Abstract
In multicellular organisms, the control of genome duplication and cell division must be tightly coordinated. Essential roles of the minichromosome maintenance (MCM) proteins for genome duplication have been well established. However, no genetic model has been available to address the function of MCM proteins in the context of vertebrate organogenesis. Here, we present positional cloning of a zebrafish mcm5 mutation and characterization of its retina phenotype. In the retina, mcm5 expression correlates closely with the pattern of cell proliferation. By the third day of development, mcm5 is down-regulated in differentiated cells but is maintained in regions containing retinal stem cells. We demonstrate that a gradual depletion of maternally derived MCM5 protein leads to a prolonged S phase, cell-cycle-exit failure, apoptosis, and reduction in cell number in mcm5m850 mutant embryos. Interestingly, by the third day of development, increased apoptosis is detectable only in the retina, tectum, and hindbrain but not in other late-proliferating tissues, suggesting that different tissues may employ distinct cellular programs in responding to the depletion of MCM5.
Keywords: stem cell, cell proliferation, ciliary marginal zone, embryogenesis, development
Proper duplication of the genome during the cell cycle is of paramount importance for the life of an organism. In eukaryotes, each replication origin is “licensed” to fire once per cell cycle through the cell-cycle-dependent formation and destruction of a prereplication complex (preRC) (1–3). A key component of this preRC is a family of six structurally related proteins, MCM2 through -7, which are evolutionarily conserved in all eukaryotes. The MCM proteins were originally identified as proteins required for minichromosome maintenance in Saccharomyces cerevisiae (4). MCM2 through -7 belong to a distinct subgroup of the large AAA+ ATPase family (5) and share a conserved central region of ≈200 amino acids (MCM box). Biochemical studies in Xenopus have established the role of MCM2 through -7 as replication-licensing factors (6–8). Subsequent studies illustrate that proper orchestration of the functional interactions among MCM2 through -7 proteins and other components of the preRC by cell-cycle-dependent protein kinases results in initiation of DNA synthesis once every cell cycle (9, 10). The MCM2 through -7 proteins appear to form heterohexamers and play important roles in initiation and elongation during DNA replication (4, 11, 12). Furthermore, recent evidence supports involvement of MCMs in many other chromosome transactions, including transcription, chromatin remodeling, and genome stability (13). However, in vivo analysis of MCM-protein functions in multicellular organism has been scarce. Here, we report the isolation, positional cloning, and in-depth characterization of a vertebrate mutant in an MCM-family protein. We demonstrate that the zebrafish m850 allele harbors a mutation in the mcm5 gene and exhibits developmental defects in late-proliferating tissues, including the retina and the brain. We focus on the analysis of retinal development because of its well described cell types and proliferation pattern and show that mcm5 is expressed in the region where retinal stem cells and progenitor cells are found. Furthermore, we find that the reduction in MCM5 level leads to S-phase prolongation, defects in cell-cycle progression, and a strong activation of apoptosis in the zebrafish retina. The activation of apoptosis is also observed in tectum and hindbrain but not in other proliferative tissues, indicating the existence of tissue-specific responses to the reduction of MCM5 level.
Materials and Methods
Fish Maintenance and Mutagenesis Screen. Zebrafish maintenance and breeding were carried out under standard conditions at 28.5°C (14). Embryos were staged and fixed at desired time points [hours or days postfertilization (hpf or dpf, respectively)] in 4% paraformaldehyde in PBS. To avoid formation of melanin pigments, embryos were incubated in 0.2 mM 1-phenyl-2 thiourea (Sigma). mcm5m850 was isolated during an ethyl-nitrosourea mutagenesis screen performed in our laboratory (15).
In Situ Hybridization, Immunohistochemistry, Cartilage Staining, and Apoptosis Assay. Whole-mount in situ hybridization was performed as described in ref. 16. Digoxigenin- or fluorescein-labeled antisense RNA probes were prepared by using RNA-labeling reagents (Roche Biochemicals). We prepared RNA probes for the following genes: th (17), ath5 (18), and rhodopsin (a gift of K. Dürr, University of Freiburg). To generate an antisense probe for mcm5, the N-terminal 725-bp fragment of the mcm5 gene was PCR-amplified. Immunohistochemistry was performed as described in ref. 15. Anti-Zn5 (19), anti-BrdUrd (Becton Dickinson), and anti-phospho histone H3 (Upstate Biotechnology, Lake Placid, NY) were used at 1:1,000 dilution. Whole-mount stained embryos were cut into 15-μm sections by using a vibratome. Unstained 3-dpf embryos were embedded in plastic (Technovit, Heraeus), cut into 6-μm sections, and stained for a few minutes with methylene blue solution. For S-phase analysis, 48-hpf embryos were incubated for 15 min at room temperature in 10 mM BrdUrd solution (containing 15% DMSO in egg water), fixed immediately in 4% paraformaldehyde in PBS, and embedded in paraffin. Transverse sections of 6-μm thickness were subjected to antigen heat retrieval. A biotin–avidin horseradish peroxidase system (Vectastain ABC-kit, Vector Laboratories) was used for detection. TUNEL assay was performed by using the Apoptag peroxidase in situ apoptosis-detection kit (Intergen). Fixed and permeabilized embryos were incubated in ethanol/acetic acid mixture for 15 min and incubated in equilibration buffer, followed by terminal transferase enzyme solution. digoxigenin-labeled DNA fragments were detected by using antidigoxigenin antibody conjugated to alkaline phosphatase. Anti-active caspase-3 immunohistochemistry and detection of MCM5 protein are described in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
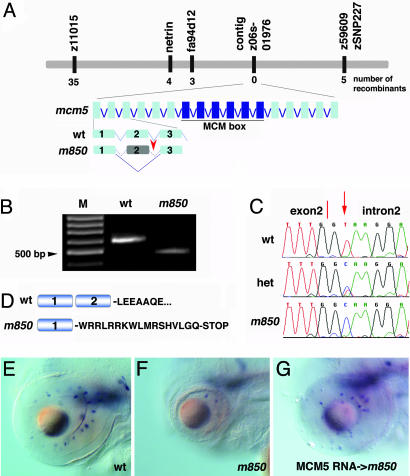
Mapping and Cloning of mcm5m850. We mapped mcm5m850 using bulked-segregant analysis (20) with pooled DNA from F2 homozygous mutants and F2 wild-type siblings from a mcm5m850 AB × India map cross. We linked mcm5m850 to linkage group 3 between simple-sequence-polymorphism markers z11015 and z59609. We generated additional polymorphic markers based on sequences from EST fa94d12 and the netrin gene. z59609 was used to identify bacterial artificial chromosome (BAC) zC100E2 from the Well-come Trust Sanger Center BAC fingerprinting project (www.sanger.ac.uk/Projects/D_rerio/WebFPC/zebrafish). An end sequence of BAC zC100E2 was used to identify contig z06s019176 from the Wellcome Trust Sanger zebrafish genome project assembly (www.ensembl.org/Danio_rerio). The mcm5 sequence was identified within this contig and represented the sequence identical to EST BC044460. mcm5 isolated from cDNA generated from seven dpf m850 mutants and their sibling embryos was sequenced, revealing the lack of exon 2 in transcripts derived from mcm5m850 mutant embryos. An ≈500-bp region surrounding exon 2 was amplified from genomic DNA isolated from individual mcm5m850 mutants and their sibling embryos. Sequencing of these PCR products revealed a T-to-C base change within the splice donor site at the end of the second exon in the mutant embryos.
RNA Injection. To generate mcm5 mRNA, full-length mcm5 was PCR-amplified from cDNA from wild-type embryos and cloned into pCS2+ vector (Clontech). Synthetic capped mRNA was generated by using the mMessage mMachine kit (Ambion), and 200 pg of mRNA was injected into single one-cell-stage embryos. The morpholino-injection procedure can be found in Supporting Materials and Methods.
DNA-Content Analysis. Retinae were dissected from anesthetized 48- to 50-hpf mutant and wild-type embryos. Single-cell suspension was achieved after room-temperature incubation for 1 hour in 20 units/ml papain in L15 tissue culture media (Sigma) and repeated trituration using fire-polished glass pipettes. Cells were resuspended in 4 mM citrate buffer, pH 6.5, containing 0.1 mg/ml propidium iodide (Sigma), 200 μg/ml RNase, and 0.1% Triton X-100 and were stored in the dark at 4°C until analysis. Data acquisition was performed by using a Becton Dickinson FACS-Calibur machine and analyzed by using the program cellquest.
Results
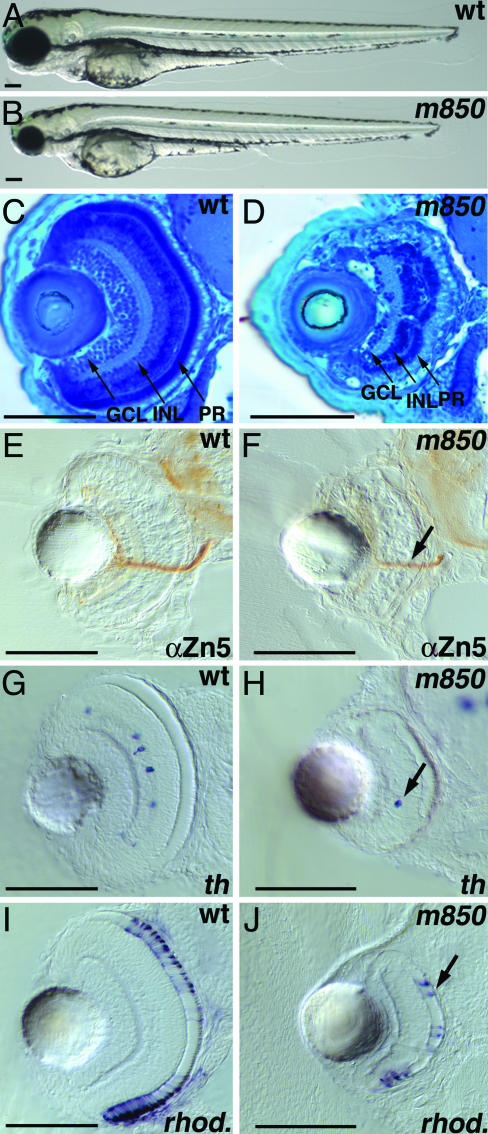
m850 Mutant Embryos Display Reduction in Cell Number in the Retina. We isolated the m850 mutant allele in a genetic screen for mutations affecting the formation of the catecholaminergic system, based on the severe reduction in the number of tyrosine hydroxylase (th)-expressing cells in the CNS of mutant embryos. Furthermore, m850 mutant embryos develop morphological defects during dpf 3, including a smaller retina and head compared with wild-type embryos (Fig. 1 A and B). The jaw and branchial arches remain small and do not develop properly in mutant embryos (see Fig. 6, which is published as supporting information on the PNAS web site). In contrast, the overall body size is similar in mutant and wild-type embryos, and early-forming tissues, including somites and notochord, are largely normal during the first 4 days of development. To better understand the defects in m850 mutants, we focused our phenotypic analysis on the retina, because it contains several well characterized cell types, and its cell-proliferation pattern has been well described (21). The zebrafish retina, as is typical for vertebrates, consists of three major cell-body layers. The innermost ganglion cell layer consists of ganglion cells whose processes form the optic nerve. The amacrine, bipolar, and horizontal cells are found in the intermediate inner nuclear layer, and photoreceptor cells are found in the outermost photoreceptor layer (Fig. 1C). In m850 mutant retinae, at dpf 3, the relative positioning of the individual layers appears to be preserved in the central retina; however, the number of cells in each layer is severely reduced (Fig. 1D).
Fig. 1.
m850 mutant embryos display reduction in cell number in the retina. Live wild-type (A) and m850 mutant (B) embryos at 3 dpf. A smaller head and smaller eyes characterize the m850 mutant. (C and D) Transverse sections through the eye at 3 dpf, stained with methylene blue. (C) The wild-type retina shows characteristic stratification in three nuclear layers (GCL, INL, and PR) and two plexiform layers. (D) In m850 mutant embryos, the stratification of retinal layers occurs, although the number of cells across all three retinal nuclear layers is severely reduced. (E and F) Transverse sections of 3-dpf embryos stained with anti-Zn5 (α-Zn5) antibody exhibit the presence of optic nerves in wild-type (E) and m850 mutant (F) embryos, albeit here with reduced intensity. (G and H) Transverse sections through the retinae of wild-type embryos (G) display dopaminergic amacrine cells, as detected by the expression of tyrosine hydroxylase (th) at 3 dpf. Dopaminergic amacrine cells can also be detected in mutant embryos (H). (I and J) Transverse sections through the retinae of 3-dpf embryos showing rod photoreceptors, as detected by the expression of rhodopsin (rhod.). The presence of differentiated rod photoreceptors is clearly visible in mutants (I). (A and B) Lateral views, anterior left, dorsal up; (C–J) transversal sections, dorsal up. (Scale bars, 100 μm.) Black arrows indicate the optic nerve (F), a dopaminergic amacrine cell (H), or a photoreceptor cell (J). GCL, ganglion cell layer; INL, inner nuclear layer; PR, photoreceptor layer.
To elucidate whether differentiation and cell-type specification are affected in m850 mutant retinae, we used markers for differentiated cell types present in each of the three retinal layers. In zebrafish, the Zn5 monoclonal antibody recognizes a surface adhesion molecule of the Ig superfamily neurolin/DM-GRASP (22) and marks retinal ganglion cells and their projections. Albeit reduced in its intensity, the projection of retinal ganglion cells is clearly visible in the mutant retinae (Fig. 1 E and F). To test whether the differentiation of cells from other retinal layers is affected, we used th expression to mark the dopaminergic subset of amacrine cells within the inner nuclear layer (17) and rhodopsin expression to label photoreceptors. We detected both of these neuronal types in mutant retinae, although their cell number was severely reduced (Fig. 1 G–J). These results suggest that patterning and cell-type specification occur in m850 mutant embryos similar to wild-type embryos and that the primary defect in the m850 mutant retina may involve cell proliferation or cell survival.
The m850 Mutation Disrupts the Zebrafish mcm5 Gene. To elucidate the molecular basis of the m850 mutation, we identified the affected gene by using a positional cloning strategy. Using the simple-sequence-polymorphism-marker-based map (23) and EST-derived markers, we defined a critical interval of 0.45 cM flanked by markers z59609 and EST fa94d12 (Fig. 2A). Using z59609 sequences, we isolated BAC zC110E2, whose end sequence overlapped with the contig z06s019176 from the Wellcome Trust Sanger zebrafish genomic sequence database (www.emsembl.org/Danio_rerio). A polymorphic marker generated from this contig sequence had no recombination event in 1,786 meioses, suggesting close proximity to the mutation. The contig z06s019176 contained two ORFs: an ORF with 80% identity to human MCM5 at the protein level and the heme oxygenase 1(hmox1) gene. In vertebrates, Hmox1 catabolizes cellular heme to biliverdin, carbon monoxide, and free iron (24) and, in zebrafish, is expressed in the extraembryonic yolk syncytial layer, lens, and a small population of blood cells (Thisse et al., 2004 at www.zfin.org). Because hmox1 is not expressed in the tissues where we observe strongest mutant phenotypes, including retina and the brain, we decided to focus on the mcm5 gene for our mutation search. Sequencing of the genomic mcm5 locus from the mutant embryos revealed a T-to-C base change within the highly conserved splice donor site at the end of the second exon (Fig. 2C). RT-PCR using mRNA from mutants and wild-type embryos revealed the presence of a shorter mcm5 transcript in mutant embryos that lacks the entire second exon (Fig. 2B). The mutant transcript contains a frame shift and a premature stop codon at amino acid 74, resulting in a protein that lacks ≈90% of the wild-type sequence, including the highly conserved MCM box, and, thus, is likely to be a null allele (Fig. 2D). To provide further evidence that mcm5 is the gene disrupted in the m850 allele, we rescued the retinal phenotypes by mcm5 mRNA injection into mutant embryos and phenocopied m850 by morpholino-knockdown experiments (Figs. 2 E–G; and see Figs. 7 and 8, which are published as supporting information on the PNAS web site). Taken together, our results demonstrate that m850 is an amorphic mutant allele at the zebrafish mcm5 locus. Thus, we will refer to the m850 mutation as mcm5m850 from here onward.
Fig. 2.
The zebrafish mcm5 gene is disrupted in m850. (A) Schematic representation of the genetic map of linkage group 3 (LG 3) and genomic organization of the mcm5 locus showing the position of the mutation. Some of the closest simple-sequence-polymorphism and SNP markers are listed, and their genetic distances relative to the mutation are indicated as the number of recombinants in a total of 1,786 meioses. (B) RT-PCR from wild-type and mutant RNA using primers from exons 1 and 3 reveals that the mutant transcript is shorter. M, 100-bp DNA ladder. (C) Chromatogram of genomic sequences from the 3′ end of exon 2 from wild-type, heterozygous, and mutant embryos. The T-to-C base change in the mutant embryo affects the first of the two core bases, highly conserved in eukaryotic splice donor sites. (D) Skipping exon 2 produces a frame shift, which introduces a stop codon at amino acid residue 74 in mutant embryos. (E–G) RNA rescue of the m850 mutant phenotype. The mutant phenotype was judged by the eye and head size and the presence of dopaminergic amacrine cells, as detected by th whole-mount in situ hybridization at 3 dpf. All pictures show lateral views, dorsal up. (E) Wild-type retina. (F) m850 mutant retina. (G) Injection of 200 pg of mcm5 RNA into m850 mutant embryos rescued the mutant phenotype (n = 19 of 26).
Although mcm5m850 is likely a null allele, the presence of maternally provided mcm5 mRNA and MCM5 protein may partially substitute for zygotic loss of mcm5 during early embryonic development. By generating antibody against MCM5 protein, we demonstrated the existence of maternally derived MCM5 protein, which persists beyond 3 dpf in zebrafish embryos (see Fig. 9, which is published as supporting information on the PNAS web site).
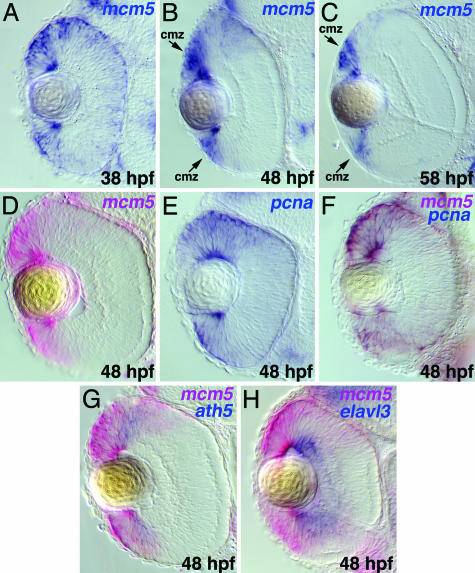
mcm5 Expression Correlates with the Pattern of Cell Proliferation. To learn more about the function of mcm5 in the zebrafish retina, we analyzed its expression pattern during development by using whole-mount in situ hybridization. Until ≈28 hpf, zebrafish retinae consist of an apparently uniform population of rapidly proliferating neuroepithelial cells (25, 26). The onset of terminal mitoses begins ≈28 hpf and continues in three major bursts separated by two 10-hour intervals (27). Most retinal cell proliferation has slowed down by the end of this period (58 hpf); however, it continues in the ciliary marginal zone (CMZ), where retinal stem cells are maintained, even in adult fish (28, 29). The pattern of mcm5 expression follows closely this pattern of proliferation in the zebrafish retina. Early on, mcm5 is ubiquitously expressed in the entire retina (data not shown). However, by 38 hpf, mcm5 expression has begun to decrease where differentiation occurs in the retina but is maintained in a region that includes the CMZ (Fig. 3A). By 58 hpf, mcm5 expression remains detectable only in the CMZ (Fig. 3C).
Fig. 3.
In the retina, mcm5 expression is down-regulated in regions containing differentiated cells but is maintained in the region containing retinal stem cells. (A–C) Transverse sections of the retina of wild-type embryos after whole-mount in situ hybridization for mcm5. (A) At 38 hpf, whereas mcm5 expression in the retina is broad, it is stronger in the marginal zone. At 48 (B) and 58 (C) hpf, strong mcm5 expression is maintained in the ciliary marginal zone (cmz) but is down-regulated in other areas of the retina. (D–F) Expression of pcna (blue) (E), marking noncommitted proliferating cells, overlaps exactly with that of mcm5 (red) (D), resulting in brown staining (F). (G) ath5 (blue), marking neurons before full differentiation, overlaps with the lateral extent of mcm5 (red). (H) elavl3 (blue), marking fully differentiated neurons, shows no overlap with mcm5 (red). Anterior is up.
To elucidate the cell types in which mcm5 is expressed, we compared the expression pattern of mcm5 with that of proliferating cell nuclear antigen (pcna), ath5, and elavl3 at 48 hpf (Fig. 3 D–H). Whereas pcna is expressed in noncommitted proliferating cells, the expression of the bHLH transcription factor ath5 serves as one of the first markers of neurogenesis in the zebrafish retina. ath5 is first expressed in a small group of cells in the ventronasal retina at 25 hpf, and its expression spreads in a manner preceding differentiation (18). By 48 hpf, ath5 is expressed in retinoblast cells and, possibly, postmitotic neurons before full differentiation but not in differentiated cells in the central retina. In contrast, the zebrafish elavl3 gene encodes an RNA-binding protein homologous to Drosophila elav that serves as an early marker for differentiated neurons (30). In vertebrate retinae, elavl3 is expressed in ganglion cells and the majority of amacrine cells in the inner nuclear layer (31). Our analysis shows that mcm5 and pcna are expressed in identical populations of proliferating cells (Fig. 3F). mcm5 and elavl3 are expressed in a complimentary manner with virtually no overlap, whereas mcm5- and ath5-expression domains partially overlap (Fig. 3 G and H), revealing that mcm5 expression is maintained in proliferative retinal cells in the CMZ region but is down-regulated in differentiated cells in the central retina.
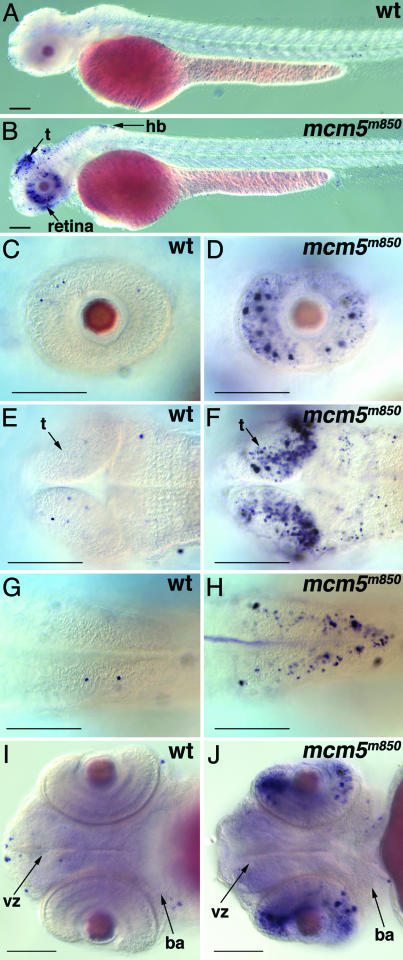
Activation of Apoptosis in CNS Tissues in mcm5m850 Mutant Embryos. mcm5m850 mutant embryos die as larvae between 5 and 10 dpf with small head and eyes. To test whether this is because of the lack of cell proliferation or whether activation of programmed cell death is involved, we performed TUNEL assays in mcm5m850 mutants at 48 hpf, after the maternally derived MCM5 levels have significantly decreased. We observed a strong activation of apoptosis in the retina, tectum, and hindbrain (Fig. 4). This activation was most dramatic at 2 dpf and continued beyond 3 dpf (Fig. 4 and data not shown). The same pattern of apoptosis was detected by immunohistochemistry using anti-active caspase-3 (see Fig. 10, which is published as supporting information on the PNAS web site). Caspase-3 is a key protease that is activated during the early stages of apoptosis and is, therefore, a sensitive and specific marker for cells undergoing apoptosis (32). Together, these results suggest that activation of programmed cell death might be the prominent cause of reduction in head and eye size in mcm5m850 mutant embryos. Interestingly, retina and tectum are only some of the proliferative tissues in the zebrafish embryo at 2–3 dpf. Strong mcm5 expression is detected in highly proliferative tissues such as forebrain ventricular zones, the branchial arches, and endoderm (Thisse et al., 2004 at www.zfin.org; S.R. and W.D., unpublished data), which do not show activation of apoptosis at 2 dpf (Fig. 4 A and J) or at 3 dpf (data not shown). This result suggests that different proliferative tissues may employ distinct cellular programs in responding to the reduction in MCM5 level.
Fig. 4.
Apoptosis is activated in CNS tissues in m850 mutant embryos. (A–J) TUNEL assay for apoptosis in whole-mount wild-type and mcm5m850 mutant embryos at 2 dpf. (A and B) Apoptosis is activated in retina, tectum, and hindbrain in the mutant embryos. (C–H) mcm5m850 mutant embryos show more apoptotic cells in the retina (C and D), the tectum (E and F), and the hindbrain (G and H) but not in the branchial arches and the brain ventricular zone (I and J), compared with wild-type embryos. (A–D) Lateral view; (E–H) dorsal views; (I and J) ventral views. Anterior toward the left. (Scale bars, 100 μm.) ba, branchial arches; hb, hindbrain; t, tectum; vz, ventricular zone.
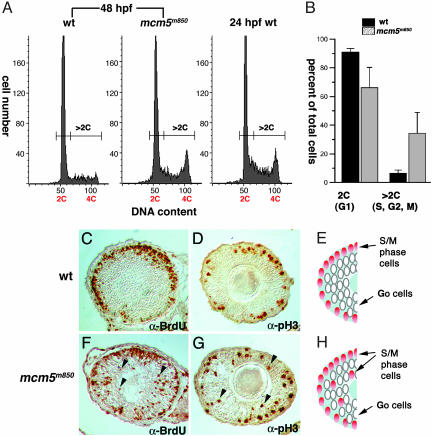
Cell-Cycle-Progression Defects in the Retinas of mcm5m850 Mutant Embryos. We next investigated how retinal cells respond to reduced levels of MCM5. To detect potential defects in replication and cell-cycle progression, we performed FACS analysis of propidium-iodide-stained retinal cells to measure DNA content per cell. For this experiment, we used embryos between 48 and 50 hpf, when the maternally derived MCM5 protein level in mcm5m850 mutants is already significantly reduced, and the mutant embryos can be distinguished morphologically. A typical DNA-content distribution is shown for mcm5m850 mutant and wild-type cells (Fig. 5A). At 48 hpf, the majority of retinal cells of wild-type embryos are in G1 phase. In contrast, retinae of mcm5m850 mutant embryos contain more cells with >2C DNA content (1C = one haploid genome equivalent). To assess the location of cells with 4C DNA content, we analyzed the retinal cells from wild-type embryos at 24 hpf (Fig. 5A). We reasoned that a G2/M peak would be readily detectable in these cells, because the zebrafish retina proliferates rapidly at this stage. The DNA content of the second peak in mutant embryos is very similar to those of the wild-type 24-hpf embryos, suggesting that many mutant cells may have finished replicating the bulk of their DNA. Summary analysis of four independent experiments revealed that the proportion of cells with >2C DNA content is <10% in wild-type retinae and >30% in mcm5m850 mutant retinae (Fig. 5B), demonstrating that cells in S/G2/M phase accumulate in mcm5m850 mutant embryos and suggesting that cell-cycle progression in mutant embryos is delayed or arrested, and many cells do not exit the cell cycle.
Fig. 5.
S-phase prolongation and cell-cycle-progression defects can be detected in the retinae of mcm5m850 mutant embryos. (A) DNA content of dissociated retinal cells at 48–50 hpf from wild-type (wt) (Left) and mcm5m850 mutant (Center) embryos and at 24 hpf from wild-type embryos, as measured by quantitative FACS analysis of propidium-iodide-labeled cells. Rapidly proliferating retina cells at 24 hpf allow determination of the location of the 4C peak (Right). In contrast, at 48–50 hpf, retinae from wild-type embryos contain mostly cells with 2C DNA content (1C = one haploid genome equivalent). The mutant retinae contained an increased portion of cells with DNA content >2C. (B) Bar graph summarizing results from four independent experiments. Retinae from mcm5m850mutant embryos contained fewer cells in G1 phase and more cells in S, G2, or M phase. The error bars represent the standard deviation. (C, D, F, and G) Lateral sections through the eye of paraffin-embedded wild-type or mutant embryos, probed with antibodies against Br-dUrd or phosphorylated histone H3 (pH3) to label S or M phase cells, respectively. At 48 hpf, in wild-type retinae, the great majority of cells in the central region have already exited the cell cycle. In contrast, mcm5m850 mutant retinae contain cells in S or M phase in the central region, which are thus either delayed or arrested in S or M phase (black arrowheads). (E and H) Summary diagrams representing the results from C and D and F and G, respectively.
To investigate whether retinal cells in mcm5m850 mutant embryos arrest in late S phase, we compared retinae from wild-type and mutant embryos by using markers for S or M phase. A short BrdUrd incubation, followed by fixation and detection with anti-BrdUrd antibody, was used to mark those cells that are in S phase. The presence of the phosphorylated form of histone H3 serves as a marker for late G2 or M phase (33). In zebrafish retinae, histogenesis and the pattern of cell-cycle exit are spatially and temporally stereotyped, following a center-to-periphery pattern of progression (21). Accordingly, at 48 hpf in wild-type retinae, most of the cells in the central region of the retina have already exited the cell cycle, whereas cells in S or M phase are present as a ring of cells located in the periphery. These cells can be detected by BrdUrd incorporation and the presence of phosphorylated histone H3 (Fig. 5 C and D). In contrast, in mcm5m850 mutant retinae, many cells in the central region still show BrdUrd incorporation and are antiphospho-histone-H3 immunoreactive and, thus, have not yet exited the cell cycle (Fig. 5 F and G).
Discussion
Mutants in three Drosophila mcm and one Arabidopsis mcm gene have been characterized (34–37). In Drosophila mcm2 mutants, proliferation of cells in the imaginal discs and the CNS is inhibited, and an apparent prolongation of S phase in the embryonic and larval CNS is seen (34). Similar defects can be observed in disk-proliferation abnormal (dpa), a mutant in the Drosophila mcm4 gene, and its effect was shown to be limited to mitotic replication but not to endoreplication (36). The lethal mutations in Drosophila mcm6 affect both mitotic cycle and endocycles and cause severe defects in cell proliferation within the brain and imaginal discs (35). In vertebrates, no mouse mutant in an MCM protein has been reported. Furthermore, although three additional zebrafish mutations in MCM-complex genes exist (mcm2, mcm3, and mcm7) (38), their phenotypes have not yet been analyzed. Our characterization of the phenotype of mcm5m850 mutant embryos is a detailed analysis of an MCM-complex mutant in vertebrates.
To begin to understand the in vivo function of mcm5 in zebrafish, we have analyzed its expression pattern in the retina. In human cells, a strong correlation has been observed between entry into G0 and loss of the MCM complex, prompting the notion that the presence of the replication-licensing system defines the proliferative state of a cell (39). Similarly, we find that mcm5 expression correlates remarkably closely with pcna expression and with the pattern of cell proliferation. Furthermore, we compared the mcm5-expression pattern with those of early stage (ath5) and later stage (elavl3) markers of neurogenesis. mcm5 expression overlaps partially with that of ath5 but does not overlap with that of elavl3, suggesting that mcm5 is rapidly down-regulated as differentiation proceeds. Interestingly, at 48 hpf, most mcm5 expression is in the marginal zone of the retina, although BrdUrd and phosphorylated histone H3 immunoreactivity indicate the presence of cycling cells in the periphery, representing cells that will soon undergo cell-cycle exit and differentiation. The retinae of many fish and amphibians grow throughout life, and new retinal cells are continually added at the CMZ. Furthermore, retinal regeneration in these animals occurs through retinal stem cells present in the CMZ (40). In contrast, the other areas within the retina are largely composed of fully differentiated cells with no further potential for proliferation. Thus, our expression analysis in the retina suggests that mcm5 expression may provide a sensitive marker to define tissues that possess the capacity for further proliferation. mcm5 is expressed in cells that can continue to proliferate. However, it appears to be down-regulated, ahead of the actual cell cycle exit, in those cells that will soon differentiate.
To assay the effect of reduced MCM5 at the cellular level, we assayed DNA content by FACS analysis at 48–50 hpf, when the maternal MCM5 level is significantly reduced. Using dissociated retinal cells from mcm5m850 mutant and wild-type embryos, we observed an increased fraction of cells with >2C DNA content in the mutant retinae. This increase could indicate either an increase in the number of cells entering S phase or a prolongation of S phase in mcm5m850 mutant embryos. Our data supports the latter explanation, because we observe no increase in cells positive for BrdUrd incorporation in mcm5m850 mutant embryos at this stage (see Fig. 11, which is published as supporting information on the PNAS web site). The prolongation of S phase is most likely caused by a defect in DNA replication, because MCM2 through -7 have been suggested to be important for both initiation and elongation during DNA replication. With the reduced level of functional MCM complex in mcm5m850 mutants, fewer replication forks would likely be initiated, and the elongation of existing replication forks might slow, perhaps resulting in delayed progression through S phase. Interestingly, many mcm5m850 mutant cells accumulate near 4C peak and may have finished replicating the bulk of their DNA, reminiscent of the situation in budding yeast mcm mutants, which exhibit a cell-division-cycle arrest in S phase, with a nearly doubled DNA content (41, 42).
In both Arabidopsis and Drosophila, null mutations in MCM proteins are lethal, and the lethality has been largely attributed to defects in cell proliferation (34, 35). However, in these mutants, whether an active cell-death process is involved has not yet been described. Our data suggest that, in mcm5m850 mutant embryos, some tissues may respond to the reduced level of MCM5 by activating programmed cell death, whereas others do not. Activation of apoptosis may be a direct consequence of the replication defect which could activate the S phase checkpoint system responsible for coordinating cell-cycle progression and cellular responses to DNA damage (43, 44). For example, induction of rereplication through perturbation of prereplication-complex components activates ATM/ATR-dependent checkpoint pathways that lead to an arrest of the cell cycle and/or apoptosis (45–47). Alternatively, MCM5 may have a role in the checkpoint system independent of its replication function. MCM proteins are expressed far in excess of what is required to support normal levels of DNA replication (4). Recently, human MCM7 was shown to interact directly with ATRIP, a protein that binds and activates ATR kinase. Interestingly, MCM7 was shown to regulate S-phase checkpoint response, even under the conditions where DNA replication is not perturbed (48). It remains to be tested whether MCM5 could also regulate S-phase checkpoint response in a similar manner.
In conclusion, our study reveals the crucial role of MCM5 in ensuring efficient genomic duplication and cell-cycle progression during vertebrate development. Furthermore, we show that MCM5 is a sensitive marker to identify cells with proliferative capacity. Finally, our data point to the existence of an added level of complexity in multicellular organisms in responding to the reduction in MCM5 by activation of a tissue-specific apoptosis program.
Supplementary Material
Acknowledgments
We thank S. Götter for expert zebrafish care; R. Koppa for plastic sections; K. Dürr, F. van Eeden (The Netherlands Institute for Developmental Biology), and S. W. Wilson (University College London) for plasmids and probes; D. Meyer for helpful mapping advice; A. Würth for FACS analysis; A. Neubüser, A. Intal, and M. Wulliman for advice on paraffin sections; and A. Neubüser, J. von Lintig, K. Lunde, D. Meyer, and H. Beckmann for critical reading of the manuscript. S.R. thanks H. Beckmann for helpful discussions throughout this project. This work was supported by Human Frontiers Science Program long-term Fellowship DFG-SFB 505-B7 (to S.R.) and the European Union Integrated Project Zebrafish Models of Human Disease and Development (W.D.).
Author contributions: S.R. and W.D. designed research; S.R., J.H., S.E., and A.-K.E. performed research; S.R. and J.H. contributed new reagents/analytic tools; S.R. and W.D. analyzed data; and S.R. and W.D. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CMZ, ciliary marginal zone; dpf, days postfertilization; hpf, hours postfertilization; MCM, minichromosome maintenance.
References
- 1.Blow, J. J. (2001) EMBO J. 20, 3293–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donaldson, A. D. & Blow, J. J. (1999) Curr. Opin. Genet. Dev. 9, 62–68. [DOI] [PubMed] [Google Scholar]
- 3.Diffley, J. F. X. & Labib, K. (2002) J. Cell. Sci. 115, 869–872. [DOI] [PubMed] [Google Scholar]
- 4.Tye, B. K. (1999) Annu. Rev. Biochem. 68, 649–686. [DOI] [PubMed] [Google Scholar]
- 5.Davey, M. J., Jeruzalmi, D., Kuriyan, J. & O'Donnell, M. (2002) Nat. Rev. Mol. Cell Biol. 3, 826–835. [DOI] [PubMed] [Google Scholar]
- 6.Chong, J. P. J., Mahbubani, H. M., Khoo, C. Y. & Blow, J. J. (1995) Nature 375, 418–421. [DOI] [PubMed] [Google Scholar]
- 7.Thömmes, P., Kubota, Y., Takisawa, H. & Blow, J. J. (1997) EMBO J. 16, 3312–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kubota, Y., Mimura, S., Nishimoto, S., Masuda, T., Nojima, H. & Takisawa, H. (1997) EMBO J. 16, 3320–3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bell, S. P. & Dutta, A. (2002) Annu. Rev. Biochem. 71, 333–374. [DOI] [PubMed] [Google Scholar]
- 10.Kelly, T. J. & Brown, G. W. (2000) Annu. Rev. Biochem. 69, 829–880. [DOI] [PubMed] [Google Scholar]
- 11.Kearsey, S. E. & Labib, K. (1998) Biochim. Biophys. Acta 1398, 113–136. [DOI] [PubMed] [Google Scholar]
- 12.Labib, K., Tercero, J. A. & Diffley, J. F. X. (2000) Science 288, 1643–1647. [DOI] [PubMed] [Google Scholar]
- 13.Forsburg, S. L. (2004) Microbiol. Mol. Biol. Rev. 68, 109–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Westerfield, M. (1995) The Zebrafish Book (University of Oregon Press, Eugene, OR).
- 15.Holzschuh, J., Barrallo-Gimeno, A., Ettl, A. K., Dürr, K., Knapik, E. W. & Driever, W. (2003) Development (Cambridge) 130, 5741–5754. [DOI] [PubMed] [Google Scholar]
- 16.Hauptmann, G. & Gerster, T. (1994) Trends Genet. 10, 266. [DOI] [PubMed] [Google Scholar]
- 17.Holzschuh, J., Ryu, S., Aberger, F. & Driever, W. (2001) Mech. Dev. 101, 237–243. [DOI] [PubMed] [Google Scholar]
- 18.Masai, I., Stemple, D., Okamoto, H. & Wilson, S. W. (2000) Neuron 27, 251–263. [DOI] [PubMed] [Google Scholar]
- 19.Trevarrow, B., Marks, D. L. & Kimmel, C. B. (1990) Neuron 4, 669–679. [DOI] [PubMed] [Google Scholar]
- 20.Michelmore, R. W., Paran, I. & Kesseli, R. V. (1991) Proc. Natl. Acad. Sci. USA 88, 9828–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Easter, S. S. J. & Malicki, J. J. (2002) in Pattern Formation in Zebrafish, ed. Solnica-Krezel, L. (Springer, Berlin, Heidelberg), Vol. 40, pp. 346–70. [Google Scholar]
- 22.Fashena, D. & Westerfield, M. (1999) J. Comp. Neurol. 406, 415–424. [DOI] [PubMed] [Google Scholar]
- 23.Knapik, E. W., Goodman, A., Ekker, M., Chevrette, M., Delgado, J., Neuhauss, S., Shimoda, N., Driever, W., Fishman, M. C. & Jacob, H. J. (1998) Nat. Genet. 18, 338–343. [DOI] [PubMed] [Google Scholar]
- 24.Poss, K. D. & Tonegawa, S. (1997) Proc. Natl. Acad. Sci. USA 94, 10919–10924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, Z., Joseph, N. M. & Easter, S. S., Jr. (2000) Dev. Dyn. 218, 175–188. [DOI] [PubMed] [Google Scholar]
- 26.Schmitt, E. A. & Dowling, J. E. (1994) J. Comp. Neurol. 344, 532–542. [DOI] [PubMed] [Google Scholar]
- 27.Hu, M. & Easter, S. S. J. (1999) Dev. Biol. 207, 309–321. [DOI] [PubMed] [Google Scholar]
- 28.Raymond, P. A. & Hitchcock, P. F. (1997) Adv. Neurol. 72, 171–184. [PubMed] [Google Scholar]
- 29.Perron, M. & Harris, W. A. (2000) BioEssays 22, 685–688. [DOI] [PubMed] [Google Scholar]
- 30.Kim, C. H., Ueshima, E., Muraoka, O., Tanaka, H., Yeo, S. Y., Huh, T. L. & Miki, N. (1996) Neurosci. Lett. 216, 109–112. [DOI] [PubMed] [Google Scholar]
- 31.Ekstrom, P. & Johansson, K. (2003) Brain Res. Dev. Brain Res. 145, 1–8. [DOI] [PubMed] [Google Scholar]
- 32.Dai, C. & Krantz, S. B. (1999) Blood 93, 3309–3316. [PubMed] [Google Scholar]
- 33.Hendzel, M. J., Wei, Y., Mancini, M. A., Van Hooser, A., Ranalli, T., Brinkley, B. R., Bazett-Jones, D. P. & Allis, C. D. (1997) Chromosoma 106, 348–360. [DOI] [PubMed] [Google Scholar]
- 34.Treisman, J. E., Follette, P. J., O'Farrell, P. H. & Rubin, G. M. (1995) Genes Dev. 9, 1709–1715. [DOI] [PubMed] [Google Scholar]
- 35.Schwed, G., May, N., Pechersky, Y. & Calvi, B. R. (2002) Mol. Biol. Cell 13, 607–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feger, G., Vaessin, H., Su, T. T., Wolff, E., Jan, L. Y. & Jan, Y. N. (1995) EMBO J. 14, 5387–5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Springer, P. S., McCombie, W. R., Sundaresan, V. & Martienssen, R. A. (1995) Science 268, 877–880. [DOI] [PubMed] [Google Scholar]
- 38.Golling, G., Amsterdam, A., Sun, Z., Antonelli, M., Maldonado, E., Chen, W., Burgess, S., Haldi, M., Artzt, K., Farrington, S., et al. (2002) Nat. Genet. 31, 135–140. [DOI] [PubMed] [Google Scholar]
- 39.Blow, J. J. & Hodgson, B. (2002) Trends Cell Biol. 12, 72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reh, T. A. & Fischer, A. J. (2001) Brain Behav. Evol. 58, 296–305. [DOI] [PubMed] [Google Scholar]
- 41.Gibson, S. I., Surosky, R. T. & Tye, B. K. (1990) Mol. Cell. Biol. 10, 5707–5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lei, M., Kawasaki, Y., Young, M. R., Kihara, M., Sugino, A. & Tye, B. K. (1997) Genes Dev. 11, 3365–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nyberg, K. A., Michelson, R. J., Putnam, C. W. & Weinert, T. A. (2002) Annu. Rev. Genet. 36, 617–656. [DOI] [PubMed] [Google Scholar]
- 44.Zhou, B. B. & Elledge, S. J. (2000) Nature 408, 433–439. [DOI] [PubMed] [Google Scholar]
- 45.Vaziri, C., Saxena, S., Jeon, Y., Lee, C., Murata, K., Machida, Y., Wagle, N., Hwang, D. S. & Dutta, A. (2003) Mol. Cell 11, 997–1008. [DOI] [PubMed] [Google Scholar]
- 46.Zhu, W., Chen, W. & Dutta, A. (2004) Mol. Cell. Biol. 24, 7140–7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mellixetian, M., Ballabeni, A., Masiero, L., Gasparini, P., Zamponi, R., Bartek, J., Lukas, J. & Helin, K. (2004) J. Cell Biol. 165, 473–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cortez, D., Glick, G. & Elledge, S. J. (2004) Proc. Natl. Acad. Sci. USA 101, 10078–10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.