Abstract
Clostridium difficile is a major nosocomial pathogen that produces two large protein toxins [toxin A (TcdA) and toxin B (TcdB)] capable of disrupting intestinal epithelial cells. Both belong to the family of large clostridial cytotoxins, which are characterized by the presence of a repetitive C-terminal repetitive domain (CRD). In TcdA, the CRD is composed of 39 repeats that are responsible for binding to cell surface carbohydrates. To understand the molecular structural basis of cell binding by the toxins from C. difficile, we have determined a 1.85-Å resolution crystal structure of a 127-aa fragment from the C terminus of the toxin A CRD. This structure reveals a β-solenoid fold containing five repeats, with each repeat consisting of a β-hairpin followed by a loop of 7–10 residues in short repeats (SRs) or 18 residues in long repeats (LRs). Adjacent pairs of β-hairpins are related to each other by either 90° or 120° screw–axis rotational relationships, depending on the nature of the amino acids at key positions in adjacent β-hairpins. Models of the complete CRDs of toxins A and B suggest that each CRD contains straight stretches of β-solenoid composed of three to five SRs that are punctuated by kinks introduced by the presence of a single LR. These structural features provide a framework for understanding how large clostridial cytotoxins bind to cell surfaces and suggest approaches for developing novel treatments for C. difficile-associated diseases by blocking the binding of toxins to cell surfaces.
Clostridium difficile is a Gram-positive bacterium responsible for a variety of gastrointestinal diseases that range in severity from antibiotic-associated diarrhea to pseudomembranous colitis (1). Every year in the United States, C. difficile causes ≈250,000 clinically diagnosed cases of disease, contributing to the cause of thousands of deaths and costing the health-care system over $1 billion (2, 3). Healthy individuals rarely develop C. difficile-associated diseases, but patients whose normal gut flora have been disrupted by treatment with common broad-spectrum antibiotics are highly susceptible. Because C. difficile forms spores that are extremely difficult to remove from institutional settings such as hospitals and nursing homes, a source of new infections is nearly always present. It is estimated that 1–3% of all hospitalized patients treated with antibiotics become infected with C. difficile (4). Most disturbingly, a new highly virulent strain appears to be causing outbreaks with increased disease incidence, severity, and mortality in North America since 2001 (5). Common treatments for C. difficile-associated diseases include terminating the original antibiotic treatment and administering either metronidazole or vancomycin. Unfortunately, C. difficile strains resistant to these antibiotics are beginning to emerge (6, 7). Both metronidazole and vancomycin are also broad-spectrum antibiotics that continue to disrupt normal colonic bacterial populations and, as a result, a relapse of C. difficile infection after termination of antibiotic treatment is quite common and can be very difficult to treat. Several new approaches to treating C. difficile-associated diseases are currently being developed, but there is a clear need for more effective therapeutics specifically targeting the pathological mechanisms of C. difficile (8, 9).
Toward this end, we have initiated studies aimed at understanding the molecular structural basis of C. difficile virulence factors. Although the bacterium can produce three toxins, toxin A (TcdA) and toxin B (TcdB) are recognized as the main virulence factors (10–12). Like all members of the group of large clostridial toxins, TcdA and TcdB are large (250- to 308-kDa) single-subunit polypeptides, with structures that can be organized into three regions: (i) the N-terminal region, which contains glucosyltransferase activity; (ii) the hydrophobic central region, important for translocating the toxins across the cell membrane; and (iii) the highly repetitive C-terminal region, which appears to be primarily responsible for receptor binding (13–16).
The C-terminal region of TcdA interacts with cell-surface carbohydrates, including Gal-α1,3-Gal-β1,4-GlcNAc, as an initial step in pathogenesis (17–20). The C-terminal and central regions of the toxin then help mediate entry into the cell through receptor-mediated endocytosis (21). Once internalized, both toxins use UDP-glucose to glucosylate small Ras-like GTPases at a threonine residue (Thr-37 in Rho) in the effector domain (22–25). Glucosylation inhibits downstream signaling through effector molecules, leading to the depolymerization of the actin cytoskeleton, disruption of tight junctions, and apoptosis in colonic epithelial cells (10, 26). This cytotoxic effect, action on intestinal neurons, and modulation of immune system in vivo result in inflammation and diarrhea.
The most striking feature of the receptor-binding C-terminal region of TcdA and TcdB is the presence of repeating units of 21-, 30-, or 50-aa residues (13–15, 27). Different approaches to analyzing the sequence of TcdA reveal that this region contains between 30 and 38 contiguous repeats, whereas in TcdB there appear to be between 19 and 24 repeats. The repeats in TcdA show a low level of sequence similarity with repeats that have been found in a number of extracellular bacterial proteins, most of which bind to bacterial cell walls (28). Recently, a number of fragments containing 5–15 repeats from TcdA have been shown to form stable folded secondary structures independently of other structures in the intact toxin (29, 30). It is likely that this region's modular design and multiple repeats help to amplify binding affinity through an avidity effect seen in many carbohydrate-binding proteins that bind to cell surfaces (15, 31).
Here, we report the crystal structure of the C-terminal 127 residues of TcdA (TcdA-f1). This is the first 3D structural information on TcdA or large clostridial toxins in general and provides a basis for understanding the architecture of the entire C-terminal receptor-binding domain. The structure of TcdA-f1 provides a framework for understanding receptor binding, which will ultimately lead to the development of novel therapeutic approaches aimed at interfering with interactions between toxins and cell surfaces.
Experimental Procedures
Cloning, Expression, and Purification. PCR (forward primer = 5′ GGA ATT CCA TAT GCA TCA TCA TCA TCA TCA CAC TGG TTG GGT AAC TAT TGA T and reverse primer = 5′ CGG GAT CCC TAA TAT ATC CCA GGG GCT TTT ACT CC) was used to amplify the coding region for residues 2573–2709 of TcdA (C. difficile strain 48489, toxinotype VI, numbering according to strain VPI 10463, toxinotype 0) by using clone pA3-48489. pA3-48489 was constructed by cloning the amplified A3 PCR fragment, which was prepared as described in ref. 32, into vector pQE11A18. After restriction enzyme digestion with NdeI and BamHI, this fragment was ligated into pET-3a (Novagen) and transformed into Escherichia coli JM109. Dideoxy chain-termination sequencing was used to verify the sequence of the expression clone.
The recombinant TcdA fragment (TcdA-f0) was expressed in E. coli BL21 (DE3) pLysS after induction with 0.5 mM isopropyl β-d-thiogalactoside and growth in LB medium at 25°C for 18 h. Cells from 1 liter of culture were harvested by centrifugation and resuspended in 35 ml of lysis buffer (100 mM sodium phosphate/200 mM sodium chloride/5 mM imidazole, pH 8.0). The cells were treated with 0.5 mg of DNase I and 0.1 mM PMSF for 10 min, lysed by sonication, and clarified by centrifugation. The clarified extract was chromatographed on Nickel-NTA-Sepharose (1 × 5-cm column, Qiagen, Valencia, CA), yielding 15 ml at 3.0 mg/ml. After dialysis against buffer B (20 mM phosphate/20 mM NaCl/5% glycerol, pH 6.0) overnight at 4°C, bovine pancreatic trypsin (Sigma, lyophilized) was added at a mass ratio of 80:1 TcdA-f0/trypsin and incubated for 4 h at 25°C. Trypsin and small proteolytic fragments were removed by cation exchange chromatography by using a Vivapure S Maxi H spin column (Vivascience, Hannover, Germany) (elution at 30 mM NaCl, 20 mM sodium-Hepes, pH 7.0). Undigested protein containing the N-terminal histidine tag was removed by chromatography on nickel-NTA-Sepharose. The major proteolytic fragment (TcdA-f1) eluted during the wash (20 mM phosphate/5 mM imidazole, pH 8.0) was dialyzed overnight in 20 mM Tris, pH 8.0/140 mM NaCl/1 mM EDTA and concentrated to 10 mg/ml by using Vivaspin 15R concentrators, 5000 MWCO (Vivascience). MALDI-MS revealed a mass of 14,194 Da, which is consistent with a predicted mass of 14,175 Da for residues 2,583–2,709 of TcdA48489. Selenomethionine-substituted TcdA-f0 was expressed in M9 minimal medium supplemented with all of the natural α-amino acids except methionine, and with selenomethionine added at 50 μg/ml. Selenomethionine-substituted TcdA-f1 was purified according to the same procedure as the native protein.
Crystallization and Data Collection. Crystals were grown by the vapor diffusion method (1 μl of protein + 1 μl of reservoir equilibrated against 0.5 ml of reservoir solution) at 21°C. Initial crystallization conditions were obtained from sparse matrix screens (Index-HT and Crystal-HT, Hampton, San Diego). Diffraction-quality crystals could not be obtained from TcdA-f0, but TcdA-f1 yielded well-diffracting crystals after the removal of the purification tag before crystallization. The optimized crystallization condition for both native and selenomethionine-substituted forms of TcdA-f1 was 250 mM ammonium tartrate, 20% glycerol, 100 mM sodium acetate, pH 5.5. Diffraction data were measured from crystals that were transferred from mother liquor directly to a nitrogen gas stream at ≈110 K. Diffraction data were initially measured by using a MAR 345 image plate and x-rays produced with a rotating copper anode (Rigaku, Tokyo, RUH3R). Higher-resolution data and data for multiwavelength anomalous diffraction (MAD) experiments were measured by using an ADSC (Poway, CA) Quantum-315 charge-coupled device detector at the Advanced Light Source (Lawrence Berkeley Laboratory, Berkeley, CA) on beamline 8.3.1. Data were processed and scaled by using denzo, scalepack, and programs from ccp4 (Version 5.0.2) (33, 34). The space group was determined to be P41212 by examining systematic absences, as well as the electron density maps from MAD phases calculated in space groups P41212 and P43212. Crystallographic statistics are summarized in Table 1.
Table 1. Crystallographic statistics.
| Statistics | Edge | Peak | Remote |
|---|---|---|---|
| Data collection | |||
| Crystal/space group | TcdA-f1/P41212 | ||
| Unit cell lengths, Å | 42.05 × 42.05 × 132.11 | ||
| Unit cell angles,° | 90, 90, 90 | ||
| Wavelength, Å | 0.979741 | 0.979571 | 1.019867 |
| Resolution, Å | 40.16-1.85 | 40.16-1.85 | 40.16-1.85 |
| High resolution, Å | 1.92-1.85 | 1.92-1.85 | 1.92-1.85 |
| Total reflections* | 69,718 (6,383) | 69,632 (6,321) | 70,319 (6,608) |
| Unique reflections* | 10,802 (1,020) | 10,798 (1,015) | 10,812 (1,031) |
| Completeness, %* | 99.2 (96.1) | 99.2 (95.7) | 99.3 (97.2) |
| I/σ* | 26.5 (8.7) | 21.4 (7.4) | 31.6 (11.0) |
| Rsym*† | 0.046 (0.182) | 0.057 (0.187) | 0.038 (0.158) |
| Refinement | |||
| Resolution, Å | 40.16-1.85 (1.90-1.85) | ||
| Rwork‡ | 0.160 (0.135) | ||
| Rfree§ | 0.205 (0.175) | ||
| Number of atoms | |||
| Protein | 988 | ||
| Solvent and Ions | 218 | ||
| rms deviations from ideal geometry | |||
| Bond lengths, Å | 0.007 | ||
| Bond angles, ° | 1.00 | ||
| Average B factor, Å2 | 13.8 | ||
Values from the outermost resolution shell are given in parentheses.
Rsym = ΣhΣi(|Ii(h) — 〈I(h) 〉|)/ΣhΣiIi(h), where Ii(h) is the ith integrated intensity of a given reflection, and 〈I(h) 〉 is the weighted mean of all measurements of I(h).
Rwork = Σh∥F(h)o|—|F(h)c∥/Σh|F(h)o| for the 95% of reflection data used in refinement.
Rfree = Σh∥F(h)o|—|F(h)c∥/Σh|F(h)o| for the 5% of reflection data excluded from ref inement.
Experimental phases were obtained by using a three-wavelength MAD experiment with a selenomethionine derivative. A single copy of TcdA-f1 was present in the asymmetric unit (calculated solvent content = 0.40), and the positions for all three Se atoms were determined by using solve (35). An excellent electron-density map was obtained by using all data to 1.85-Å resolution (z score = 13.8, figure of merit = 0.61). Density modification and a partial structure consisting of 80 residues of a total of 127 were generated by resolve (36). An additional 45 residues were built manually by using xfit (Version 4.0), and iterative rounds of refinement and model building were performed by using refmac (Version 5.1) and xfit (37, 38). Refinement was carried out against the native amplitude data set obtained from an analysis of the MAD data scaled and analyzed by solve. Five percent of the data was randomly selected for crossvalidation calculations before the commencement of refinement. All but two of the 127 residues were well-defined by electron density; the first residue at the N terminus and the last residue at the C terminus appear to be disordered. The quality of stereochemical parameters in the model was assessed by using procheck (39) and whatcheck (40).
Results and Discussion
Structure of TcdA-f1. As an initial step in understanding the structure of TcdA, we have determined the 3D structure of a fragment from the C terminus of the TcdA C-terminal repetitive domain (CRD) from C. difficile strain 48489, toxinotype VI (TcdA-f1). This fragment corresponds to residues 2582–2709 in the type strain VPI 10463, toxinotype 0, and the sequence of TcdA48489-f1 is 94% identical to the sequence in the type strain, with no gaps and only structurally neutral substitutions appearing in a sequence alignment. TcdA-f1 was obtained by expressing a slightly longer fragment (TcdA-f0, residues 2573–2709) in E. coli and isolating a subfragment generated by limited proteolysis with bovine trypsin. Only needle-like crystals could be obtained with TcdA-f0, but well-diffracting prisms were readily obtained from TcdA-f1.
The structure of TcdA-f1 is highly repetitive, as predicted from sequence analysis. To understand the structure of TcdA-f1 and how it relates to the overall structure of the TcdA CRD, it is useful to define two types of repeats appearing in both the structure and the sequence. These definitions are based on the 3D structural motifs observed in TcdA-f1 and differ slightly from previous definitions based on sequence analysis alone (13–15, 27). Both the structure and sequence of TcdA-f1 reveal the presence of four copies of a short repeat (SR) and one copy of a long repeat (LR) (Fig. 1 and Fig. 5, which is published as supporting information on the PNAS web site). Sequence analysis of the entire C-terminal domain of TcdA10463 reveals the presence of a C-terminal region containing 32 SRs of 15–21 residues and seven LRs of 30 residues. These repeats are similar to the consensus repeats of oligopeptides that have been identified earlier (13–15, 27).
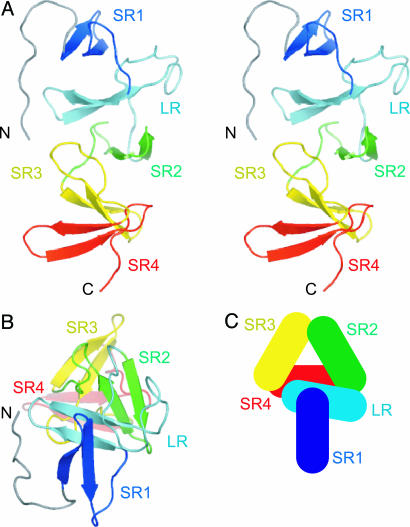
Fig. 1.
Overall structure of TcdA-f1. (A) Stereoscopic view of a ribbon representation of the structure of TcdA-f1, with each β-hairpin motif colored separately (N terminus to C terminus): blue, cyan, green, yellow, and red. (B) View of TcdA-f1 ribbon representation from the N-terminal end and down the axis of the β-solenoid. (C) Schematic representation of β-hairpins (rectangles) in the β-solenoid fold. All figures were prepared by using pymol (52).
The crystal structure of TcdA-f1 reveals that each SR or LR contains a single β-hairpin consisting of a pair of five- to six-residue antiparallel β-strands connected by a tight turn (usually type I′) (Fig. 1). The 3D structure of TcdA-f1 suggests that the boundaries of each SR or LR should be defined to coincide with the beginning of the β-hairpin and the end of the connecting loop preceding the following β-hairpin. Each β-hairpin interacts with both the preceding and following β-hairpins, except for the N- and C-terminal hairpins. The N-terminal end of TcdA-f1 adopts a nonnatural structure due to the artificial truncation of the protein, and the N-terminal hairpin contains some exposed hydrophobic residues that form a nonnatural intermolecular interface with a few exposed hydrophobic residues near the natural C terminus of a different molecule in the crystal lattice. It is likely that the structure of the N-terminal end of TcdA-f1 differs from that seen in intact TcdA, because an additional series of β-hairpin repeats normally precedes this part of the structure. Residues 1–11 of TcdA-f1 adopt an extended structure that packs against SR1 (Fig. 1). In intact TcdA-f1, it is likely that these residues form part of the preceding SR.
The structure of each β-hairpin is highly conserved, with the residues at positions 2 and 3 of strand 1 and positions 3, 4, and 5 of strand 2 forming a small hydrophobic cluster that brings consecutive pairs of β-hairpins together in a regularly repeating manner (Fig. 2). The high degree of sequence conservation for these hydrophobic residues in C. difficile toxins and other related proteins with repetitive structure has been noted previously, and the structural importance of this high level of sequence conservation has also been postulated (15). Specific hydrophobic packing interactions or hydrogen-bonding interactions between adjacent pairs of β-hairpins dictate the regular arrangement of these secondary structural elements. Specifically, each adjacent pair of β-hairpins is related to the previous β-hairpin by a 31 screw–axis transformation, in which adjacent β-hairpins are related by a 120° rotation and a translation of ≈10 Å, thus creating a left-handed β-solenoid helix. This fold is predicted to be found in a wide range of bacterial cell-surface-binding proteins and falls in the more general class of repeating solenoid fold proteins (28, 41).
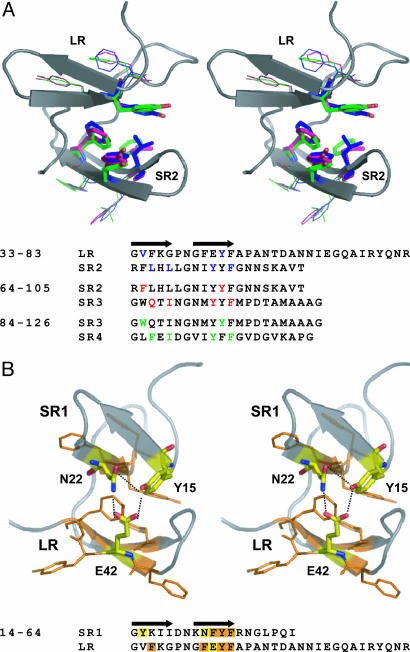
Fig. 2.
Packing interfaces between LRs and SRs. (A) Stereoscopic view of key residues forming the LR:SR2, SR2:SR3, and SR3:SR4 hydrophobic packing interfaces. The backbone traces of LR and SR2 are drawn in a cartoon representation. (B) Stereoscopic view of the hydrogen-bonding network formed at the interface between the LR and preceding SR in TcdA-f1. The highly conserved residues forming hydrogen bonds (dashed lines) are highlighted in both the primary and tertiary structure diagrams.
Following the second strand of each β-hairpin is a loop of 7–18 residues. For each SR, the second strand is followed by a loop of 7–10 residues that connects consecutive pairs of β-hairpins. The three instances of connecting loops in the SRs found in TcdA-f1 adopt differing structures and appear to play a neutral role in the overall arrangement of β-hairpins. That is, differences in the structures of these connecting loops do not appear to affect how the two flanking β-hairpins are arranged relative to each other. The primary role of interhairpin packing interactions and the secondary role of loop structure in dictating the overall structure of the solenoid fold are similar to that seen in the structures of other β-solenoid fold proteins (28, 42, 43).
In contrast, a much longer loop of 18 residues follows the second β-strand in the only LR found in TcdA-f1. As described below, the sequence of this loop is one of the most highly conserved elements in the CRD, and the most highly conserved sequence positions appear to play key roles in stabilizing the 3D fold of this loop. In TcdA-f1, the presence of this longer loop and insertion of a perfectly conserved glutamic acid residue in the second β-strand of the hairpin induces a strikingly different packing relationship between the hairpin belonging to the LR and the preceding SR. This altered packing arrangement introduces a kink in the overall path of the β-solenoid.
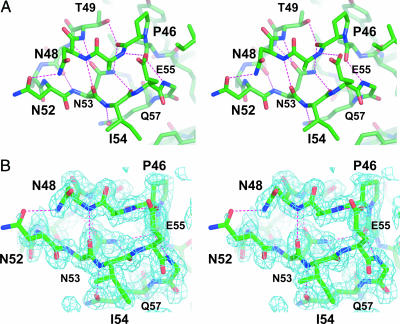
Structural Features of the Highly Conserved Hairpin and Loop in LRs. Sequence analysis reveals that the 18-residue loop following the β-hairpin in each LR has a highly conserved sequence (Fig. 5). The structure of TcdA-f1 reveals how this highly conserved sequence promotes the formation of a compact tightly folded 3D structure (Fig. 3). Most prominently seen are a series of hydrogen bonds involving the side chains of highly conserved amino acids. A key hydrogen-bonding network is formed between the side chain of Asn-53 and main-chain amide and carbonyl groups at the center of the loop. The amide nitrogen of the Asn-53 side chain donates hydrogen bonds to the carbonyl oxygen atoms of Pro-46 and Ile-54, whereas the carbonyl oxygen accepts hydrogen bonds from the amide nitrogen atoms of Thr-49 and Asp-50. The side chains of Thr-49, Glu-55, and Gln-57 also form hydrogen bonds with the main-chain peptide groups of Pro-46, Ala-47, and Ile-54. Finally, hydrogen bonds are formed between the side chains of Asn-48 and Asn-52, as well as between the side chains of Asp-50 and Arg-60. In addition to these key hydrogen bonds, Ala-45, Pro-46, Thr-49, and Ile-54 form a hydrophobic interface that packs against the aromatic ring of Tyr-43 in the β-hairpin motif preceding the loop. The high degree of sequence conservation in this loop and the intricate network of interactions stabilizing the 3D structure strongly suggest that this structure is found in each of the seven instances of LRs seen in TcdA.
Fig. 3.
LR loop structure. (A) Stereoscopic view of the highly conserved loop region found within the LRs from TcdA (residues 45–64 in TcdA-f1). Hydrogen bonds are drawn as dashed lines. (B) Stereoscopic view of the experimentally phased electron density map (contoured at 1 σ) in the region of the highly conserved loop. The map is calculated by using phases following MAD analysis with solve and density modification with resolve.
The β-hairpin in the LR also contains unique structural features. In the second β-strand of each SR, three aromatic residues (F or Y, followed by Y and then F) occupy the third, fourth, and fifth positions of the strand, whereas in the second β-strand of each LR, the second, third, fourth, and fifth positions always contain the sequence FEYF. The structure of TcdA-f1 reveals that the glutamate residue in this highly conserved sequence motif accepts hydrogen bonds from the hydroxyl group of a tyrosine residue at the start of the first strand in the preceding SR, as well as an asparagine residue at the beginning of the second strand in that same SR (Fig. 2a). Close inspection of the repeat sequences in TcdA reveals that for each SR immediately preceding each LR, the first position of the first strand is always occupied by Tyr, and the second position of the second strand is occupied by either His or Asn (Fig. 5). Because the side chains of each of these residues would be expected to form hydrogen bonds with the side-chain hydroxyl group of Tyr, we predict that the same hydrogen-bonding network seen in TcdA-f1 is preserved for each of the seven LRs in TcdA. It is notable that adjacent pairs of SRs have a mainly hydrophobic interface involving aromatic residues that is very different from this highly conserved hydrogen-bonding network (Fig. 2b).
An important effect of the packing seen between the hairpin in the LR and the preceding hairpin is that the relative orientation of these two hairpins differs significantly from the orientation adopted by the two hairpins of adjacent SRs. The hydrogen-bonding interactions formed by the LR and the preceding SR give rise to an ≈90° rotational relationship between the β-hairpins in these two repeats (Fig. 1). In contrast, the hydrophobic packing interactions formed between pairs of adjacent SRs result in an ≈120° rotational relationship. The high degree of sequence conservation in the residues participating in these packing interactions throughout the entire CRD suggests that each LR induces a ≈30° kink in the regular repeating solenoid arrangement of hairpins formed by SRs.
The presence of regularly interspersed LRs amidst a larger number of SRs in the large clostridial toxins (LCT group) appears to be unique in the β-solenoid family. Other members of the family appear to have much less variation in the sequence and length of the repeat segments (44). It is possible to speculate that the kinks introduced by the LRs may play important roles related to CRD interactions with cell-surface receptors, as well as related to pore formation and cell entry. Although pore formation has been shown directly only for TcdB (45), it is well established that both TcdA and TcdB enter the cytosol from the endosomal compartment. Unlike toxins that enter the cytosol from the endoplasmic reticulum, which can use endogenous transporters for entering the cytoplasm, toxins using the endosomal pathway usually encode the translocation machinery by themselves. For TcdB, pore-forming potential has been mapped to the C-terminal portion of the protein (residues 547-2366) although not directly to the repeat region (45). The CRD of TcdA has also been shown to be critical for endocytosis, in addition to cell-surface receptor binding (21).
Receptor-Binding Sites Formed by SRs and LRs. TcdA binds to carbohydrate structures (particularly Gal-α1,3-Gal-β1,4-GlcNAc) that are present on a diverse range of molecules including bovine thyroglobulin, rabbit erythrocytes, and both Ig and non-Ig components of human milk (17, 46, 47). Although the binding of the C-terminal receptor-binding domain of TcdA to cell-surface carbohydrates is believed to be the key initial step toward entry into intestinal epithelial cells, a specific, functional receptor in humans has yet to be positively identified. Plate-binding and frontal affinity chromatography-binding assays suggest that TcdA-f1 may not bind strongly to the Gal-α1,3-Gal-β1,4-GlcNAc trisaccharide that longer subfragments of the CRD or the entire CRD bind to (unpublished observations). Attempts to crystallize a complex composed of TcdA-f1 and Gal-α1,3-Gal-β1,4-GlcNAc also have not been successful.
At least three explanations can be proposed to account for the low affinity of TcdA-f1 for Gal-α1,3-Gal-β1,4-GlcNAc. First, higher-affinity binding sites for this trisaccharide may reside in other repeats of the CRD. Sequence and structure variation in different repeats may be related to variations in binding affinity or specificity. Second, a weak binding site for carbohydrates may reside in TcdA-f1, but this isolated binding site may be too weak to detect without using a multivalent ligand that interacts with other sites outside of TcdA-f1. It is likely that the CRD interacts with cell-surface carbohydrates through a multivalent mechanism, as seen in many other toxins and lectins. In nearly all of these cases, the binding affinity of a single site for a univalent ligand is extremely weak, with dissociation constants in the millimolar range (31). Also, studies by Frisch et al. (21) have shown that the middle part of TcdA (region A2, residues 900-1750) significantly increases the binding and internalization of the C-terminal part. This suggests that the native binding mode of TcdA may be fairly complex and may involve multiple interactions involving parts of the C-terminal repeats and the central part of the toxin. Third, a recent study suggests that Gal-α1,3-Gal-β1,4-GlcNAc may not be the receptor required to mediate toxin endocytosis in glycosylation-deficient CHO cell mutants, because this cell line is still toxin-sensitive.¶ This study suggests that the C-terminal receptor-binding domain may recognize other cell-surface receptors, in addition to the previously identified trisaccharide.
It may be possible to speculate on the location of potential binding pockets in TcdA-f1 and TcdA by comparing the structure of TcdA-f1 with other proteins containing β-solenoid folds. The closest structural homologues to TcdA-f1 are the cell wall-binding domains of phage Cp-1 endolysin and pneumococcal autolysin. Although these proteins share a very low degree of sequence identity with TcdA-f1 (≈10% and 12%, respectively), 67 Cα pairs can be aligned with an rms deviation (rmsd) 1.42 Å for endolysin, and 76 Cα pairs can be aligned with an rmsd of 1.61 Å for autolysin (Fig. 6, which is published as supporting information on the PNAS web site). The arrangement of pairs of adjacent β-hairpins is very similar in all three proteins, except for the unusual packing arrangement seen between the LR and preceding SR in TcdA-f1, which is not seen in either endolysin or autolysin.
In addition to the similarities in folds between TcdA-f1 and the cell-wall-binding proteins, the hydrophobic choline-binding pockets appear to be conserved in structure. Structures of autolysin and endolysin bound to choline, DDAO, and ofloxacin reveal a common binding site formed by two Trp residues and a Tyr residue at the packing interface between two pairs of adjacent β-hairpins (28, 42, 43, 48). This pocket is less hydrophobic in TcdA, because the Trp residues are replaced by Tyr and Phe (Fig. 6d). As expected, choline has been shown to bind to and stabilize TcdA, presumably at this same pocket (30), and the binding of TcdA to bacterial cells of noncytotoxic C. difficile strains has also been shown (10). Because the hydrophobic face of galactose residues is commonly found to pack against hydrophobic aromatic residues in carbohydrate-binding proteins, it is possible that this pocket may form at least part of the carbohydrate-binding site in TcdA (31).
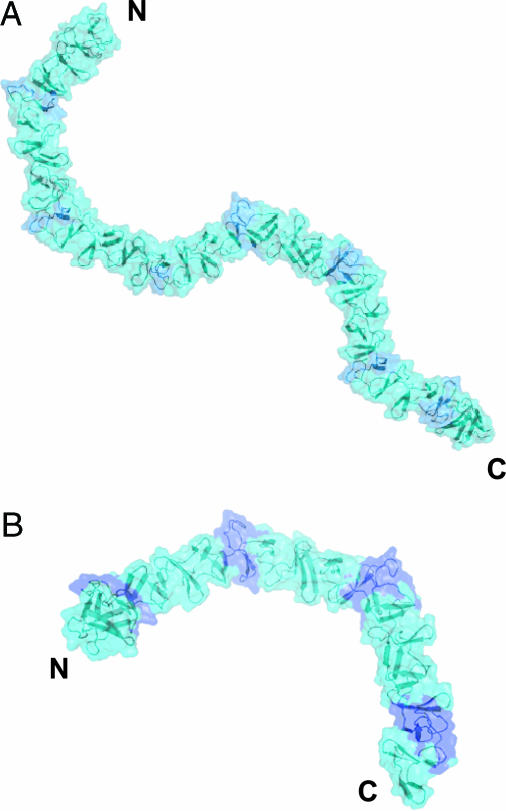
Models for the Complete CRDs of TcdA and TcdB. The structure of TcdA-f1 reveals that SRs and LRs are arranged in a very regular manner dictated by two types of highly conserved interfaces formed by adjacent pairs of β-hairpins (Fig. 2). Sequence analysis of the C-terminal receptor-binding domain of TcdA and TcdB reveals that clusters of three to five SRs are followed by a single LR. The same sequence (or pattern) of SRs and LRs also is conserved in variant C. difficile strains with deletions in tcdA, further suggesting the potential functional importance of the structural variations introduced by the insertion of LRs into stretches of SRs (49–51). The structure of TcdA-f1 suggests a likely arrangement for all of the β-hairpins in the CRDs of TcdA and TcdB (Fig. 4). As described above, the interface formed between adjacent pairs of SRs leads to a 120° screw–axis transformation, whereas an SR followed by an LR leads to a 90° screw–axis transformation. As a result, the overall structure of the CRD is predicted to consist of straight segments of β-solenoid structure composed of SRs that are broken up by the kinks introduced by the altered packing arrangement of single LRs. The overall features of this model are likely correct, but variations from the model are likely introduced by slight sequence differences in interface residues, especially in TcdB. The differences in receptor-binding specificity between TcdA and TcdB are also likely reflected in differences in each toxin's CRD structures.
Fig. 4.
Models of the C-terminal receptor-binding domains of (A) TcdA and (B) TcdB. LRs are colored dark blue, and the SRs are colored light blue. The N and C termini of each fragment are indicated.
It is difficult to speculate about the functional consequences of the kinks introduced by the LRs without additional information about the nature of the carbohydrate-binding sites. It is likely that these kinks may affect how multiple carbohydrate-binding sites within the CRD may interact with cell-surface carbohydrates. Mutagenesis experiments that remove or replace LRs with SRs and test the relative binding affinities of mutant CRDs may be particularly informative for testing the importance of the kinks introduced by the LRs.
Supplementary Material
Acknowledgments
We thank I. Zec for performing sequence analysis on the pA3–48489 clone and Dr. Isabelle Barrette-Ng for helpful discussions on this manuscript. This work was supported by the Alberta Ingenuity Centre for Carbohydrate Sciences, Canadian Institutes for Health Research New Investigator Award (to K.N.), and Alberta Heritage Foundation for Medical Research (AHFMR) Medical Scholar Award (to K.N.). J.G.S.H. was supported by a fellowship from the Alberta Ingenuity Fund. M.R. was supported by the Slovenian Research Agency (project J1-6456). X-ray diffraction data were collected at beamline 8.3.1 of the Advanced Light Source (ALS) at Lawrence Berkeley Laboratory, Berkeley, CA, under an agreement with the Alberta Synchrotron Institute (ASI). The ALS is operated by the U.S. Department of Energy and supported by the National Institutes of Health. Beamline 8.3.1 was funded by the National Science Foundation, the University of California, and Henry Wheeler. The ASI synchrotron access program is supported by grants from the Alberta Science and Research Authority and AHFMR.
Author contributions: K.K.-S.N. designed research; J.G.S.H. and A.G. performed research; M.R. contributed new reagents/analytic tools; J.G.S.H., A.G., M.R., and K.K.-S.N. analyzed data; and J.G.S.H., M.R., and K.K.-S.N. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CRD, C-terminal repetitive domain; SR, short repeat; LR, long repeat; TcdA, Clostridium difficile toxin A; TcdB, Clostridium difficile toxin B; MAD, multiwavelength anomalous diffraction.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2F6E).
Footnotes
Hofmann, F., Stieglitz, L., Gerardy-Schahn, R., Eckhardt, M. & Just, I. (2004) First International Clostridium difficile Symposium, May 5–8, 2004, Gozd Martuljck, Slovenia.
References
- 1.Kelly, C. P. & LaMont, J. T. (1998) Annu. Rev. Med. 49, 375–390. [DOI] [PubMed] [Google Scholar]
- 2.Kyne, L., Hamel, M. B., Polavaram, R. & Kelly, C. P. (2002) Clin. Infect. Dis. 34, 346–353. [DOI] [PubMed] [Google Scholar]
- 3.Wilkins, T. D. & Lyerly, D. M. (2003) J. Clin. Microbiol. 41, 531–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beaugerie, L., Flahault, A., Barbut, F., Atlan, P., Lalande, V., Cousin, P., Cadilhac, M. & Petit, J. C. (2003) Aliment. Pharmacol. Ther. 17, 905–912. [DOI] [PubMed] [Google Scholar]
- 5.Pepin, J., Valiquette, L., Alary, M. E., Villemure, P., Pelletier, A., Forget, K., Pepin, K. & Chouinard, D. (2004) CMAJ 171, 466–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pelaez, T., Alcala, L., Alonso, R., Rodriguez-Creixems, M., Garcia-Lechuz, J. M. & Bouza, E. (2002) Antimicrob. Agents Chemother. 46, 1647–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerding, D. N. (2005) Clin. Infect. Dis. 40, 1598–1600. [DOI] [PubMed] [Google Scholar]
- 8.Braunlin, W., Xu, Q., Hook, P., Fitzpatrick, R., Klinger, J. D., Burrier, R. & Kurtz, C. B. (2004) Biophys. J. 87, 534–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McFarland, L. V. (2005) J. Med. Microbiol. 54, 101–111. [DOI] [PubMed] [Google Scholar]
- 10.Just, I. & Gerhard, R. (2004) Rev. Physiol. Biochem. Pharmacol. 152, 23–47. [DOI] [PubMed] [Google Scholar]
- 11.Rupnik, M., Dupuy, B., Fairweather, N. F., Gerding, D. N., Johnson, S., Just, I., Lyerly, D. M., Popoff, M. R., Rood, J. I., Sonenshein, A. L., et al. (2005) J. Med. Microbiol. 54, 113–117. [DOI] [PubMed] [Google Scholar]
- 12.Voth, D. E. & Ballard, J. D. (2005) Clin. Microbiol. Rev. 18, 247–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dove, C. H., Wang, S. Z., Price, S. B., Phelps, C. J., Lyerly, D. M., Wilkins, T. D. & Johnson, J. L. (1990) Infect. Immun. 58, 480–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.von Eichel-Streiber, C., Laufenberg-Feldmann, R., Sartingen, S., Schulze, J. & Sauerborn, M. (1992) Mol. Gen. Genet. 233, 260–268. [DOI] [PubMed] [Google Scholar]
- 15.von Eichel-Streiber, C., Sauerborn, M. & Kuramitsu, H. K. (1992) J. Bacteriol. 174, 6707–6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.von Eichel-Streiber, C., Boquet, P., Sauerborn, M. & Thelestam, M. (1996) Trends Microbiol. 4, 375–382. [DOI] [PubMed] [Google Scholar]
- 17.Krivan, H. C., Clark, G. F., Smith, D. F. & Wilkins, T. D. (1986) Infect. Immun. 53, 573–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tucker, K. D. & Wilkins, T. D. (1991) Infect. Immun. 59, 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pothoulakis, C., Gilbert, R. J., Cladaras, C., Castagliuolo, I., Semenza, G., Hitti, Y., Montcrief, J. S., Linevsky, J., Kelly, C. P., Nikulasson, S., et al. (1996) J. Clin. Invest. 98, 641–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pothoulakis, C., Galili, U., Castagliuolo, I., Kelly, C. P., Nikulasson, S., Dudeja, P. K., Brasitus, T. A. & LaMont, J. T. (1996) Gastroenterology 110, 1704–1712. [DOI] [PubMed] [Google Scholar]
- 21.Frisch, C., Gerhard, R., Aktories, K., Hofmann, F. & Just, I. (2003) Biochem. Biophys. Res. Commun. 300, 706–711. [DOI] [PubMed] [Google Scholar]
- 22.Just, I., Fritz, G., Aktories, K., Giry, M., Popoff, M. R., Boquet, P., Hegenbarth, S. & von Eichel-Streiber, C. (1994) J. Biol. Chem. 269, 10706–10712. [PubMed] [Google Scholar]
- 23.Just, I., Selzer, J., Wilm, M., von Eichel-Streiber, C., Mann, M. & Aktories, K. (1995) Nature 375, 500–503. [DOI] [PubMed] [Google Scholar]
- 24.Just, I., Wilm, M., Selzer, J., Rex, G., von Eichel-Streiber, C., Mann, M. & Aktories, K. (1995) J. Biol. Chem. 270, 13932–13936. [DOI] [PubMed] [Google Scholar]
- 25.Hofmann, F., Busch, C., Prepens, U., Just, I. & Aktories, K. (1997) J. Biol. Chem. 272, 11074–11078. [DOI] [PubMed] [Google Scholar]
- 26.Aktories, K. & Barbieri, J. T. (2005) Nat. Rev. Microbiol. 3, 397–410. [DOI] [PubMed] [Google Scholar]
- 27.von Eichel-Streiber, C. & Sauerborn, M. (1990) Gene 96, 107–113. [DOI] [PubMed] [Google Scholar]
- 28.Fernandez-Tornero, C., Lopez, R., Garcia, E., Gimenez-Gallego, G. & Romero, A. (2001) Nat. Struct. Biol. 8, 1020–1024. [DOI] [PubMed] [Google Scholar]
- 29.Mathieu, R., Lim, J., Simpson, P., Prasannan, S., Fairweather, N. & Matthews, S. (2003) J. Biomol. NMR 25, 83–84. [DOI] [PubMed] [Google Scholar]
- 30.Demarest, S. J., Salbato, J., Elia, M., Zhong, J., Morrow, T., Holland, T., Kline, K., Woodnutt, G., Kimmel, B. E. & Hansen, G. (2005) J. Mol. Biol. 346, 1197–1206. [DOI] [PubMed] [Google Scholar]
- 31.Weis, W. I. & Drickamer, K. (1996) Annu. Rev. Biochem. 65, 441–473. [DOI] [PubMed] [Google Scholar]
- 32.Rupnik, M., Braun, V., Soehn, F., Janc, M., Hofstetter, M., Laufenberg-Feldmann, R. & von Eichel-Streiber, C. (1997) FEMS Microbiol. Lett. 148, 197–202. [DOI] [PubMed] [Google Scholar]
- 33.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 34.Collaborative Computational Project (1994) Acta Crystallogr. D 50, 760–763.15299374 [Google Scholar]
- 35.Terwilliger, T. C. & Berendzen, J. (1999) Acta Crystallogr. D (Pt 4), 849–861. [DOI] [PMC free article] [PubMed]
- 36.Terwilliger, T. C. (2002) Acta Crystallogr. D 58, 1937–1940. [DOI] [PubMed] [Google Scholar]
- 37.McRee, D. E. (1999) J. Struct. Biol. 125, 156–165. [DOI] [PubMed] [Google Scholar]
- 38.Winn, M. D., Isupov, M. N. & Murshudov, G. N. (2001) Acta Crystallogr. D 57, 122–133. [DOI] [PubMed] [Google Scholar]
- 39.Morris, A. L., MacArthur, M. W., Hutchinson, E. G. & Thornton, J. M. (1992) Proteins 12, 345–364. [DOI] [PubMed] [Google Scholar]
- 40.Hooft, R. W., Vriend, G., Sander, C. & Abola, E. E. (1996) Nature 381, 272. [DOI] [PubMed] [Google Scholar]
- 41.Kobe, B. & Kajava, A. V. (2000) Trends Biochem. Sci. 25, 509–515. [DOI] [PubMed] [Google Scholar]
- 42.Fernandez-Tornero, C., Garcia, E., Lopez, R., Gimenez-Gallego, G. & Romero, A. (2002) J. Mol. Biol. 321, 163–173. [DOI] [PubMed] [Google Scholar]
- 43.Hermoso, J. A., Monterroso, B., Albert, A., Galan, B., Ahrazem, O., Garcia, P., Martinez-Ripoll, M., Garcia, J. L. & Menendez, M. (2003) Structure (Cambridge) 11, 1239–1249. [DOI] [PubMed] [Google Scholar]
- 44.Wren, B. W. (1991) Mol. Microbiol. 5, 797–803. [DOI] [PubMed] [Google Scholar]
- 45.Barth, H., Pfeifer, G., Hofmann, F., Maier, E., Benz, R. & Aktories, K. (2001) J. Biol. Chem. 276, 10670–10676. [DOI] [PubMed] [Google Scholar]
- 46.Rolfe, R. D. & Song, W. (1995) J. Med. Microbiol. 42, 10–19. [DOI] [PubMed] [Google Scholar]
- 47.Dallas, S. D. & Rolfe, R. D. (1998) J. Med. Microbiol. 47, 879–888. [DOI] [PubMed] [Google Scholar]
- 48.Fernandez-Tornero, C., Garcia, E., de Pascual-Teresa, B., Lopez, R., Gimenez-Gallego, G. & Romero, A. (2005) J. Biol. Chem. 280, 19948–19957. [DOI] [PubMed] [Google Scholar]
- 49.von Eichel-Streiber, C., Zec-Pirnat, I., Grabnar, M. & Rupnik, M. (1999) FEMS Microbiol. Lett. 178, 163–168. [DOI] [PubMed] [Google Scholar]
- 50.Kato, H., Kato, N., Katow, S., Maegawa, T., Nakamura, S. & Lyerly, D. M. (1999) FEMS Microbiol. Lett. 175, 197–203. [DOI] [PubMed] [Google Scholar]
- 51.Sambol, S. P., Merrigan, M. M., Lyerly, D., Gerding, D. N. & Johnson, S. (2000) Infect. Immun. 68, 5480–5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DeLano, W. L. (2002) pymol (DeLano Scientific, San Carlos, CA).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.