Abstract
The human La autoantigen can bind to nascent RNA transcripts and has also been postulated to act as an RNA polymerase III (pol III) transcription initiation and termination factor. Here, we show by chromatin immunoprecipitation (ChIP) that La is associated with pol III-transcribed genes in vivo. In contrast, the Ro autoantigen, which can also bind pol III transcripts, is not found at these genes. The putative pol III transcription factors NF1 and TFIIA are also not detected at class III genes. Binding of La remains when transcription is repressed at mitosis and does not correlate with the presence of polymerase at the gene. However, gene occupancy depends on the phosphorylation status of La, with the less prevalent, unphosphorylated form being found selectively on pol III templates.
Keywords: La, NF1, TFIIA, chromatin immunoprecipitation, RNA polymerase III transcription
RNA polymerase III (pol III) is responsible for the production of a number of short, untranslated RNAs that are essential for cellular growth; pol III transcription is therefore a major determinant of the biosynthetic capacity of cells (reviewed in refs. 1 and 2). The key to understanding how this influence is regulated is the identification of factors that can control expression. Pol III promoters can be divided into three types that utilize different transcription factors for their activity. Type 1 and 2 promoters, exemplified by the 5S rRNA and tRNA promoters, respectively, recruit TFIIIA and/or TFIIIC, through binding to sequence-specific elements within the gene (reviewed in refs. 3 and 4). TFIIIC in turn recruits a TFIIIB complex containing Brf1, TBP, and Bdp1, which is able to recruit the polymerase through protein–protein interactions. In contrast, type 3 promoters, such as that for U6 small nuclear RNA (snRNA), contain gene external elements that are recognized by the SNAPc complex and TFIIIB (4); however, the TFIIIB in this case differs from that used for type 1 and type 2 promoters in utilizing Brf2 instead of Brf1 (4, 5). In addition to these basal transcription factors, a number of other proteins have been found to influence pol III transcription in mammals. Some of these regulators have been shown to associate with pol III templates in vivo, including c-Myc (6), RB (7), β-actin (8), and the protein kinase CK2 (9, 10). However, there are a number of other proteins, including La, TFIIA and NF1, for which a role in pol III transcription has been suggested from experiments in vitro, but which has not been confirmed in vivo.
The human autoantigen La is a highly abundant protein found associated with newly synthesized pol III products (11, 12). La binds to nascent pol III transcripts through the UUU-OH at their 3′ ends, which corresponds to the pol III termination signal, and also has affinity for their 5′ ends (13–15). One of the functions of La is in the stabilization of these transcripts, which promotes their processing to the mature forms (16). However, La has also been reported to have effects on pol III transcription. Immunodepletion of La from cell extracts was found to reduce pol III output in vitro, which led to the suggestion that La could act as a transcriptional termination factor that mediates nascent transcript release (17, 18). Indeed, addition of human recombinant La to isolated pol III transcription complexes assembled from mammalian cell extracts led to increases in transcription, apparently due to enhanced pol III recycling and reinitiation (14, 19, 20). However, a number of other studies contradict these observations. Saccharomyces cerevisiae strains, which are null for the La homologue, are viable and have no reduction of pol III transcription (21, 22). Immunodepletion of La from Xenopus or HeLa extracts did not result in any discernible decrease in pol III transcription (23, 24). Furthermore, a highly purified human system that is fully active for U6 gene transcription did not contain any detectable La (9).
Two differentially phosphorylated pools of La have been identified in human cells, with the abundant phosphorylated form (pLa) being found exclusively in the nucleoplasm, associated with nascent pol III transcripts. Although the nonphosphorylated form (npLa) is also found in the nucleoplasm, it is much less abundant there than pLa, and it associates with only trace amounts of pol III transcripts (25). In contrast to pLa, a significant fraction of npLa is found in the nucleolus, associated with nucleolin at sites of rRNA biogenesis (26), and in the cytoplasm, associated with mRNAs that bear 5′ terminal oligopyrimidine tracts that encode ribosome subunits and general translation factors (25, 26). Human La is phosphorylated on residue S366 by CK2 (26, 27). Phosphorylation of S366 was found to inhibit the effect of La on pol III transcription in vitro (27), although it is the pLa form that associates with newly synthesized pol III transcripts and facilitates their maturation (25, 28, 29).
La was detected as part of a putative human pol III holoenzyme complex that was purified by using a FLAG-tagged subunit of the polymerase and shown to be competent for transcription (30). The DNA-binding factor NF1 was also identified as part of this complex and was reported to play a role in transcriptional termination in vitro (31). Another putative pol III factor is TFIIA, a component of the general pol II machinery, which was reported to stimulate transcription of a range of class III genes in a partially purified, reconstituted human system (32, 33). The functions of TFIIA, La, and NF1 in pol III transcription were postulated on the basis of experiments carried out in vitro and have yet to be tested in living mammalian cells. We have used ChIP analyses to examine whether these proteins are associated with class III genes in vivo, as would be expected of bona fide pol III transcription factors.
Materials and Methods
Cell Culture. HeLa cells were cultured and synchronized in mitosis, as described (34). Cell synchronization was confirmed by FACS analysis of DNA content.
Chromatin Immunoprecipitation (ChIP). ChIP assays were carried out as described (6). Antibodies used were as follows: C-21 against Oct-1, N-20 against NF1, FL-109 against TFIIA, and C-18 against TFIIB (Santa Cruz Biotechnology); 128 against Brf1 (35); 2663 against Bdp1, and 1900 against pol III subunit RPC155 (36); Go, La 1st, anti-pLa and anti-npLa against La (25); SS/A, which recognizes Ro60 (ref. 25 and data not shown), was obtained from the Centers for Disease Control (Atlanta).
Quantitative PCRs on input and bound DNA were carried out by using the following primers: 5′-GGT AGG AGG ATT GCT TGA G-3′ and 5′-CCT AAC TGA TTT AGA GTA GCC-3′ for hY4; and 5′-CGT TGT CTA CTT CTG TTA-3′and 5′-CGA TCA TGG CAT AGG CTC T-3′ for hY5. Cycling parameters were 95°C for 3 min, 25 cycles of 95°C for 30 s, 50°C for 30 s, 72°C for 30 s and 72°C for 5 min. For colony-stimulating factor 1 (CSF1), 5′-CAA AGG ATT TCC CTC CCT TC-3′ and 5′-CTT CCA AGC CTT CAG CAA AC-3′ and for 7SK 5′-CAA AGC GCC AGG TCA GCG GTC CCG GCT G-3′ and 5′-CGT TCT CCT ACA AAT GGA C-3′, with cycling conditions of 95°C for 3 min, 30 cycles of 95°C for 30 s, 55°C for 30 s, 72°C for 30 s followed by 5 min at 72°C. Other primers and amplification conditions have been described (6, 37, 38). Serial dilutions of chromatin were used to determine whether the PCRs were within a linear range.
Immunoprecipitation of Autoantigen-Associated RNAs. Immunoprecipitation of Ro and La and Northern analyses of associated U6 and hY RNAs were carried out as described (25), except that the washes were with RIPA buffer. Pretreatment with formaldehyde was the same as for ChIP assays.
Results
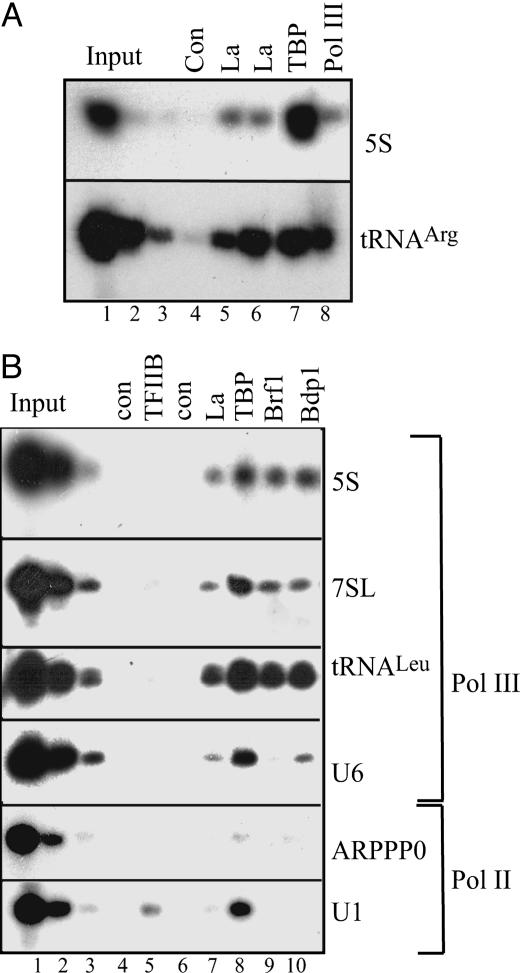
La Associates with Pol III-Transcribed Genes in Vivo. To test whether La is associated with pol III–transcribed genes in vivo, we used formaldehyde-crosslinked ChIP assays. We carried out immunoprecipitations with crosslinked chromatin from cycling HeLa cells using both an autoimmune serum (Go) that recognizes La and an affinity-purified antipeptide antibody (1st) against La, along with positive control antibodies against pol III and the TFIIIB subunit TBP; we then examined whether the protein is bound to a particular region of DNA by PCR analysis (Fig. 1A). A fraction of the input chromatin was used to test that the PCR was quantitative. Both antibodies detected the presence of La at tRNAArg and 5S rRNA genes in vivo.
Fig. 1.
La is present at pol III-transcribed genes in cycling HeLa cells. (A) ChIP assay using antibodies against La [anti-serum Go (lane 5) and the affinity-purified anti-peptide serum 1st (lane 6)], anti-TBP (lane 7), anti-pol III RPC155 subunit (lane 8), or no antibody control (lane 4). Immunoprecipitated material was amplified by using primers against 5S rRNA and tRNAArg. Lanes 1, 2, and 3, respectively, show product intensities obtained by using 2%, 0.4%, and 0.08% of input (Upper) or 10%, 2%, and 0.4% of input (Lower). (B) Crosslinked chromatin was immunoprecipitated by using antibodies against TFIIB (lane 5), La (lane 7), TBP (lane 8), Brf1 (lane 9), Bdp1 (lane 10), or no antibody (lanes 4 and 6). Immunoprecipitated material was analyzed by using primers against 5S rRNA, 7SL, tRNALeu, U6, ARPPP0, and U1 genes, as indicated. Lanes 1, 2, and 3 show product intensities obtained by using 10%, 2%, and 0.4% of input, respectively.
To test the generality of these effects, primers were used against the class III genes encoding 5S rRNA, 7SL RNA, tRNALeu, and U6 snRNA. These genes represent all three types of pol III promoter, with 5S rRNA having type I, tRNAs type II, and U6 a type III promoter arrangement (3, 4). To control for the specificity of binding to pol III-transcribed genes, primers were also used against a pol II-transcribed U1 snRNA gene and the gene encoding acidic ribosomal phosphoprotein (ARPP) P0, with the primers for the latter being located in the main body of the gene, away from the promoter (Fig. 1B). As positive controls, antibodies were used against the TFIIIB subunits TBP, Brf1, and Bdp1. The pol II general transcription factor TFIIB was used as a negative control. As expected, the Bdp1 subunit of TFIIIB was found at all of the class III genes examined, but not at U1 or ARPP P0. The Brf1 subunit of TFIIIB was also found at all of the class III genes except U6, which uses Brf2 instead of Brf1 (5, 38). TBP was associated with all of the promoters tested but was close to background within the coding region for ARPP P0. In contrast, the pol II transcription factor TFIIB gave a clear signal at the U1 gene, but not at any of the pol III-transcribed genes. These controls therefore all showed the behavior anticipated. When the autoimmune serum (Go) was used for La, DNA was amplified from all of the pol III-transcribed genes, whereas amplification of U1 and ARPP P0 DNA was close to background. For example, the signal obtained for U1 with the La antibody was <1% of the signal with the TBP antibody, whereas the La/TBP ratio was ≈12% for U6. This ratio cannot be used to infer the stoichiometry of interaction but does demonstrate the specificity of La binding. These data suggest that La associates selectively in vivo with a variety of class III genes, representing all three types of promoter arrangement.
The human Autoantigen Ro Is Not Found Associated with Pol III-Transcribed Genes. Like La, the human autoantigen Ro binds to pol III transcripts to form ribonucleoprotein particles. In the case of Ro, however, binding is specific for the pol III-transcribed Y RNAs and defective 5S rRNA molecules (39, 40). The function of Ro is unclear, although the observation that it binds specifically to nonfunctional 5S transcripts suggests it may have a role in 5S rRNA quality control (40). We tested whether Ro would ChIP with class III genes, to determine whether this is a feature shared by proteins that bind to pol III transcripts.
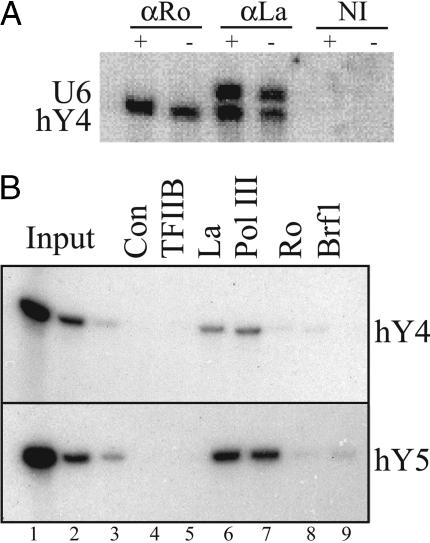
Northern analysis was used to confirm that the anti-Ro serum used for ChIP could immunoprecipitate Ro-associated hY RNAs. As expected, anti-La immunoprecipitated hY4 and U6 RNAs, whereas anti-Ro immunoprecipitated hY4, but not U6 RNA (Fig. 2A). This was the case whether or not the cells were pretreated with formaldehyde. hY5 RNA was also found in immunoprecipitations with anti-Ro and anti-La, but not with nonimmune serum (data not shown). In contrast to the transcripts, we were unable to detect any association of Ro with the hY4 or hY5 genes using this autoimmune anti-Ro serum (Fig. 2B). However, La was detected at these genes, along with pol III. Brf1 was not found, consistent with the prediction, based on sequence, that these genes have type III promoters (41, 42). We have also tested 5S rRNA and tRNALeu genes and again saw no association of Ro, although La is clearly detected (Fig. 5A). Control experiments (data not shown) confirmed that the anti-Ro antiserum can immunoprecipitate the Ro/hY4 RNA complex under the conditions used for ChIP. Taken together, these data show that the association of La with class III genes is a specific effect not seen using an autoimmune serum against another RNP component that binds to pol III transcripts.
Fig. 2.
ChIP assays detect La but not Ro at class III genes in vivo.(A) Anti-Ro can immunoprecipitate Ro-associated hY RNAs. Shown are Northern blots to detect U6 RNA and hY4 RNA after immunoprecipitation with anti-Ro anti-serum (lanes 1 and 2), anti-La antiserum (lanes 3 and 4), or nonimmune serum (lanes 5 and 6). Cells were either formaldehyde-treated (+ lanes) or mock-treated (–lanes) before harvesting. (B) ChIP assay using antibodies against TFIIB (lane 5), La (lane 6), pol III RPC155 subunit (lane 7), Ro (lane 8), Brf1 (lane 9), or no antibody control (lane 4). Primers specific to hY4 and hY5 were used to amplify the immunoprecipitated material.
Fig. 5.
ChIP assays detect La but not NF1 or TFIIA at class III genes in vivo.(A) ChIP assay using antibodies against Oct-1 (lane 5), La (lane 6), pol III RPC155 subunit (lane 7), Brf1 (lane 8), the 110-kDa subunit of TFIIIC (lane 9), Ro (lane 10), NF1 (lane 11), or no antibody control (lane 4). Immunoprecipitated DNA was analyzed by using primers against tRNALeu, 5S rRNA, and CSF1 genes, as indicated. (B) ChIP assay using antibodies against Brf1 (lane 4) or TFIIA (lane 5). Immunoprecipitated DNA was analyzed by using primers against tRNALeu or the cyclin D2 promoter region. (C) ChIP assays using antibodies against TFIIA (lane 5) or TBP (lane 6) or no antibody control (lane 4). Immunoprecipitated DNA was analyzed by using primers against the indicated genes.
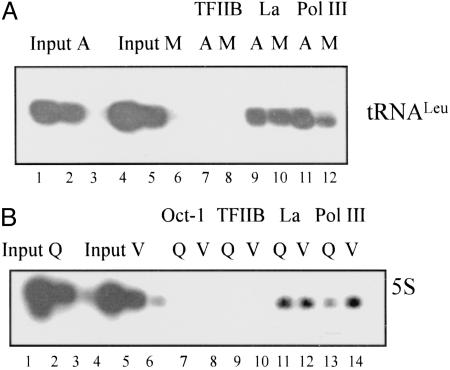
La Remains at Pol III-Transcribed Genes When Transcription Is Repressed. As La binds to nascent pol III transcripts, its presence at the promoter might be explained by its binding to the RNA, making it accessible to be crosslinked to the chromatin. The failure to crosslink Ro to hY genes argues against this possibility. Nevertheless, to test it further, ChIP analysis was carried out on HeLa cells arrested in mitosis. There is a general repression of transcription during mitosis, including that of pol III (43). We and others have shown that pol III and the TFIIIB subunit Bdp1 are selectively released from promoters in mitotic HeLa cells, although other components of the transcription complex (TBP, Brf1, Brf2, TFIIIC, and SNAPc) remain bound (36, 44). The levels of La at the tRNALeu gene change little in mitotic cells (M) compared with asynchronous (A), despite the occupancy of pol III decreasing (Fig. 3A). Therefore, the association of La with tRNA genes seems not to correlate with ongoing transcription, which suggests that its presence is not simply due to tethering to nascent transcripts. In addition, the fact that La occupancy is unchanged when pol III occupancy falls suggests that La is not associated with the tRNA gene purely through its presence as part of a holoenzyme complex.
Fig. 3.
La remains associated with class III genes when transcription is repressed. (A) ChIP assay showing occupancy levels of TFIIB, La, and pol III at tRNALeu genes in asynchronous (A) or mitotic (M) HeLa cells. Serial dilutions of input DNA were used to confirm that the PCRs are within the linear range and that A and M samples utilize equivalent amounts of input chromatin. (B) ChIP assay showing levels of Oct-1, TFIIB, La, and pol III at 5S rRNA genes in vehicle (V) or quercetin-treated (Q) HeLa cells. Cells were treated overnight with 50 μM quercetin or vehicle.
As a further test of the effect of decreasing pol III transcription on occupancy by La, ChIP analysis was carried out on HeLa cells treated with the CK2 inhibitor quercetin. CK2 is a positive regulator of pol III activity in HeLa cells (9, 10). Consistent with this role, the amount of pol III associated with 5S rRNA genes is decreased in quercetin-treated cells (Q) compared with those treated with vehicle (V) (Fig. 3B). The levels of La, however, remain unchanged. These data provide further evidence that detection of La does not correlate with the presence of the polymerase.
The Non-Phospho from of La Is Preferentially Associated with Pol III Promoters. CK2 phosphorylates La at residue S366 in its C-terminal domain, and this phosphorylation was reported to inhibit the transcriptional activity of La in vitro (26, 27). Phosphorylation of S366 also attenuates its binding to the 5′ termini of nascent RNAs (28). npLa binds more stably in vitro than pLa, and this interaction interferes with the 5′ processing of tRNAs. Nevertheless, specific antibodies raised against pLa and npLa have been used to demonstrate that pol III transcripts from cells preferentially immunoprecipitate with pLa compared with npLa (25).
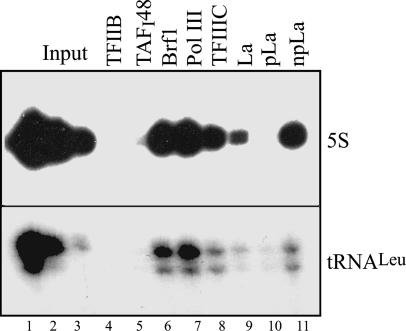
To test whether a particular form of La associates with class III genes, we utilized these phosphorylation-specific antibodies in a ChIP with cycling HeLa cells. The antibodies were used in quantities that immunoprecipitate comparable amounts of La protein. Despite the fact that npLa is much less abundant than pLa (25), it was found to be preferentially associated with the genes for tRNALeu and 5S rRNA, whereas little or no pLa was detected (Fig. 4).
Fig. 4.
The nonphosphorylated pLa is preferentially associated with pol III-transcribed genes. ChIP analysis was carried out using antibodies against TFIIB (lane 4), TAFI48 (lane 5), Brf1 (lane 6), Pol III RPC155 subunit (lane 7), TFIIIC110 (lane 8), total La (GoLa, lane 9), phospho-La (pLa, lane 10), and non-phospho-La (npLa, lane 11). Immunoprecipitated DNA was analyzed using primers against the indicated genes.
NF1 and TFIIA Are Not Detected at Pol III-Transcribed Genes in Vivo. La has been detected as a component of a pol III holoenzyme complex along with TFIIIB and TFIIIC (30). Also purified as part of this complex was an activity that binds the termination region of a VA template and that was identified as NF1 (31). On the basis of further experiments carried out in vitro, it was suggested that NF1 can function as a general pol III transcription factor, influencing expression of a range of class III genes (31). The study, however, ignored an earlier paper from the same laboratory that had concluded that NF1 has no effect on VA gene transcription (45). We used ChIP assay to test whether NF1 is present at class III genes in vivo. Antibodies directed against pol III, Brf1, TFIIIC, and La were all able to immunoprecipitate the 5S rRNA and tRNALeu genes. However, these genes were not amplified in the immunoprecipitates obtained with NF1 antibodies (Fig. 5A). The human CSF1 promoter is regulated by NF1 and has been shown by ChIP to bind NF1 in vivo (46). We therefore used this promoter as a positive control. Consistent with the previous study, NF1 was found associated with the CSF1 promoter, although La was not detected. These data suggest that NF1 is present at the CSF1 promoter but not at cellular class III genes in growing HeLa cells.
The pol II general transcription factor TFIIA has also been postulated to function as a regulator of mammalian pol III transcription (32). Addition of purified or recombinant TFIIA to a partially purified system was reported to stimulate pol III transcription from U6 snRNA, 5S rRNA, VAI, and tRNA gene templates (32, 33). Recent experiments with extensively purified factors suggested that TFIIA is not required for U6 gene transcription, but did not rule out the possibility of an auxiliary role (9). We used ChIP analysis to test whether TFIIA can be found at pol III-transcribed genes in living cells. Immunoprecipitates using antibody against the Brf1 subunit of TFIIIB contained DNA corresponding to the tRNALeu gene but not the pol II-transcribed cyclin D2 promoter (Fig. 5B, lane 4). In contrast, antibody against TFIIA immunoprecipitated the cyclin D2 promoter, but not the tRNALeu gene (Fig. 5B, lane 5). We also examined various other class III genes and could demonstrate the presence of TBP (Fig. 5C, lane 5), but not TFIIA (Fig. 5C, lane 4) at the pol III-transcribed 5S rRNA, tRNATyr, U6, and 7SK genes. In contrast, both TBP and TFIIA were present at the pol II-transcribed U1 gene. These data argue against a direct role for TFIIA in pol III transcription in vivo, at least under the conditions examined.
Discussion
ChIP experiments with three alternative antibodies indicate that La is present at class III genes in living HeLa cells. Furthermore, it is the npLa but not the pLa form that is detected. This specificity is striking, given that pLa is in considerable excess over npLa in the nucleus (25). The specificity of La binding to these genes is further demonstrated by the fact that we do not see La at the pol II templates we have looked at. Moreover, Ro, TFIIA, and NF1 were not detected at class III genes, when tested in parallel. This failure to detect TFIIA and NF1 contrasts with reports that these factors can stimulate pol III transcription from purified systems in vitro (31–33). The discrepancy should not reflect a difference in cell type because the previous studies used HeLa cells as their source of transcription factors, but variations in cell properties or culture conditions cannot be excluded. There is also the possibility that these proteins are inaccessible to our antibodies when assembled in a pol III transcription complex. We confirmed that the antibodies are capable of recognizing TFIIA and NF1 when bound to chromatin in vivo, using class II genes as positive controls, but these factors might conceivably be masked in the context of different transcription complexes. Such uncertainties are inherent to ChIP experiments, but the assay nevertheless provides a valuable means of examining models in vivo. Another limitation of these assays is that their resolution is limited to ≈500 bp, the size of sheared chromatin fragments, so we cannot determine where a protein binds in relation to short class III genes.
The stoichiometry of La binding cannot be ascertained from our data. The intensity of a ChIP signal reflects the quality of antibody and accessibility of its epitopes, as well as the amount of target protein that is actually present in vivo. This is clearly illustrated in Fig. 1B, where TBP consistently gives a stronger signal than Bdp1 and Brf1, although these three polypeptides are believed to function in equimolar ratios in the TFIIIB complex. Although the La signal is generally of comparable intensity to that of pol III in our experiments, one cannot infer that they occupy class III genes with similar stoichiometries. Indeed, our primers for 5S, tRNA, and 7SL will detect some pseudogenes as well as active genes, and it is possible that the former contribute to the La signal in these cases. However, we consider it unlikely that pseudogenes are solely responsible for the La signal detected at these gene families, and the primers for U6, hY4, and hY5 recognize specific flanking sequences of individual genes with active promoters.
Our data suggest that the presence of La at class III genes is not due to tethering by nascent transcripts. Ro interacts stably with hY RNAs but was not detected at the hY genes in our ChIP assays. Occupancy of La does not decrease when cells are in mitosis, despite the fact that transcription ceases and pol III dissociates from promoters (36, 44). This observation, together with the CK2 inhibitor data, where again La remains constant as pol III occupancy decreases, suggests that La is retained at class III genes independently of the polymerase and therefore, presumably, of the nascent transcript.
That npLa associates with pol III templates in cells is consistent with the report that phosphorylation of La on S366 by CK2 inhibits its transcriptional activity in vitro (27). CK2 phosphorylates human La in vivo (26), but treatment of cells with quercetin to reduce CK2 activity did not alter the occupancy of La at these genes. This finding may indicate that there is sufficient npLa to saturate binding sites at class III genes even when CK2 is fully active. Although the ratio of npLa to pLa is expected to increase after quercetin treatment, this increase in itself cannot explain its retention at class III genes. Because CK2 associates stably with the pol III machinery in HeLa cells (9, 10), it may be well positioned to phosphorylate La at some stage in the transcription cycle, perhaps triggering its release.
It is unclear what feature of class III genes is recognized selectively by La. A shared DNA motif is unlikely to be responsible because there is little sequence conservation between the various types of pol III template, apart from the oligo(dT) termination sequence, which also occurs frequently in class II genes. The obvious alternative would involve specific protein/protein interactions between La and the pol III transcription machinery. The fact that Bdp1 and pol III dissociate during mitosis, when La remains, argues against either of these anchoring La, although a role in recruitment remains plausible. SNAPc and TBP are not good candidates because they are also used by pol II templates, where La is not found. TFIIIA, TFIIIC, Brf1, and Brf2 are each used only by some pol III promoter types, whereas La seems to be present at all. However, interactions with a combination of these factors might suffice to cover the entire class; for example, recognition of epitopes shared between Brf1 and Brf2 could potentially provide a docking site for La at every class III gene. Such possibilities have yet to be addressed.
Although recombinant human La was found to influence pol III transcription under certain conditions in vitro, other work in several systems found no effect of La on transcription (23, 24). Nevertheless, our results show that La can be found at all three types of pol III template in living cells. A possible explanation for the conflicting data is that La might contribute to transcription only under specific circumstances that have yet to be defined. Alternatively, its presence at class III genes might reflect some role unrelated to transcription. Although the functional significance of these observations remains to be determined, the consistent presence of La is striking and seems unlikely to be entirely fortuitous.
Acknowledgments
We are grateful to an anonymous reviewer for helpful comments. We also thank the Centers for Disease Control for reference serum and K. Reinisch (Yale University School of Medicine, New Haven, CT) for purified Ro60. This research was supported by the Biotechnology and Biological Sciences Research Council, the Wellcome Trust, and the Medical Research Council (U.K.), as well as the Intramural Research Program of the National Institute of Child Health and Development, National Institutes of Health. R.J.M. served as a Commissioned Officer in the U.S. Public Health Service.
Author contributions: J.A.F. and R.J.W. designed research; J.A.F., T.K., N.S.K., and R.V.I. performed research; R.J.M. contributed new reagents/analytic tools; J.A.F. and R.J.W. analyzed data; and J.A.F. and R.J.W. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: snRNA, small nuclear RNA; pol III, RNA polymerase III; pLa, phosphorylated La; npLa, nonphosphorylated La; ChIP, chromatin immunoprecipitation; ARPP, acidic ribosomal phosphoprotein; CSF1, colony-stimulating factor 1.
References
- 1.White, R. J. (2004) Oncogene 23, 3208–3216. [DOI] [PubMed] [Google Scholar]
- 2.White, R. J. (2005) Nat. Rev. Mol. Cell Biol. 6, 69–78. [DOI] [PubMed] [Google Scholar]
- 3.Geiduschek, E. P. & Kassavetis, G. A. (2001) J. Mol. Biol. 310, 1–26. [DOI] [PubMed] [Google Scholar]
- 4.Schramm, L. & Hernandez, N. (2002) Genes Dev. 16, 2593–2620. [DOI] [PubMed] [Google Scholar]
- 5.Teichmann, M., Wang, Z. X. & Roeder, R. G. (2000) Proc. Natl. Acad. Sci. USA 97, 14200–14205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gomez-Roman, N., Grandori, C., Eisenman, R. N. & White, R. J. (2003) Nature 421, 290–294. [DOI] [PubMed] [Google Scholar]
- 7.Hirsch, H. A., Jawdekar, G. W., Lee, K.-A., Gu, L. & Henry, R. W. (2004) Mol. Cell. Biol. 24, 5989–5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu, P., Wu, S. & Hernandez, N. (2004) Genes Dev. 18, 3010–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu, P., Wu, S. & Hernandez, N. (2003) Mol. Cell 12, 699–709. [DOI] [PubMed] [Google Scholar]
- 10.Johnston, I. M., Allison, S. J., Morton, J. P., Schramm, L., Scott, P. H. & White, R. J. (2002) Mol. Cell. Biol. 22, 3757–3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lerner, M. R., Boyle, J. A., Hardin, J. A. & Steitz, J. A. (1981) Science 211, 400–402. [DOI] [PubMed] [Google Scholar]
- 12.Rinke, J. & Steitz, J. A. (1982) Cell 29, 149–159. [DOI] [PubMed] [Google Scholar]
- 13.Stefano, J. E. (1984) Cell 36, 145–154. [DOI] [PubMed] [Google Scholar]
- 14.Maraia, R. J. & Intine, R. V. A. (2001) Mol. Cell. Biol. 21, 367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bogenhagen, D. F. & Brown, D. D. (1981) Cell 24, 261–270. [DOI] [PubMed] [Google Scholar]
- 16.Wolin, S. L. & Cedervall, T. (2002) Annu. Rev. Biochem. 71, 375–403. [DOI] [PubMed] [Google Scholar]
- 17.Gottlieb, E. L. & Steitz, J. A. (1989) EMBO J. 8, 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gottlieb, E. L. & Steitz, J. A. (1989) EMBO J. 8, 841–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodier, J. L., Fan, H. & Maraia, R. J. (1997) Mol. Cell. Biol. 17, 5823–5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maraia, R. J. (1996) Proc. Natl. Acad. Sci. USA 93, 3383–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pannone, B. K., Xue, D. H. & Wolin, S. L. (1998) EMBO J. 17, 7442–7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoo, C. J. & Wolin, S. L. (1997) Cell 89, 393–402. [DOI] [PubMed] [Google Scholar]
- 23.Lin-Marq, N. & Clarkson, S. G. (1998) EMBO J. 17, 2033–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weser, S., Bachmann, M., Seifart, K. H. & Meibner, W. (2000) Nucleic Acids Res. 28, 3935–3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Intine, R. V., Tenenbaum, S. A., Sakulich, A. L., Keene, J. D. & Maraia, R. J. (2003) Mol. Cell 12, 1301–1307. [DOI] [PubMed] [Google Scholar]
- 26.Schwartz, E. I., Intine, R. V. & Maraia, R. J. (2004) Mol. Cell. Biol. 24, 9580–9591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan, H., Sakulich, A. L., Goodier, J. L., Zhang, X. L., Qin, J. & Maraia, R. J. (1997) Cell 88, 707–715. [DOI] [PubMed] [Google Scholar]
- 28.Fan, H., Goodier, J. L., Chamberlain, J. R., Engelke, D. R. & Maraia, R. J. (1998) Mol. Cell. Biol. 18, 3201–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Intine, R. V. A., Sakulich, A. L., Koduru, S. B., Huang, Y., Pierstorff, E., Goodier, J. L., Phan, L. & Maraia, R. J. (2000) Mol. Cell 6, 339–348. [DOI] [PubMed] [Google Scholar]
- 30.Wang, Z. X., Luo, T. & Roeder, R. G. (1997) Genes Dev. 11, 2371–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, Z. X., Bai, L., Hsieh, Y. J. & Roeder, R. G. (2000) EMBO J. 19, 6823–6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waldschmidt, R. & Seifart, K. H. (1992) J. Biol. Chem. 267, 16359–16364. [PubMed] [Google Scholar]
- 33.Meissner, W., Holland, R., Waldschmidt, R. & Seifart, K. H. (1993) Nucleic Acids Res. 21, 1013–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.White, R. J., Gottlieb, T. M., Downes, C. S. & Jackson, S. P. (1995) Mol. Cell. Biol. 15, 1983–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sutcliffe, J. E., Brown, T. R. P., Allison, S. J., Scott, P. H. & White, R. J. (2000) Mol. Cell. Biol. 20, 9192–9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fairley, J. A., Scott, P. H. & White, R. J. (2003) EMBO J. 22, 5841–5850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winter, A. G., Sourvinos, G., Allison, S. J., Tosh, K., Scott, P. H., Spandidos, D. A. & White, R. J. (2000) Proc. Natl. Acad. Sci. USA 97, 12619–12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schramm, L., Pendergrast, P. S., Sun, Y. L. & Hernandez, N. (2000) Genes Dev. 14, 2650–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hendrick, J. P., Wolin, S. L., Rinke, J., Lerner, M. R. & Steitz, J. A. (1981) Mol. Cell. Biol. 1, 1138–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Brien, C. A. & Wolin, S. L. (1994) Genes Dev. 8, 2891–2903. [DOI] [PubMed] [Google Scholar]
- 41.Maraia, R., Sakulich, A. L., Brinkmann, E. & Green, E. D. (1996) Nucleic Acids Res. 24, 3552–3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maraia, R. J., Sasakitozawa, N., Driscoll, C. T., Green, E. D. & Darlington, G. J. (1994) Nucleic Acids Res. 22, 3045–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gottesfeld, J. M. & Forbes, D. J. (1997) Trends Biochem. Sci. 22, 197–202. [DOI] [PubMed] [Google Scholar]
- 44.Hu, P., Samudre, K., Wu, S., Sun, Y. L. & Hernandez, N. (2004) Mol. Cell 16, 81–92. [DOI] [PubMed] [Google Scholar]
- 45.Van Dyke, M. W. & Roeder, R. G. (1987) Mol. Cell. Biol. 7, 1021–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu, R., Liu, H., Chen, X., Kirby, M., Brown, P. O. & Zhao, K. J. (2001) Cell 106, 309–318. [DOI] [PubMed] [Google Scholar]