Abstract
Highly ordered arrays of stretched DNA molecules were generated over the millimeter scale by using a modified molecular combing method and soft lithography. Topological micropatterning on polydimethyl siloxane stamps was used to mediate the dynamic assembly of DNA molecules into arranged nonostrand arrays. These arrays consisted of either short nanostrands of several micrometers with fixed length and orientation or long nanostrands up to several hundred micrometers in length. The nanostrand arrays were transferred onto flat solid surfaces by contact printing, allowing for the creation of more complex patterns. This technique has potential applications for the construction of next-generation DNA chips and functional circuits of DNA-based 1D nanostructures.
Keywords: molecular combing, soft lithography
Patterning DNA molecules at the micrometer scale forms the basis of DNA chips, a widely used technology for genetic analysis and diagnosis (1, 2). At the molecular level, single DNA molecules have been stretched for physical mapping of genes and molecular diagnosis of diseases (3). If stretched DNA molecules can be patterned into a well defined array, large-scale and highly automated analysis may be realized. On the other hand, 1D nanostructures are of great interest for the construction of future devices (4–6). With its high aspect ratio, unique base-pairing ability, designable base sequence, and availability of various techniques for functionalization, DNA is a very attractive material for preparing 1D nanostructures for electronic, magnetic, photonic, and chemical sensing applications (7–13). The ability to position a large number of 1D nanostructures with well defined linear arrangements is a prerequisite for integrating them into functional devices. The lack of this ability is currently hindering the realization of functional devices built on DNA-based 1D nanostructures.
Molecular combing is a technique for stretching, aligning, and immobilizing coiled DNA molecules in a solution onto a flat surface through a dewetting process (3). By creating a pattern of surface structures or properties, combing can be further controlled (14–18). A number of studies in molecular combing have been reported in the literature, but none are able to demonstrate well defined arrays. For example, a single nanofiber has been combed and placed between two electrodes in a nanojunction (14). But this method cannot control the size of nanofibers and has not demonstrated any ability to pattern nanofiber arrays covering a large area. By flowing DNA solution through microchannels, stretched DNA molecules confined in the microchannels were obtained. Their orientation and curvature were directed by controlling the geometry of the air–water interface (15). However, the DNA molecules were randomly distributed in the microchannels. In a separate study, polystyrene lines were lithographically patterned on a substrate for end-specific binding of DNA molecules. Combing of the DNA on this substrate created lines of stretched single DNA molecules (16). A line-patterned polydimethyl siloxane (PDMS) stamp, which contained a positively charged surface as a substrate for combing, also produced an array of stretched DNA molecules (17). The stretched DNA molecules could be transferred onto other solid surfaces by contact printing, allowing for the generation of more complex patterns (17, 18). However, the position and length of the DNA molecules in the arrays prepared by these two methods could not be tightly controlled. In another study, an array of stretched DNA molecules was created by combing the DNA tethered to positively charged microdots, but the length and distribution of DNA molecules in the array were not uniform (19). In this paper, we describe a modified molecular combing method combined with soft lithography (20). This method is capable of micropatterning stretched and aligned DNA molecules into highly ordered nanostrand arrays of different lengths and transferring them onto other surfaces.
Materials and Methods
Materials. To prepare the DNA solution, λ-DNA (New England Biolabs, 48,502 bp, 500 μg/ml in 10 mM Tris·HCl/1 mM EDTA, pH 8) was diluted in a buffer solution (10 mM Tris·HCl/2 mM EDTA/10 mM NaCl, pH 8) to 100 μg/ml and labeled with fluorescent dye (YOYO-1, Invitrogen) at a dye/base-pair ratio of 1:5. Incubation was conducted in the dark at room temperature for a minimum of 2 h. The solution was then further diluted to 20 or 50 μg/ml in a 1 wt% glycerin solution in the same buffer. The solutions were used for preparing nanostrand samples characterized by fluorescence microscopy and atomic force microscopy (AFM). A different DNA solution without fluorescent dye was also prepared by diluting the original 500 μg/ml DNA solution into 20 μg/ml in the buffer solution. This solution was used for the preparation of samples for scanning electron microscopy (SEM). DNA solutions with a concentration of 20 μg/ml were used throughout this study unless otherwise noted. PDMS (Silastic T2, Dow-Corning) stamps with two types of microfeatures were prepared by soft lithography (20). One consisted of microwells 5 μm in diameter, 10 μm in center-to-center distance, and 4 μm in depth. Stamps with this microfeature were used throughout this study unless otherwise noted. The other type of microfeature was composed of microwells 5 μm in diameter, 8 μm in center-to-center distance, and 1.9 μm in depth. A flat PDMS stamp was also used as a control. The stamps were cut into 1 × 1-cm pieces.
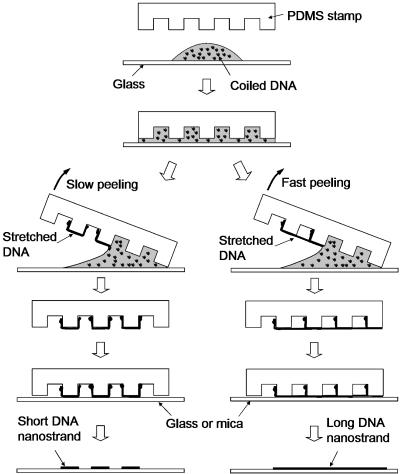
Generating Nanostrand Array. Fig. 1 is a schematic representation of the procedure for generating and transferring the DNA nanostrand arrays. First, 10 μl of DNA solution was dropped on a glass coverslip (Corning brand cover glass, Fisher Scientific). A PDMS stamp was then placed on the solution. The DNA solution instantly spread over the entire surface of the stamp. A pressure of ≈45 kPa (force was measured by a balance placed under the glass coverslip) was applied on the stamp manually for 5 sec, followed by peeling up the stamp from one end with the other end remaining in contact with the glass surface. The peeling was controlled manually at either low (rotating 90° in ≈1 sec) or high (rotating 90° in <0.1 sec) speed, which was measured from video images of the peeling process recorded by a JVC (Wayne, NJ) DVL9800 digital video camera. After peeling, the stamp surface was dry under the naked eye, and the DNA solution remained on the glass coverslip. To transfer the formed nanostrands onto a solid surface, the stamp with nanostrands was brought in contact with the surface for 1 min without external pressure and then peeled away. To make more complex patterns, a second contact printing was performed on the already printed array.
Fig. 1.
Schematic of generating and transferring DNA nanostrand arrays.
Fluorescence Imaging. The stamp with nanostrands was placed on a glass coverslip with the nanostrands in contact with the coverslip. The coverslip was mounted and imaged under an inverted Nikon TMS epifluorescence microscope equipped with a ×100/1.3-numerical aperture oil immersion objective lens.
SEM Imaging. The stamp with DNA nanostrands was sputtercoated with 20-nm-thick Pt/Pd and imaged at 1 kV accelerating voltage in a Hitachi (Tokyo) S-4300 SEM.
AFM Imaging. AFM images of DNA nanostrands on freshly cleaved mica were obtained on a MultiMode microscope with a Nanoscope IIIa controller (Digital Instruments, Santa Barbara, CA) in tapping mode operated in air.
Results
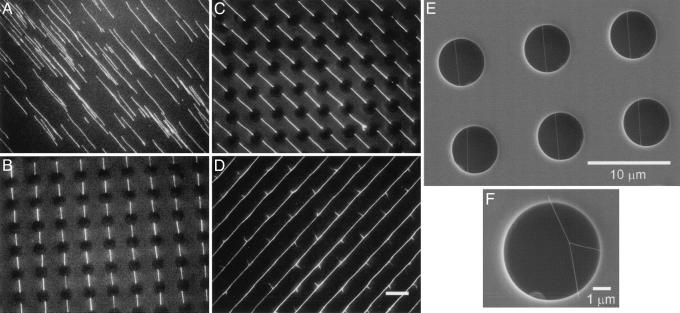
Fig. 2A shows stretched DNA molecules on a flat PDMS stamp generated at a low peeling speed. The molecules are well aligned but randomly distributed with different lengths. In contrast, highly ordered arrays of DNA strands were obtained on the PDMS stamps with microwells as shown in Fig. 2 B–D. Depending on the peeling speed, either short (Fig. 2 B and C) or long (Fig. 2D) strands can be obtained at low and high speed, respectively and repeatedly. Fig. 2 B and C show two different arrays of short strands connecting two vertically and diagonally adjacent microwells, respectively. In each picture, the bright strands of the same length and orientation are precisely positioned between adjacent microwells that are vaguely visible. Such arrays covering an area of several millimeters were obtained repeatedly with scattered defects such as misoriented DNA strands and strands with either one or both ends not in the microwells. An array of long and parallel DNA strands on the stamp is shown in Fig. 2D. The strands were up to several hundred micrometers long and cover an area up to 1 × 1 mm2. Two compounded fluorescence images showing long nanostrands over a large area are shown in supporting information, which is published on the PNAS web site. Many long strands have multiple “thorn”-like structures extending aside from the “stems” as shown in Fig. 2D. SEM images (Fig. 2 E and F) further display that the long DNA nanostrands and the “thorns” are suspended over the microwells.
Fig. 2.
Fluorescence micrographs of (A) DNA molecules combed on a flat PDMS stamp, (B) vertically and (C) diagonally aligned DNA strands on the PDMS stamps with microwells, and (D) long DNA strands on the PDMS stamp with microwells. SEM images of (E) an array of long DNA strands and (F) a “thorn”-like structure over microwells of a PDMS stamp. The portions of DNA strands on the top surface of the stamp are not visible because of their small thickness. [Scale bar, 10 μm (A–D).]
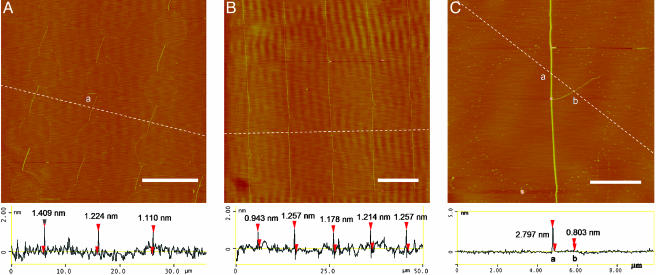
Fig. 3 shows AFM images of the DNA nanostrands on a mica surface. The height of the short nanostrands on the mica is fairly uniform along the length of individual strands. For example, the heights of the nanostrand marked as “a” in Fig. 3A at its upper, center, and lower positions are 0.97, 1.22, and 1.27 nm, respectively. However, the heights among different nanostrands vary considerably. Measurements on 10 nanostrands at their centers in Fig. 3A gave an average height of 1.48 nm (±0.73 nm) and a range from 0.88 to 3.31 nm. The heights of the stretched single DNA molecules that were printed on mica from a flat PDMS stamp are typically <0.4 nm (9). Thus the nanostrands shown in this work are likely composed of a bundle of stretched DNA molecules with different numbers of strands. Fig. 3B shows similar AFM measurements of long DNA nanostrands. Measurements on 33 nanostrands gave an average height of 1.19 nm (±0.52 nm) and a range from 0.58 to 2.80 nm. A “thorn” extending aside from a long DNA nanostrand is shown in Fig. 3C.
Fig. 3.
Tapping-mode AFM height images and section profiles of short DNA nanostrands (A), long DNA nanostrands (B), and a long DNA nanostrand with a “thorn” on the mica surface (C). [Scale bars, 10 μm(A and B) and 2 μm(C).]
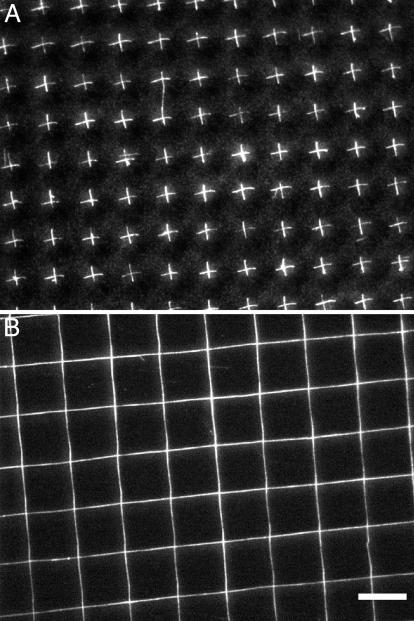
Fig. 4A demonstrates an array of discrete nanostrand “crosses.” These were prepared by printing the vertically aligned short DNA nanostrands on a glass slide twice in a perpendicular fashion by using the stamp with microwells 5 μm in diameter, 8 μm in center-to-center distance, and 1.9 μm in depth. Fig. 4B shows a crossbar structure prepared by double printing the long nanostrands produced by using a 50 μg/ml DNA solution.
Fig. 4.
Fluorescence micrographs of arrays of short (A) and long (B) DNA nanostrands prepared by double printing. (Scale bar, 10 μm.)
Discussion
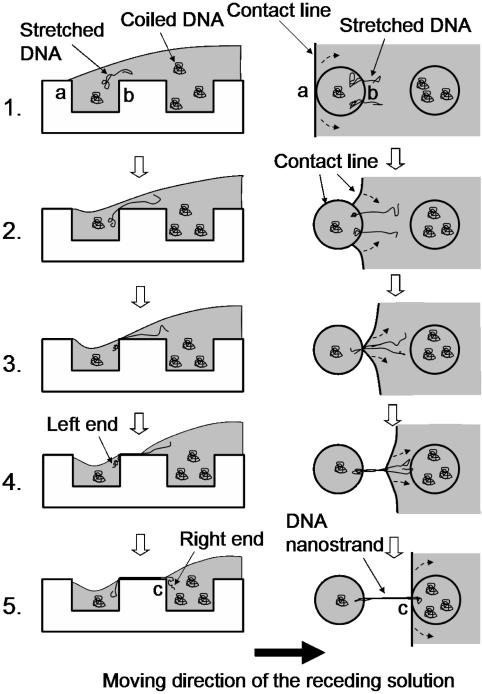
To generate a highly ordered DNA nanostrand array covering a large area, it is important to preload DNA molecules in every microwell by pressing the stamp against the glass coverslip before peeling it off. Otherwise, significantly fewer DNA molecules remain on the stamp. Separating the stamp and the glass coverslip from one end leads to the receding of the DNA solution toward the opposite end due to the dewetting of the aqueous solution from hydrophobic surface of the PDMS stamp. We hypothesize that formation of the short DNA nanostrands follows a molecular combing-based process as shown in Fig. 5. The microwells not only act as starting and ending points of the combing of individual nanostrands but also facilitate assembly of originally separated DNA molecules into bundled strands. The formation of long nanostrands is believed to follow a similar process, but the combing of a nanostrand on the top surface of the stamp does not stop due to the high-speed peeling when the contact line reaches the next microwell. Combing of the nanostrand thus continues over the microwell, where formation of another nanostrand is initiated. As a result, the two consecutive nanostrands bundle together. This process leads to a DNA nanostrand much longer than the contour length of λ-DNA (16.3 μm) (21). The high shear force generated near the solid–liquid interface during rapid peeling-off may “fracture” the surface of the suspended nanostrands causing a split of DNA bundles forming the “thorns.” By optimizing the stamp microfeature, DNA concentration, and peeling process, it is possible to generate continuous DNA nanostrands over a larger area. The orientation of the DNA strands is the same as the moving direction of the receding meniscus, which is generally also the peeling direction of the stamp. The patterns in Fig. 2 B and C were created by manually controlling the peeling direction to drive the meniscus to move vertically and diagonally relative to the microfeature lattice of the stamp, respectively.
Fig. 5.
Schematic of the hypothetical process for the formation of the short nanostrands. The side and corresponding top views are shown on Left and Right, respectively. The dashed arrows (Right) indicate the moving directions of the contact line around the microwells. (1) The moving contact line of the receding DNA solution reaches the left edge (marked by “a”) of a microwell on the hydrophobic PDMS stamp. The coiled DNA molecules close to the right edge (marked by “b”) of the microwell are stretched due to the velocity gradient of the liquid flow at the edge. (2) The microwell acts as a wetting defect to temporarily pin the moving contact line. Consequently, the liquid film above the microwell thins, and the contact line moves around the edge of the circular microwell. (3) With the thinning of the liquid film, the liquid–air interface reaches the lip of edge “b,” where the liquid film breaks (22). At the same time, the moving contact line around the microwell drives the originally separated DNA molecules together. (4) After the liquid film breaks at the lip of edge “b,” the left ends of the DNA molecules are trapped in the microwell and thereby anchored. A small amount of water is also left in the microwells, and it evaporates rapidly due to the small depth of the wells. The released contact line moves on the top surface of the stamp in the receding direction, combing the anchored DNA molecules into a bundled nanostrand. (5) At a low peeling-off speed, the combing process of the nanostrand stops when the contact line reaches the left edge (marked by “c”) of the next microwell and is pinned again. The right end of the nanostrand thus stays in the microwell, from which another nanostrand starts to form following the same process.
The concentration of the DNA solution plays an important role in the formation of nanostrand arrays. A DNA solution of 0.01-μg/ml concentration produces sparsely scattered short DNA nanostrands at both low and high peeling speeds (data not shown). Increasing the concentration leads to more coverage of the nanostrand array. At 20 μg/ml, a complete array of nanostrands covering an area of several millimeters wide can be generated repeatedly. Moreover, high-speed peeling using a 20 μg/ml DNA solution leads to the formation of long strands covering a significant fraction of the whole stamping area and a higher DNA concentration correlates with a higher area fraction of the long strands. The DNA nanostrands show different brightness in the fluorescence micrographs, implying that they are bundles composed of different numbers of strands rather than single stretched DNA molecules. Microwell size may affect the number of molecules in the nanostrands. Smaller microwells would anchor fewer DNA molecules. By tailoring DNA concentration and microwell size, it is possible to create single DNA nanostrand arrays.
The DNA nanostrands can be transferred onto other solid surfaces by contact printing (17, 18, 20). For short nanostrands, we found that the stretched DNA molecules broke at the edges of the microwells, resulting in portions in the microwells being untransferred. As a result, we obtained DNA nanostrands with the monodispersed length determined by the geometry of the microfeature on the stamp. For long nanostrands, the segments suspended over the microwells were also transferred as those on the top surface of the stamp. A small amount of glycerin (concentration <1 wt %) was added in the DNA solution to enhance transfer efficiency. Without glycerin, many nanostrands did not transfer or broke during transfer. We hypothesize that the viscous glycerin served as a “glue” to stick the nanostrands onto the solid surface. Multiple printing of the DNA nanostrand arrays onto a solid surface allows for the formation of more complex patterns. Various functional nanocircuits may therefore be created by designing the pattern of microwells on the stamp; by performing multiple contact printings with alignment; and by functionalizing the DNA with electronically, magnetically, photonically, or chemically active materials.
Conclusion
A simple and robust method based on molecular combing and soft lithography has been developed to produce highly ordered arrays of DNA nanostrands that have well defined length and orientation and are precisely positioned over a millimeter-scale area. The array of DNA nanostrands can be transferred onto other flat surfaces, allowing for the generation of more complex patterns by multiple printing. This technique may be used for the construction of next-generation DNA chips and functional circuits of DNA-based 1D nanostructures. Further extension of the technique may lead to micropatterning of other nanofibers and polymers, e.g., conducting polymers, at the level of a few or even single molecules with a variety of potential applications such as molecular electronics.
Supplementary Material
Acknowledgments
We thank Nick Ferrell at Ohio State University for technical support. This work was supported by National Science Foundation Grant DMI-0425626.
Author contributions: J.G. and L.J.L. designed research; J.G. performed research; J.G. and L.J.L. analyzed data; and J.G. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PDMS, polydimethyl siloxane; AFM, atomic force microscopy/microscope; SEM, scanning electron microscopy/microscope.
References
- 1.Gerhold, D., Rushmore, T. & Caskey, C. T. (1999) Trends Biochem. Sci. 24, 168–173. [DOI] [PubMed] [Google Scholar]
- 2.Cuzin, M. (2001) Transfus. Clin. Biol. 8, 291–296. [DOI] [PubMed] [Google Scholar]
- 3.Michalet, X., Ekong, R., Fougerousse, F., Rousseaux, S., Schurra, C., Hornigold, N., van Slegtenhorst, M., Wolfe, J., Povey, S., Beckmann, J. S., et al. (1997) Science 277, 1518–1523. [DOI] [PubMed] [Google Scholar]
- 4.Patolsky, F. & Lieber, C. M. (2005) Mater. Today 8, 20–28. [Google Scholar]
- 5.Huang, Y., Duan, X., Cui, Y., Lauhon, L. J., Kim, K. & Lieber, C. M. (2001) Science 294, 1313–1317. [DOI] [PubMed] [Google Scholar]
- 6.Huang, Y., Duan, X. & Lieber, C. M. (2005) Small 1, 142–147. [DOI] [PubMed] [Google Scholar]
- 7.Hopkins, D. S., Pekker, D., Goldbart, P. M. & Bezryadin, A. (2005) Science 308, 1762–1765. [DOI] [PubMed] [Google Scholar]
- 8.Yan, H., Park, S. H., Finkelstein, G., Reif, J. H. & LaBean, T. H. (2003) Science 301, 1882–1884. [DOI] [PubMed] [Google Scholar]
- 9.Nyamjav, D., Kinsella, J. M. & Ivanisevic, A. (2005) Appl. Phys. Lett. 86, 093107-1–093107-3. [Google Scholar]
- 10.Heilemann, M., Tinnefeld, P., Mosteiro, G. S., Parajo, M. G., Van Hulst, N. F. & Sauer, M. (2004) J. Am. Chem. Soc. 126, 6514–6515. [DOI] [PubMed] [Google Scholar]
- 11.Ma, Y., Zhang, J., Zhang, G. & He, H. (2004) J. Am. Chem. Soc. 126, 7097–7101. [DOI] [PubMed] [Google Scholar]
- 12.Ito, Y. & Fukusaki, E. (2004) J. Mol. Catal. B 28, 155–166. [Google Scholar]
- 13.Stoltenberg, R. M. & Woolley, A. T. (2004) J. Biomed. Microdevices 6, 105–111. [DOI] [PubMed] [Google Scholar]
- 14.Ondarçuhu, T. & Joachim, C. (1999) Nanotechnology 10, 39–44. [Google Scholar]
- 15.Petit, C. A. P. & Carbeck, J. D. (2003) Nano Lett. 3, 1141–1146. [Google Scholar]
- 16.Klein, D. C. G., Gurevich, L., Janssen, J. W., Kouwenhoven, L. P., Carbeck, J. D. & Sohn, L. L. (2001) Appl. Phys. Lett. 78, 2396–2398. [Google Scholar]
- 17.Gad, M., Sugiyama, S. & Ohtani, T. (2003) J. Biomol. Struct. Dyn. 21, 387–393. [DOI] [PubMed] [Google Scholar]
- 18.Nakao, H., Gad, M., Sugiyama, S., Otobe, K. & Ohtani, T. (2003) J. Am. Chem. Soc. 125, 7162–7163. [DOI] [PubMed] [Google Scholar]
- 19.Opitz, J., Braun, F., Seidel, R., Pompe, W., Voit, B. & Mertig, M. (2004) Nanotechnology 15, 717–723. [Google Scholar]
- 20.Xia, Y. & Whitesides, G. M. (1998) Angew. Chem. Int. Ed. 37, 550–575. [DOI] [PubMed] [Google Scholar]
- 21.Smith, D. E., Perkins, T. T. & Chu, S. (1996) Macromolecules 29, 1372–1373. [Google Scholar]
- 22.Jackman, R. J., Duffy, D. C., Ostuni, E., Willmore, N. D. & Whitesides, G. M. (1998) Anal. Chem. 70, 2280–2287. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.