 284, 292, and 298 (in the C-terminal domain) cannot respond to cCF10 (8, 10). (b) Topology diagram for PrgX. N-terminal DNA-binding domain is shown in cyan. The dimerization/cCF10-binding domain is shown in green. The C-terminal regulatory domain is shown in red. The cCF10-binding domain is formed by two layers of α-helices arranged as a left-handed superhelix.
Methods
Protein Purification. His-PrgX protein was expressed and purified according to the method described in Bae et al. (9). Escherichia coli BL21(DEB) carrying pET28b-PrgX or pET28b containing mutant (Y1533C or ΔCT) alleles was grown in LB overnight at 30°C with shaking. Protein expression was induced with 0.1 mM isopropyl β-d-thiogalactoside when cells reached OD600 of 0.5-0.7. Subsequently, cells were resuspended in nickel column binding buffer (20 mM Tris·HCl, pH 7.9/5 mM imidazole/0.5 M NaCl/1 ppm Triton X-100) containing 10 μg/ml lysozyme and disrupted by sonication. The soluble fraction of the cell lysate collected after centrifugation was purified by nickel affinity chromatography. His-PrgX was eluted with a 200 mM imidazole elution buffer (200 mM imidazole/20 mM Tris·HCl, pH 7.9/0.5 M NaCl/1 ppm Triton X-100). The eluted fractions were analyzed for protein content by using Ponceau S (Sigma) staining.
Crystallization, Structure Determination, and Refinement. Crystals were grown by the hanging-drop vapor-diffusion method by using 2-μl droplets of protein solution in 10 mM Hepes (pH 7.2) to which 2 μl of reservoir solution was added. The crystals appeared at polyethylene glycol 4000 concentrations of 10-23% in 50 mM citrate/phosphate, pH 5-7. The sequence of pheromone is Leu-Val-Thr-Leu-Val-Phe-Val, making it very hydrophobic. To grow the crystals, 0.5 mg of cCF10 was mixed with 1 ml of 7 mg/ml solution of PrgX in 0.005 ppm Triton X-100/20 mM Hepes (pH 7.2) and incubated overnight. From this solution, crystals of complex were grown by the hanging-drop vapor diffusion method using a reservoir of 10-28% polyethylene glycol 4000/50 mM Tris malate, pH 5.6-8.
Owing to the intrinsic difficulties in solving the WT and WT/cCF10 complex multiwavelength anomalous dispersion (MAD) data, three-wavelength anomalous dispersion data for the Y153C mutant were collected to 3.1 Å with a space group of P21. The data sets were processed with either hkl2000 (15) or d*trek (16) programs. solve (17) found 52 of 72 possible selenium sites from the Y153C P21 MAD data set, and from these, resolve (17) identified 11 of 12 sets of sites related by the local 222 noncrystallographic symmetry. Initial phases to 3.0-Å resolution were calculated and subsequently extended to 2.5 Å when the higher-resolution data were available. resolve was used to automatically trace 40-50% of the model. Those fragments with correct sequences were brought together, building a monomer of 75% completeness. The noncrystallographic symmetry operators were used to propagate the monomer into the remaining 11 locations. Subsequent refinement using cns (18), map averaging, and model building using o (19) gave an Rwork and Rfree of 21.81% and 27.12%, respectively. All other structures were solved by molecular replacement method epmr (20) and refined by cns.
Results
The crystal data and refinement statistics for the five structures we determined are summarized in Table 1. A ribbon drawing of PrgX is shown in Fig. 2a. Fig. 2b is a topology drawing showing how these elements are arranged and assembled into the three domains of PrgX as well as the formation of the PrgX-binding cleft. The mutation of Tyr-153 into Cys does not cause significant structural changes. The rms deviation between the P21 form of WT and Y153C is only 0.35 Å over the Cα atoms in one PrgX monomer. In the WT/cCF10 complex, the α-helix of the C-terminal domain (helix 17) changes to two β-strands to better interact with cCF10 molecule.
Table 1. Data collection and refinement statistics.
| WT | Y153C | Y153C | WT/cCF10 | ΔCT/cCF10 | |
|---|---|---|---|---|---|
| Space group | P21 | P21 | P212121 | P212121 | P6122 |
| a, Å | 92.65 | 92.53 | 82.08 | 71.08 | 82.55 |
| b, Å | 134.72 | 134.37 | 82.08 | 83.9 | 82.55 |
| c, Å | 195.93 | 195.2 | 263.49 | 286.8 | 380.16 |
| α, ° | 90 | 90 | 90 | 90 | 90 |
| β, ° | 100.3 | 100.28 | 90 | 90 | 90 |
| γ, ° | 90 | 90 | 90 | 90 | 120 |
| Struct. sol.* | Iso | MAD | MR | MR | MR |
| Resolution, Å | 3.0 | 2.5 | 3.0 | 3.0 | 3.0 |
| Rwork/Rfree, % | 19.88/26.56 | 21.81/27.12 | 25.51/31.61 | 22.88/28.80 | 22.48/29.27 |
| No. of molecules in asymmetric unit | 12 | 12 | 4 | 4 | 2 |
| rms deviation | |||||
| Bond length, Å | 0.007 | 0.007 | 0.008 | 0.008 | 0.008 |
| Bond angle, ° | 1.216 | 1.128 | 1.232 | 1.244 | 1.242 |
| Dihedral, ° | 18.924 | 18.325 | 20.267 | 19.649 | 18.437 |
| Improper, ° | 0.822 | 0.750 | 0.909 | 0.873 | 0.740 |
| PDB ID code | 2AXU | 2AWI | 2AXV | 2AXZ | 2AW6 |
Struct. sol., structured solution; MAD, multiwavelength anomalous dispersion; MR, molecular replacement; Iso, isomorphous with Y153C
The N-terminal domain (residues 1-68) consists of five α-helices containing a canonical helix-turn-helix DNA-binding motif. The helix-turn-helix DNA-binding motif consists of helices 2 (residues 18-24) and 3 (residues 28-37) connected by a loop. Helices 1 (residues 3-15) and 4 (residues 43-55) are antiparallel, forming a base for the helix-turn-helix motif. Helix 5 (residues 57-65) crosses over helices 1 and 4 (≈120°), stabilizing the interactions between helix 1 and helix 5.
The homodimer is formed by two side-by-side monomers with their N-terminal domains swapped. The density for the amino acids connecting the N-terminal and the dimerization domains is clear in the Y153C P212121 form and in the cCF10 complexes but not in the two P21 form crystals (WT and Y153C), although the geometry requires domain swapping. Furthermore, the structures indicate that PrgX monomers are in an “open monomeric state,” which is defined in Kundu and Jernigan (21). Domain swapping with an “open interface” (21) reveals that the smallest functional unit for PrgX is a dimer.
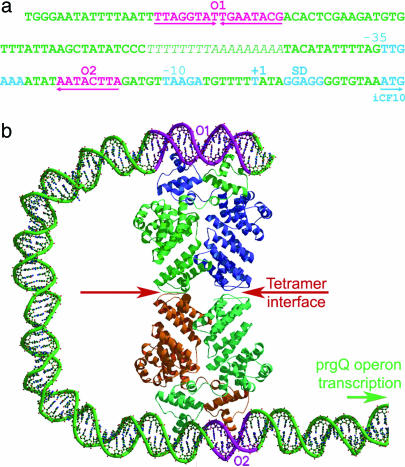
The formation of the dimeric PrgX is consistent with the palindromic sequence of operator 1 (O1) of the prgQ operon (Fig. 3a) and similar to other prokaryotic transcription regulators (22). Superposition of both N-terminal domains onto the phage 434 Cro protein dimer in the Cro/DNA complex (23) reveals the similar conformations between them and an rms deviation of 2.5 Å over 64 Cα atoms despite having only 17% sequence identity as discussed below. By using this structural homology, a PrgX/DNA complex was modeled that suggests that amino acids 12, 19, and 28 directly interact with the DNA molecule. This finding is consistent with the observation that R12S, Q19R, and S28F are all dominant negative substitution mutations (Fig. 2a; ref. 8). Cro and other homologous helix-turn-helix motifs have been shown to bend the DNA (24) upon binding, as might be expected through charge neutralization (25). This bending could add stability to the PrgX/DNA loop model.
Fig. 3.
DNA-binding of PrgX. (a) The DNA-binding sites for PrgX. The primary site is pseudopalindromic with higher binding affinity (O1); the secondary site is only half of the primary site with lower binding affinity (O2) (9). The O2 is within the PQ (promoter of prgQ operon), and therefore, the binding of PrgX to O2 will reduce the initiation of prgQ operon transcription. (b) The PrgX tetramer/DNA loop model for controlling expression of the prgQ operon. The DNA molecule is shown in green. The two operators are shown in magenta and labeled as O1 and O2. The green arrow represents the promoter region and the transcriptional direction for prgQ operon. To our knowledge, PrgX is the first bacterial protein structurally analyzed that regulates expression of two operons (prgX and prgQ) transcribed from opposing strands of the same DNA template sequence. The formation of tetramers enables PrgX to bind to the two operators cooperatively. The binding of pheromone to PrgX weakens the stability of the tetramer, effectively decreasing the affinity of PrgX toward the second operator and increasing transcription of the prgQ operon.
The central dimerization domain (residues 69-283) is endowed with both cCF10-binding and dimerization functions. The 11 helices of the central dimerization domain are nearly antiparallel [CATH database (http://cathwww.biochem.ucl.ac.uk) designation 1.50.10] and arranged in a two-layered, left-handed superhelix (26), forming a two-layered, crescent-shaped pocket 29 × 15 × 14 Å. The amino acids responsible for the dimerization are near the C terminus of the domain. In the PrgX structures without cCF10, helices 16 and 17 and the long connecting loop form a triangle. Helix 17 is docked on the edge of the pocket, mainly through three tyrosine (298, 299, and 302) and two valine (295 and 303) residues. In all three PrgX uncomplexed structures, pairs of PrgX homodimers interact with each other in a tail-to-tail fashion. Extensive hydrogen-bond and van der Waals interactions are found between the loop comprising amino acids 288-293 and the loop comprising amino acids 249-252 on the opposite dimer stabilizing PrgX tetramers (contact surface area = 4 × 475 Å2). The relative arrangement of the monomers in the tetramer is preserved in these three structures, with an rms deviation of only 0.8 Å over all Cα atoms for the tetramer superposition.
The cCF10-binding cleft is composed of six helices (helices 6, 8, 10, 12, 14, and 16). Although the cCF10 molecule is very hydrophobic, the surface of the cleft does not show high hydrophobicity. Of all the amino acids inside the cleft, only I82, F86, I200, I283, and L282 are hydrophobic amino acids. These amino acids are located in the two sides of the binding pocket, and their side chains form hydrophobic interactions with cCF10. Helices 14 and 15 are the two helices contributing most to dimerization. Several residues (L234, I263, I266, and I267) form a hydrophobic core surrounded by hydrogen-bond interactions (Y231OH···D233OD1, 2.58 Å; D233OD1···Y231OH, 2.68 Å; and N259ND2···N259′OD1, 3.07 Å) to stabilize the PrgX dimer. The long side chain of K184 forms a salt bridge with D185 of the opposite monomer.
The C-terminal regulatory domain (residues 287-305) consists of a single loop and a nine-residue α-helix (helix 17). Our data indicate that this domain likely functions as a delicate switch that regulates the expression of the prgQ operon. In the uncomplexed PrgX structures, extensive hydrogen-bond and van der Waals interactions are formed between the loop comprising amino acids 287-294 and the loop comprising amino acids 248-255 in opposite dimers, stabilizing the tetramers found in the crystals. The hydrophobic core is formed by F289, I251, L255, and P292 surrounded by hydrogen-bonding interactions (D248···T290OG1, 2.51 Å; K249O···T290N, 2.95 Å; I251N···T290O, 3.05 Å; and D252OD2···K293N, 2.59 Å). The spatial arrangement of PrgX monomers is preserved in the three uncomplexed structures (WT, P21 Y153C, and P212121 Y153C) with the long axis (the axis dividing the dimer) of the two dimers colinear. Residues 306-317 are disordered and not seen in any crystal forms described here.
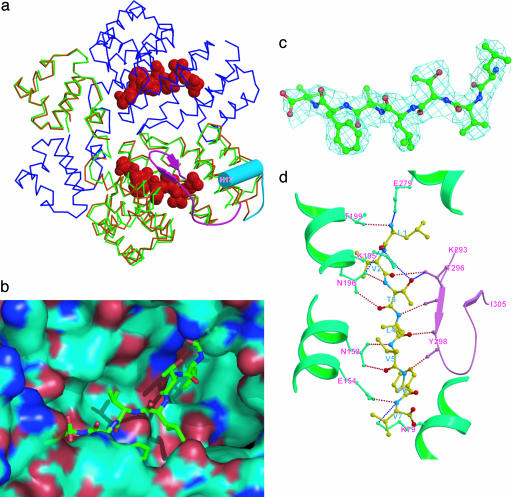
The binding of the pheromone results in a 120° rotation of the long loop of the C-terminal domain (Fig. 4a), positioning itself over the top of the pheromone. The C terminus is pulled away from the tetramer interface as the C-terminal α-helix is disrupted to form two short antiparallel β-strands (M297O···V303N, 2.92 Å; M297N···V303O, 2.78 Å; and V295O···I305N, 3.00 Å) to better interact with cCF10. The Fourier 2 Fc - Fo electron density map for cCF10 molecule is shown in Fig. 4c at the 1σ level. cCF10 molecules form a parallel β-strand interaction with residues in the first β-strand of the C-terminal domain (T3OG1···T296OG1, 3.06 Å; L4N···T296O, 3.26 Å; L4O···Y298N, 2.82 Å; and F6N···Y298O, 3.16 Å). Similar interactions are formed between cCF10 and side chains of the amino acids at the bottom of the pocket (L1O···K195OG1, 2.89 Å; L1O···T199OG1, 3.09 Å; T3O···N196OD1, 2.88 Å; V5O···N158ND2, 2.98 Å; V5N···N158OD1, 2.95 Å; and V7N···E154OE1, 3.35 Å) (Fig. 4d). The interactions made by the C-terminal loops between the two dimers in the tetramer are reduced drastically owing to the rotation of the loops (newly formed contact surface area = 4 × 200 Å2), which reduces the tetramer stability and distorts the tetramer.
Fig. 4.
Pheromone binding to PrgX. (a) Structural comparison of PrgX dimers formed by the WT uncomplexed protein (brown), WT PrgX/cCF10 complexes (green), and PrgX ΔCT/cCF10 complexes (blue). The pheromone molecules are red. The C-terminal helix of uncomplexed PrgX is a cyan cylinder. The conformation of the C terminus in the cCF10 complex is pink. Upon the pheromone binding, the C-terminal helices swing ≈120° and refold into two β-strands to cover the pheromone molecule in the binding pocket. (b) Connelly surface of the cCF10-binding pocket. A model of inhibitor peptide iCF10 is shown as colored sticks. (c) 2 Fc - Fo electron density map for the cCF10 molecule at the 1σ level in the WT/cCF10 structure. The map was calculated before the cCF10 was built. (d) Interactions between the iCF10 and PrgX. The cCF10 molecule interacts with the C-terminal amino acid through the main chain. The amino acids at the bottom of the pocket interact with cCF10 through their side chains (CH···O hydrogen bond not shown).
In the ΔCT/cCF10 complex structure, cCF10 still binds in the pocket but without interacting with C-terminal amino acids. The highly similar structures of the two PrgX/pheromone complexes (WT and ΔCT) indicate that the pocket in the central dimerization domain is the primary site for pheromone binding. In contrast, the quorum-sensing regulator TraR from Agrobacterium tumefaciens binds the acyl-homoserine lactone pheromone at the center of the dimeric interface (27).
There are four PrgX monomers in an asymmetric unit in the WT/cCF10 complex crystal structure. The four monomers form a skewed tetramer compared with the uncomplexed tetramer, with the angle between the two long axes of the dimer changed to 155°. Looking at the tetramer in Fig. 3b, the distortion is in the plane of the figure. Three of four monomers are found to be bound with cCF10 molecules, with the C-terminal domain pulled away from the tetramer interface. The fourth one does not have the cCF10 bound to it and keeps the uncomplexed conformation. Part of the loop (amino acids 285-289) of the fourth monomer is invisible, probably as a result of losing the proper contacts from the opposite dimer. Two PrgX monomers exist in the asymmetric unit in the ΔCT/cCF10 complex crystal structure, and each of the monomers is bound with a cCF10 molecule. Two homodimers also form a skewed tetramer with 165° angles between the axes of the two dimers. In the view presented in Fig. 3b, this distortion would cause the tetramer to buckle out of the plane of the drawing. Not only are the contact surfaces drastically reduced but also the hydrogen-bonding interactions are disrupted. The only possible hydrogen-bonding interaction (I251OG2M···Y254OH, 3.05 Å) is a CH···O interaction. These structural details strongly support our theory that PrgX function depends on the association/disassociation of tetramers.
Discussion
A number of repressors have been shown to exist as tetramers in crystals but to function as dimers in solutions, e.g., Cro repressor from bacteriophage λ (28). However, there are several well known examples in which dimeric repressors are understood to interact with each other. The cI repressor from bacteriophage λ binds cooperatively to two sets of three adjacent operators. Structural studies using a C-terminal fragment of the cI repressor showed that it forms a dimer by swapping a short 310 helix (29). By using this interaction, it was possible to model a tetramer with the N-binding domains positioned to allow binding to adjacent operators. Similar studies using a tryptic core fragment of the lac repressor showed that it formed tetramers through a C-terminal 16-residue helix which combine to form a four-helical bundle (30). These interactions revealed how lac repressor forms DNA loops (31). In the tet and lac repressor systems, induction changes the relative orientations of the DNA-binding domains, abolishing their ability to bind operator sequences (32, 33). However, in the PrgX system, it is the regulatory domains that undergo conformational changes when induced by the peptide pheromone cCF10. These changes are hypothesized to reduce binding to two separate operators through the deformation or dissociation of the PrgX tetramer. A similar mechanism has been proposed in the IclR transcriptional factor family member from Thermotoga maritima (34).
We propose that tetrameric PrgX represses the prgQ promoter, and that the C-terminal domain of PrgX functions as a switch controlling the association/dissociation of the tetramer in response to the presence of cCF10. There are two PrgX binding sites separated by 70 bp in the intergenic region between the prgX and prgQ genes (Fig. 3b). The primary site, O1, has a 16-bp pseudopalindromic sequence with higher PrgX binding affinity. The secondary site, O2, is within the prgQ promoter region, contains only half of the palindromic sequence, and has lower PrgX-binding affinity (Fig. 3a) (9). Each dimer in a tetramer is hypothesized to bind to one of two operators of the prgQ operon. The binding of PrgX to O1 and the formation of the tetramer should increase the binding affinity for O2 by proximity effects, as has been observed experimentally by Bae et al. (9). Similar cooperativity was observed in λ repressor (35). The dissociation of the tetramer in the presence of cCF10 reduces the affinity of PrgX toward the O2 site in the operon, increasing initiation of transcription by RNA polymerase, and therefore, expression of the prgQ operon. This finding is supported by the observation that in ΔCT, the prgQ operon is constitutively expressed (8), presumably because of a lack of stable tetramer formation (can be explained only by the lack of tetramer).
Because pheromone production is encoded by the chromosome of all E. faecalis cells (36), the plasmid encodes production of gene products that prevent its host (donor) cell from self-induction by endogenously produced cCF10 pheromone. One of these factors is the inhibitor peptide iCF10 (amino acid sequence Ala-Ile-Thr-Leu-Ile-Phe-Ile) (37). Pure cultures of donor cells excrete a mixture of these two peptides in such a way that they neutralize one another. The mechanism by which the inhibitor peptides inhibit the pheromones is not clear, and it has been suggested that they could interfere with pheromone import rather than acting intracellularly (14). From the WT/cCF10 and ΔCT/cCF10 complex structures and biochemical data (8), we propose that iCF10 (37) antagonizes cCF10 by binding to PrgX in the same pocket as cCF10. Modeling of iCF10 in place of cCF10 shows no steric clashes with the amino acids in the pocket (Fig. 4b). However, iCF10 would have close contact with Y298 if the C-terminal domain assumed the same conformation seen in the PrgX/cCF10 complex. Therefore, iCF10 should not induce the same conformational change of the C-terminal domain in PrgX that results from cCF10 binding, and thus, should not reduce tetramer stability but would compete with cCF10 for binding to PrgX. Furthermore, the similarities between the functional interactions of the TraA repressor proteins of the pAD1 and pPD1 plasmids with their cognate pheromone and inhibitor peptides (13, 14) and the interactions between PrgX and cCF10/iCF10 suggest that all enterococcal pheromone plasmids may use a similar regulatory mechanism.
Acknowledgments
We acknowledge the superb technical assistance of Mr. Edward Hoeffner (University of Minnesota). Work on this project was supported by National Institutes of Health Grants R01 AI57585 (to C.A.E.), R01 AI50607 (to D.H.O.), and R01 GM49530 (to G.M.D.). B.K.K. was supported by National Institutes of Health Grant T32 G08347 and by a Doctoral Dissertation Fellowship from the University of Minnesota. Diffraction data were collected by using the facilities of the BioCARS Beamlines 14-ID-B, 14-BM-D, and SBC 19-ID-D of the Advanced Photon Source at the Argonne National Laboratory; Beamline X25A of the National Synchrotron Light Source at Brookhaven National Laboratory; and Beamline 4.2.2 of the Advanced Light Source at Lawrence Berkeley Laboratory. Computational facilities were provided by the Basic Sciences Computer Laboratory of the Minnesota Supercomputing Institute.
Author contributions: G.M.D., D.H.O., and C.A.E. designed research., K.S., C.K.B., B.K.K., and C.A.E. performed research; Z.-Y.G., B.K.K., and C.A.E. contributed new reagents/analytic tools; K.S., B.K.K., G.M.D., D.H.O., and C.A.E. analyzed data; and K.S., C.K.B., B.K.K., G.M.D., D.H.O., and C.A.E. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: O1, operator 1; O2, operator 2.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 2AXU (PrgX), 2AXZ (PrgX/cCF10 complex), 2AWI (Y153C, P21), 2AXV (Y153C, P212121), and 2AW6 (ΔCT/cCF10 complex)].
References
- 1.Dunny, G. M. & Winans, S. C. (1999) Cell-Cell Signaling in Bacteria (Am. Soc. Microbiol., Washington, DC).
- 2.Miller, M. B. & Bassler, B. L. (2001) Annu. Rev. Microbiol. 55, 165-199. [DOI] [PubMed] [Google Scholar]
- 3.Dunny, G. M. & Leonard, B. A. (1997) Annu. Rev. Microbiol. 51, 527-564. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki, A., Mori, M., Sakagami, Y., Isogai, A., Fujino, M., Kitada, C., Craig, R. A. & Clewell, D. B. (1984) Science 226, 849-850. [DOI] [PubMed] [Google Scholar]
- 5.Chandler, J. R., Hirt, H. & Dunny, G. M. (2005) Proc. Natl. Acad. Sci. USA 102, 15617-15622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandler, J. R. & Dunny, G. M. (2004) Peptides (Tarrytown, NY) 25, 1377-1388. [DOI] [PubMed] [Google Scholar]
- 7.Bae, T., Clerc-Bardin, S. & Dunny, G. M. (2000) J. Mol. Biol. 297, 861-875. [DOI] [PubMed] [Google Scholar]
- 8.Bae, T. & Dunny, G. M. (2001) Mol. Microbiol. 39, 1307-1320. [DOI] [PubMed] [Google Scholar]
- 9.Bae, T., Kozlowicz, B. K. & Dunny, G. M. (2002) J. Mol. Biol. 315, 995-1007. [DOI] [PubMed] [Google Scholar]
- 10.Bae, T., Kozlowicz, B. K. & Dunny, G. M. (2004) Mol. Microbiol. 51, 271-281. [DOI] [PubMed] [Google Scholar]
- 11.Bensing, B. A., Meyer, B. J. & Dunny, G. M. (1996) Proc. Natl. Acad. Sci. USA 93, 7794-7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozlowicz, B. K., Bae, T. & Dunny, G. M. (2004) Mol. Microbiol. 54, 520-532. [DOI] [PubMed] [Google Scholar]
- 13.Nakayama, J., Takanami, Y., Horii, T., Sakuda, S. & Suzuki, A. (1998) J. Bacteriol. 180, 449-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujimoto, S. & Clewell, D. B. (1998) Proc. Natl. Acad. Sci. USA 95, 6430-6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307-326. [DOI] [PubMed] [Google Scholar]
- 16.Pflugrath, J. W. (1999) Acta Crystallogr. D 55, 1718-1725. [DOI] [PubMed] [Google Scholar]
- 17.Terwilliger, T. (2004) J. Synchrotron Radiat. 11, 49-52. [DOI] [PubMed] [Google Scholar]
- 18.Brünger, A. T., Kuriyan, J. & Karplus, M. (1987) Science 235, 458-460. [DOI] [PubMed] [Google Scholar]
- 19.Jones, T. A. (1978) J. Appl. Crystallogr. 11, 268-272. [Google Scholar]
- 20.Kissinger, C. R., Gehlhaar, D. K. & Fogel, D. B. (1999) Acta Crystallogr. D 55, 484-491. [DOI] [PubMed] [Google Scholar]
- 21.Kundu, S. & Jernigan, R. L. (2004) Biophys. J. 86, 3846-3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohlendorf, D. H., Anderson, W. F. & Matthews, B. W. (1983) J. Mol. Evol. 19, 109-114. [DOI] [PubMed] [Google Scholar]
- 23.Mondragon, A. & Harrison, S. C. (1991) J. Mol. Biol. 219, 321-334. [DOI] [PubMed] [Google Scholar]
- 24.Schultz, S. C., Shields, G. C. & Steitz, T. A. (1991) Science 253, 1001-1007. [DOI] [PubMed] [Google Scholar]
- 25.Matthew, J. B. & Ohlendorf, D. H. (1985) J. Biol. Chem. 260, 5860-5862. [PubMed] [Google Scholar]
- 26.Orengo, C. A., Michie, A. D., Jones, S., Jones, D. T., Swindells, M. B. & Thornton, J. M. (1997) Structure 5, 1093-1108. [DOI] [PubMed] [Google Scholar]
- 27.Zhang, R. G., Pappas, T., Brace, J. L., Miller, P. C., Oulmassov, T., Molyneaux, J. M., Anderson, J. C., Bashkin, J. K., Winans, S. C. & Joachimiak, A. (2002) Nature 417, 971-974. [DOI] [PubMed] [Google Scholar]
- 28.Anderson, W. F., Ohlendorf, D. H., Takeda, Y. & Matthews, B. W. (1981) Nature 290, 754-758. [DOI] [PubMed] [Google Scholar]
- 29.Bell, C. E., Frescura, P., Hochschild, A. & Lewis, M. (2000) Cell 101, 801-811. [DOI] [PubMed] [Google Scholar]
- 30.Friedman, A. M., Fischmann, T. O. & Steitz, T. A. (1995) Science 268, 1721-1727. [DOI] [PubMed] [Google Scholar]
- 31.Kramer, H., Niemoller, M., Amouyal, M., Revet, B., von Wilcken-Bergmann, B. & Muller-Hill, B. (1987) EMBO J. 6, 1481-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bell, C. E. & Lewis, M. (2001) J. Mol. Biol. 314, 1127-1136. [DOI] [PubMed] [Google Scholar]
- 33.Orth, P., Schnappinger, D., Hillen, W., Saenger, W. & Hinrichs, W. (2000) Nat. Struct. Biol. 7, 215-219. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, R. G., Kim, Y., Skarina, T., Beasley, S., Laskowski, R., Arrowsmith, C., Edwards, A., Joachimiak, A. & Savchenko, A. (2002) J. Biol. Chem. 277, 19183-19190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hochschild, A. & Ptashne, M. (1986) Cell 44, 681-687. [DOI] [PubMed] [Google Scholar]
- 36.Clewell, D. B., An, F. Y., Flannagan, S. E., Antiporta, M. & Dunny, G. M. (2000) Mol. Microbiol. 35, 246-247. [DOI] [PubMed] [Google Scholar]
- 37.Nakayama, J., Ruhfel, R. E., Dunny, G. M., Isogai, A. & Suzuki, A. (1994) J. Bacteriol. 176, 7405-7408. [DOI] [PMC free article] [PubMed] [Google Scholar]