Abstract
The yeast histone variant H2AZ (Htz1) is implicated in transcription activation, prevention of the ectopic spread of heterochromatin, and genome integrity. Our genome-wide localization analysis revealed that Htz1 is widely, but nonrandomly, distributed throughout the genome in an SWR1-dependent manner. We found that Htz1 is enriched in intergenic regions compared with coding regions. Its occupancy is inversely proportional to transcription rates and the enrichment of the RNA polymerase II under different growth conditions. However, Htz1 does not seem to directly regulate transcription repression genome-wide; instead, the presence of Htz1 under the inactivated condition is essential for optimal activation of a subset of genes. In addition, Htz1 is not generally responsible for nucleosome positioning, even at those promoters where Htz1 is highly enriched. Finally, using a biochemical approach, we demonstrate that incorporation of Htz1 into nucleosomes inhibits activities of histone modifiers associated with transcription, Dot1, Set2, and NuA4 and reduces the nucleosome mobilization driven by chromatin remodeling complexes. These lines of evidence collectively suggest that Htz1 may serve to mark quiescent promoters for proper activation.
Keywords: htz1, transcription, nucleosome
Chromatin structure poses an obstacle for DNA-related activities such as transcription, replication, recombination, and repair (1). The fundamental state of chromatin is generally controlled by three unique, but not mutually exclusive, mechanisms: ATP-dependent chromatin-remodeling complexes, histone covalent modification enzymes, and incorporation of histone variants. The variant histones were initially thought to be simple replacements for canonical histones; however, recent studies suggest that they have a much more profound epigenetic and structural function.
H2A.F/Z, a family of variants of the highly conserved histone H2A, constitutes ≈5–10% of total H2A protein in chromatin (2). They have been identified in a wide variety of species, including budding yeast (Htz1), fission yeast (Pht1), Tetrahymena (hv1), Drosophila (H2AvD), chicken (H2A.F), Xenopus, mice, and humans (H2A.Z) (3). Initial functional studies carried out in Saccharomyces cerevisiae suggest that Htz1 plays a dual role in transcriptional control: activation at the PHO5 and GAL1 promoters (4) and, conversely, silencing at the HMR locus and telomeres (5). These seemingly conflicting functions were reconciled by the discovery that Htz1 resides within euchromatin boundaries and prevents silencing factors from migrating into transcriptionally active regions (6). Unlike its counterpart in yeast, H2A.Z is essential for viability in mice (7), Drosophila (8, 9), and Tetrahymena (10), suggesting additional roles for H2AZ in higher eukaryotes. It is worth noting that the mammalian H2AZ (mH2AZ) has diverged from budding yeast Htz1 (61% amino acid similarity) and may not represent the functional counterpart of Htz1. Indeed, mH2AZ is enriched at pericentric regions and other heterochromatic regions (11, 12) and can work in concert with HP1α to establish a condensed higher order chromatin structure (13). These specialized chromatin conformations may explain the involvement of H2AZ in chromosome segregation, genome stability, and DNA repair (12, 14–17). The crystal structure of the H2AZ-containing nucleosome reveals that it contains a core particle similar to that of H2A (18). In a biophysical study, H2AZ has been shown to facilitate the folding of nucleosome filaments into a more compacted fiber structure, while inhibiting intra-fiber interactions and aggregation (19). In addition, using a FRET approach, Park et al. (20) demonstrated that H2AZ can stabilize the histone octamer within the nucleosome. Some models have been proposed to unify these pieces of isolated information from different organisms, yet the connection between H2AZ structure and its functional consequences remains largely elusive.
Here, using chromatin immunoprecipitation (ChIP) combined with DNA microarray technology (ChIP-chip), we mapped the distribution of Htz1 in the genome of budding yeast. We found that Htz1 is enriched in intergenic regions compared with coding regions, and its occupancy is inversely proportional to transcription rate and the occupancy of RNA polymerase II (pol II) in adjacent genes. Htz1, however, does not seem to function in transcriptional repression; instead, it is involved in the activation of a subset of genes. Biochemical studies revealed that incorporation of Htz1 into nucleosomes influences histone modifications and chromatin remodeling, supporting a role for Htz1 at inactive promoters.
Methods
Plasmids and Yeast Genetic Manipulations. pGUBdSH was constructed by removal of the sequence between SalI and HindIII sites from the pGUB plasmid (21). The coding sequence of yeast HTZ1 was PCR amplified from yeast genomic DNA and cloned into the NdeI/XhoI site of pET21a to create plasmid pBL256 (pET21a-yeast Htz1) for purification of bacterially overexpressed nontagged Htz1.
All S. cerevisiae strains (Table 1, which is published as supporting information on the PNAS web site) are derived from S288C. Htz1 genomic tagging was accomplished by a PCR-based integration method by using p3XFLAG::KANMX plasmid as templates. To generate a homozygous H2A Flag-tagged strain (YBL467), YBL326 (HTA1-Flag:LoxP) (17) was transformed with a PCR product from p3XFlag::KAN to integrate a 3xFlag tag at the HTA2 locus. Nucleosome position mapping was performed as described (22).
Protein Purification. Recombinant Dot1 and Set2 were bacterially overexpressed and purified through glutathione-Sepharose (Amersham Pharmacia) by using the manufacturer-suggested protocol, with slight modifications (23). Chromatin remodeling complexes RSC, Swi/Snf, CHD1, ISW1, Set2-TAP, and Dot1-TAP were purified through the tandem affinity purification (TAP) method as described (23).
Recombinant Histone Purification, Nucleosome Reconstitution, and Sliding Assay. Yeast recombinant histones (H3, H4, H2A, H2B, and Htz1) were individually expressed in BL21CodonPlus-RIL (Stratagene) cells and purified as described (18, 24). Histone octamers were assembled and fractionated through gel-filtration column Superdex 200. Array reconstitutions were performed as described (17). To generate mono-nucleosomes for the sliding assay, a 216-bp DNA fragment was amplified from the pGUBdSH plasmid by a using 32P end-labeled primer set. This labeled DNA was gel-purified, mixed with histone octamers in 2 M salt buffer, and subjected to serial dilution for reconstitution (25). The nucleosome sliding assay was carried out at 30°C in sliding buffer (20 mM Hepes, pH 7.9/50 mM KCl/0.5 mM PMSF/2 mM DTT/0.05% Nonidet P-40/10% glycerol/100 μg/ml BSA/10 mM MgCl2/4 mM ATP or ATP-γ-S) with 10 fmol of labeled probes and 300 fmol of cold nucleosome and ≈100 fmol of each chromatin remodeling complex. The reaction was stopped by addition of competitor mix (750 ng of Calf Thymus DNA and 500 ng of oligo-nucleosomes). Each total reaction was then directly loaded onto a 5% native polyacrylamide gel (37.5:1) and run for 4.5 h at 4°C.
Plasmid Information. The entire 216-bp DNA sequence used in the sliding assay is as follows: 5′-acattaacctataaaaataggcgtatcacgaggccctttcgtcttcaagaattcacgcgtagatctgctagcatcgatccatggactagtctcgagtttaaagatatccagctgcccgggaggccttcgcgaaatattggtaccccatggaatcgagggatcctctagacggaggacagtcctccggttaccttcgaaccacgtggccgtctagat-3′.
ChIP Assay and DNA Microarray. ChIP was performed as described (22). Sources of antibodies used in this study include anti-FLAG M2 (Sigma), anti-histone H3 C-term (Abcam, Cambridge, MA), anti-H3 di-methylated K79 and K36 (Upstate Biotechnology, Lake Placid, NY), and 8WG16 (pol II) (Covance, Richmond, CA).
DNA microarrays used in this study are polylysine-coated glass spotted with ≈14,000 features (PCR products), including all annotated ORFs; intergenic regions; and mitochondrial DNA and other noncoding regions of special interest, such as rDNA, tRNA, snoRNA, Ty transposons, LTRs, centromeres, and some introns (26). DNA amplification and labeling procedures are essentially adopted from earlier publications (26) (also see: http://derisilab.ucsf.edu/microarray/index.html). All data sets are generated from at least two independent biological samples. The raw data were normalized such that the median ratio of intensities from two channels is 1. The normalized data were then filtered by using the following criteria: the signal is >150 units over background, correlation between two channels is >0.5, and the spot quality is passed by spot-to-spot visual inspection. The median value of all experiments is used to create the final data set. The r value of the Pearson regression line between any two hybridizations is >0.8.
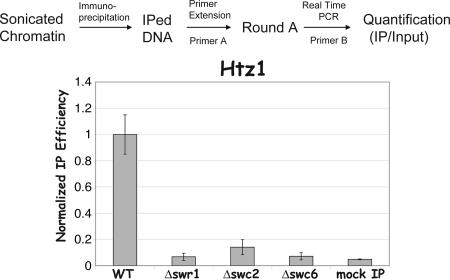
Quantitative Chromatin-Binding Assay. Chromatin-immunoprecipitated DNA and total chromatin DNA were subjected to the quantitative measurement by using two-round amplification (similar to the method used in the ChIP-chip assay) combined with real-time PCR (see Fig. 1). Round A amplification is a T7 sequenase-based primer extension reaction using a degenerate primer with an universal oligo tag at the 5′ end (primer A) (27). Two titrations of round A products were subsequently transferred into round B amplification by using the universal primer (primer B) on a real-time PCR machine (MJ Research, Cambridge, MA). The degree of chromatin binding is determined by the relative immunoprecipitation (IP) efficiency (IP/input). Preliminary experiments were conducted to ensure the linearity of the assay (data not shown).
Fig. 1.
SWR1 controls global deposition of Htz1. The flow chart of quantitative chromatin binding (QCB) assays is illustrated in Upper. Yeast strains YBL556 (WT), YBL557(Δswr1), YBL560 (Δswc2), and YBL561 (Δswc6) were subjected to QCB assay by using rabbit IgG (Sigma). The relative IP efficiency (the ratio of Htz1 IP over input) in WT was assigned as 1, and IP efficiency in mutants was normalized accordingly. Mock IP is performed by using an untagged yeast strain (BY4741).
Results
Genome-Wide Localization of Histone Variant Htz1. To determine the genome-wide localization of yeast H2AZ (Htz1), we used ChIP combined with DNA microarray analysis (ChIP-chip) using a yeast strain containing genomic epitope-tagged HTZ1. DNA microarrays used in this study cover the entire yeast genome. We PCR amplified 13,455 DNA features and spotted them on a single slide, which allows for direct comparison between intergenic regions (IGRs) and ORFs. Data showing Htz1 enrichment at all chromosomes are depicted in Fig. 6A, which is published as supporting information on the PNAS web site. From this global view, Htz1 seems to be widely, but nonrandomly, distributed throughout all 16 chromosomes, with the exception of mitochondria DNA (Q). These results are similar to those from an earlier immunolocalization study (28). As shown in the zoom-in window (Fig. 6A), Htz1 is enriched in the euchromatic region flanking the mating locus HMR (vertical dash lines), but depleted in the intervening silenced region. This finding agrees with a previous publication (6). To further validate our microarray data, we subjected the immunoprecipitated DNA directly to a semiquantitative PCR analysis using published primer sets (6). We arbitrarily picked two regions where Htz1 is enriched (P and B) and one region where H2A is more abundant (H) (6). The results (Fig. 6B) from all three regions demonstrate a very good consistency with previous reports (6, 16).
Because deposition of Htz1 to certain chromosomal loci has been shown to require SWR1, a Swi/Snf-related ATPase containing complex (17), we wondered whether the global distribution of Htz1 observed above is also controlled by SWR1. Thus, we performed ChIP-chip experiments using three loss-of-function SWR1 mutants, each of which contains epitope-tagged Htz1. Htz1 distributes randomly in all three mutants and very poorly correlates with the wild-type pattern (data not shown). This random distribution could merely reflect mislocalization of Htz1 across the whole genome (in this case, Htz1 would still be deposited onto chromatin) or be attributed to background noise in the ChIP-chip assay (in this case, Htz1 would not be bound to chromatin). To distinguish between these possibilities, we developed a quantitative chromatin-binding assay to measure the cross-linking efficiency of certain proteins to total chromatin DNA (Fig. 1). The result shows that the cross-linking efficiency of Htz1 in the mutants is reduced to background levels (Fig. 1), suggesting that SWR1 is required for global deposition of Htz1.
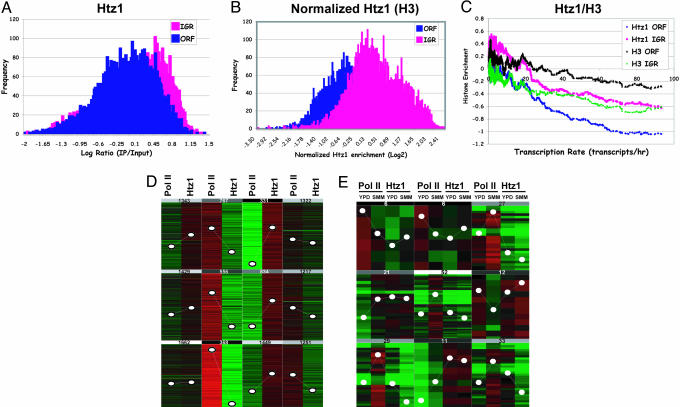
Htz1 Localization and Transcription. A genome-wide study reported that canonical histones tend to be depleted at the intergenic regions (29). We asked whether Htz1 also follows a similar trend. To this end, we performed a histogram analysis on the Htz1 data set. The result (Fig. 2A) reveals that, unlike histone H3 and its counterpart H2A, Htz1 is primarily enriched in IGRs and is rather depleted in ORFs. For direct comparison purposes, we conducted similar ChIP-chip assays using either an antibody against histone H3 or a strain carrying a FLAG tag on both copies of histone H2A genes. As expected, we reproduced the previous observations that H2A and H3 have higher occupancy at coding regions (data not shown). Given the heterogeneity of bulk nucleosome distribution, the Htz1 enrichment was normalized to the global nucleosome occupancy as measured by histone H3 ChIP-chip. As shown in Fig. 2B, Htz1 prefers to reside at IGRs over ORFs. This finding is consistent with a previous observation that Htz1 binds to the GAL1 and PHO5 promoters when the genes are inactive (4).
Fig. 2.
Htz1 is preferentially enriched at inactive promoters. (A) Histogram of Htz1 enrichment (YBL325). The frequency of Log2 ratio from Htz1 data set (same as Fig. 1) fallen into 0.02 intervals (bin size) were calculated and plotted. (B) Same as A, except that Htz1 enrichment was normalized to global nucleosomal occupancy measured by histone H3 enrichment. (C) For each ORF, the closest 5′ intergenic region was selected to represent promoter regions; thus, the transcription rate of such IGR was assigned to be the same as the downstream (3′) gene. Transcription rates used here originated from a previously published microarray analysis (34). The data set compiled with the transcription rate of each ORF and corresponding Htz1 enrichment at the ORF and IGR is first sorted by transcription rate. Next, moving averages (window size = 80, step = 1) of the Htz1 enrichment at both the ORF and IGR are calculated and plotted as a function of transcription rate. (D) K-mean cluster analysis of genome-wide distribution of Htz1 and pol II (8WG16). (E) K-mean cluster analysis of pol II and Htz1 occupancy at a group of genes whose transcription are different when grown in rich media (YPD) and synthetic minimal media (SMM) (absolute difference of pol II enrichment is >0.9 and is consistent with mRNA microarray analysis).
To determine the relationship between Htz1 and RNA pol II-dependent transcription, we calculated the moving average of Htz1 enrichment and plotted it as a function of transcription rate (29, 30). We found that Htz1 enrichment at both IGRs and ORFs negatively correlates with transcription rates of corresponding genes (Fig. 2C). Regardless of transcription frequency, the occupancy of Htz1 at IGRs seems to always be higher than that at ORFs in this analysis, which is consistent with Fig. 2 A. Two recent studies reported that active transcription can cause histone eviction from both IGRs and ORFs (29, 31). Here, we observe a similar trend with two canonical histones (H3, in Fig. 2C, and H2A, data not shown). However, when Htz1 and H3 are compared, upon increase of transcription rate, the slope of Htz1 is greater than that of H3 (Fig. 2C). These data argue that Htz1 is preferentially enriched at the inactive genes and that the role of Htz1 in transcription may be different from that of canonical histones.
Given that occupancy of RNA pol II is a reliable indicator for transcription status, we decided to compare distribution of Htz1 directly to that of pol II. We carried out a ChIP-chip experiment using an antibody against a pol II subunit. The result showed that the distribution of Htz1 is negatively correlated with pol II genome-wide (ρ =–0.251). A K-mean cluster analysis revealed that this inverse relationship becomes more evident at the regions where either pol II is highly enriched or Htz1 is very abundant (Fig. 2D and Fig. 7, which is published as supporting information on the PNAS web site). We next investigated whether this correlation still holds when transcription profiles change. Yeast strains grown in complete media (YPD) and synthetic minimal media (SMM) were collected and subjected to ChIP-chip analysis for Htz1 and pol II. We selected the cluster of genes whose transcription undergoes dramatic changes [based on pol II occupancy, and in agreement with mRNA microarray analysis (data not shown)] to compare the distribution changes of Htz1 and pol II. The K-mean cluster analysis (Fig. 2E) indicates that the trend of pol II enrichment changes is always opposite to that of Htz1. This result strengthens the notion that Htz1 is enriched at inactive genes and that high transcription is associated with less occupancy of Htz1.
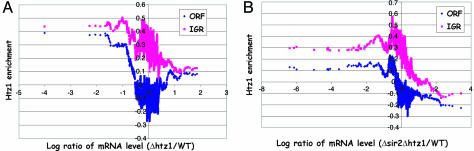
To address the relationship between Htz1 enrichment and transcription, we took advantage of a published data set that measures the transcription changes upon deletion of HTZ1 (6). Moving average analysis of Htz1 enrichment is plotted against this data set without applying any cutoff (6). As shown in Fig. 3A, the lack of Htz1 enrichment at the genes that require Htz1 for repression (right side) suggests that Htz1 may not be directly involved in transcriptional repression. Interestingly, genes that require Htz1 for activation seem to have higher Htz1 occupancy (left side). Considering the previously reported anti-silencing function of Htz1 (6), we did the similar analysis using the data set from a Δsir2/Δhtz1 to eliminate the indirect silencing effect caused by the spreading of Sir2 (6). Although genes that are strongly down-regulated by deletion of HTZ1 associate with moderate levels of Htz1 compared with the wild-type cells, Htz1 occupancy still displays a preference for the derepressed genes (Fig. 3B). Together, these data suggest that Htz1 preferentially resides at inactive promoters and may be involved in optimal transcription activation.
Fig. 3.
Htz1 enrichment and transcription dependency of Htz1. Moving average of Htz1 enrichment in WT was plotted as a function of mRNA changes in Δhtz1 (A) and Δhtz1Δsir2 (B) (6).
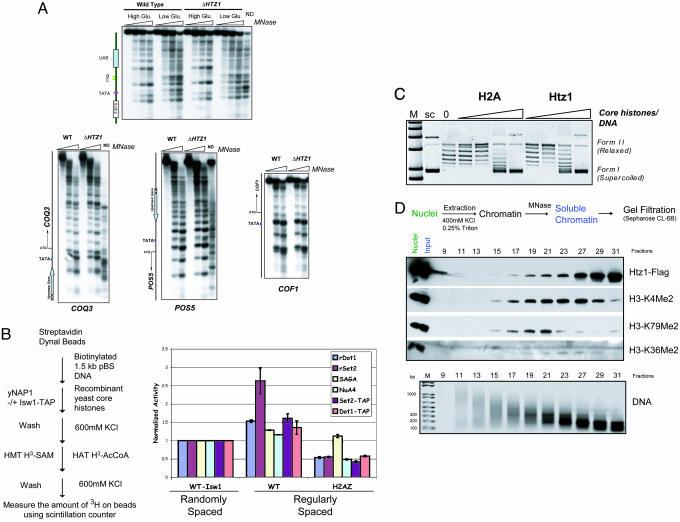
Htz1 and Chromatin Structure. Because Htz1 preferentially resides in the IGR, it might function in controlling proper nucleosome positioning at the promoter. To address this hypothesis, we used micrococcal nuclease-based chromatin mapping on SUC2 (the prototypical gene) and three Htz1-enriched genes (COQ3, POS5, and COF1) in both wild-type and Δhtz1 strains. In all four cases, deletion of HTZ1 does not alter the organized nucleosomal arrays in the regions examined (Fig. 4A), thereby suggesting that Htz1 does not play a general role in nucleosome positioning.
Fig. 4.
The influence of incorporation of Htz1 into nucleosomal arrays on chromatin structure. (A) Deletion of HTZ1 does not disrupt the nucleosome positioning at four examined genomic loci. (B) Htz1-containing nucleosomes were refractory to histone modifications. (Left) A schematic of the steps for histone modification assays using immobilized yeast recombinant nucleosomal arrays. Bacterially expressed rDot1, rSet2 and Set2-TAP and Dot1-TAP were used for histone methyltransferase assays (HMT). The number of nucleosomes used in each assay was normalized based on silver staining after reconstitution (data not shown). SAGA (Spt7-TAP) and NuA4 (Epl1-TAP) were used in histone acetyltransferase (HAT) assays. The radioactivity counts on canonical nucleosomes assembled without the spacing factors (Isw1-TAP) (which results in randomly spaced nucleosomes) after each reaction were arbitrarily denoted as 1. Error bars represent the average deviations. (C) Supercoiling assay. Recombinant Htz1 was assembled into nucleosomal arrays in a manner similar to that of canonical histones. (D) Htz1 was enriched in the nucleosomal population that did not included K79me2 and K36me2 modifications. Nuclei were prepared from YBL325. Extracted chromatin was digested by micrococcal nuclease, and the soluble portion was subsequently loaded on a gel-filtration column. Each fraction was examined by Western blotting with indicated antibodies. Purified DNA was separated on a 2% agarose gel to monitor the size of nucleosomes.
Next, we determined whether incorporation of Htz1 into nucleosomes affects histone modifications. Nucleosome arrays assembled with canonical histones (WT) or Htz1-containing core histones (H2AZ) were used to test histone acetyltransferase and histone methyltransferase activities. All four selected enzymes have been shown to be involved in active transcription (30, 32, 33). As shown in Fig. 4B, the methylation level of Htz1-containing nucleosomal arrays by Set2 is significantly reduced. We also observed a moderate decrease in histone methylation by Dot1 and histone acetylation by NuA4 on Htz1 containing nucleosomes, and very little effect on SAGA acetylation. These defects are probably not caused by the aberrant assembly of the Htz1 arrays, because a supercoiling assay revealed that Htz1 reconstituted nucleosomes to an extent similar to that of canonical histones (Fig. 4C). To demonstrate the biological relevance of these inhibition effects, we prepared native yeast chromatin. After MNase digestion, we loaded the soluble portion on a gel-filtration column. Western blotting analysis revealed that Htz1 tends to be enriched in more highly digested fractions where histone H3 K79me2 and K36me2 are not abundant, whereas methylation enriched fractions eluted as longer oligonucleosomes (Fig. 4D). This finding is consistent with our in vitro observation that Htz1 inhibits these modifications. More importantly, it is also consistent with the finding that Htz1 is enriched in IGRs, because the low density of nucleosomes in these regions may lead to an increase in MNase digestion.
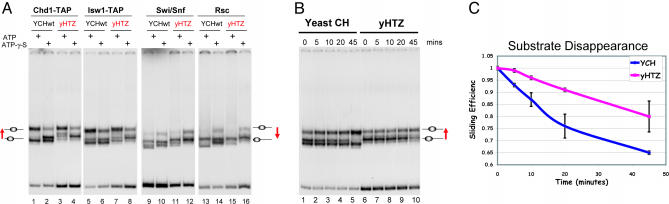
To examine the influence of Htz1 on chromatin remodeling activities, we took advantage of a nucleosome sliding assay. In this assay, the movement of histones on a 216-bp DNA fragment can be monitored by native polyacrylamide electrophoresis because migration varies depending on nucleosome position relative to DNA ends. Mono-nucleosomes containing either canonical yeast core histones or Htz1 core histones were reconstituted in parallel and tested with four chromatin remodeling complexes. All four complexes enable the mobilization of the nucleosomes along the DNA templates in an ATP-dependent manner on wild-type nucleosomes (Fig. 5A). Incorporation of Htz1, however, seemed to reduce the nucleosome sliding activity of Chd1 (Fig. 5A, lanes 3 and 4 compared with lanes 1 and 2) and have modest effects on the actions of Isw1, Swi/Snf, and Rsc. To measure these differences in a more quantitative way, we carried out a kinetics analysis of the nucleosome sliding reaction (Fig. 5B). The result reveals that Htz1 nucleosomes have a slower rate of substrate disappearance (Fig. 5C), confirming the idea that Htz1 reduces chromatin remodeling efficiency.
Fig. 5.
H2AZ interferes with chromatin remodeling in vitro. (A) Htz1 containing yeast recombinant nucleosomes reduce the sliding activity of chromatin remodeling complexes. A 216bp DNA fragment that was PCR amplified from pGUBdSH with 32P end-labeled primer set was reconstituted into mononucleosomes with yeast recombinant histone octamers that included either H2A [yeast core histone (YCH)] or Htz1 (yHTZ). The resulting nucleosomes were directly applied to nucleosome sliding assay by using four TAP-purified chromatin-remodeling complexes Chd1, Isw1, Swi/Snf, and RSC in the presence of ATP or ATP-γ-S. Native PAGE (5% acrylamide, 37.5:1) was used to resolve the mixed population of nucleosomes after remodeling. Chd1-TAP and Isw1-TAP mobilize the nucleosomes toward the center position, which migrates slower, as indicated on the Left, whereas Swi/Snf and RSC move them to the end position which migrates faster, as shown on the Right. (B) Time course analysis of sliding assay (Chd1). The sliding assay was carried out as described in A. At each time point, the remodeling reaction was stopped by addition of excess competitor DNA and nucleosomes. (C) Quantification of sliding kinetics. Each gel band was quantified by using the software imagequant tl. Sliding efficiency was defined as the intensity of the substrate over total population. Error bars represented the SD from three independent experiments.
Discussion
We report here that, at the genome-wide level, Htz1 is enriched at IGRs over ORFs (Fig. 2 A) and that its occupancy decreases upon increasing transcription frequency (Fig. 2C). This finding is consistent with an earlier observation from a gene-to-gene-based study in which it was shown that Htz1 is enriched at the GAL1 and PHO5 promoters under repressive conditions and disappears upon gene activation (4). Insertion of the variant histone at IGRs may have important biological implications. First, the enrichment of Htz1 at promoters might be advantageous in terms of preventing silencing factors (such as Sir proteins) or activators from binding (6), thereby avoiding inadvertent repression or activation. Second, we demonstrated that Htz1-containing nucleosomes are relatively immobile and refractory to chromatin remodeling (Fig. 5), which is consistent with previous findings (19, 20). Thus, once positioned to a certain location, Htz1-containing nucleosomes would be more difficult to mobilize without active chromatin remodeling or histone eviction. Such activities are thought to become available during gene activation. This situation creates a relatively static promoter conformation where the binding sites of transcription factors are either exposed or shielded by nucleosomes. Third, Htz1 at IGR might allow cells to separate the transcription regulation of two neighboring genes, presumably by preventing promoter nucleosomes from sliding. Fourth, the major histones are primarily deposited on chromosomes during S-phase in a DNA replication-dependent manner in yeast. Thus, it is difficult to link the deposition of canonical histones with transcription requirements. Replacement histones, on the other hand, can be deposited at any stage of the cell cycle, presumably by using specific loading machineries (e.g., yeast SWR1). Therefore, it may be advantageous to use replacement histones as an epigenetic mark. Finally, because Htz1 nucleosomes are relatively resistant to transcription elongation-related histone modifications (Fig. 4B), the variant histone may better serve as a “promoter marker” to ensure proper recognition by transcription initiation factors.
In this study, we demonstrated that Htz1 enrichment at IGRs and ORFs negatively correlates with transcription rates to a greater extent than histones in general (Fig. 2C). Htz1, however, does not seem to be directly involved in transcriptional repression (Fig. 3), because the genes de-repressed by deletion of HTZ1 are generally not associated with high levels of Htz1. Instead, the activation of a subset of genes seems to require the physical presence of Htz1 at promoter regions (Fig. 3). Therefore, incorporation of Htz1 during the inactive state may serve as a stable epigenetic mark but may not play a dominant role in transcription per se. Perhaps it keeps promoters in a repressed state until the appropriate activation signal is received.
Supplementary Material
Acknowledgments
We thank C. Wu (National Institutes of Health, Bethesda), T. Tsukiyama (Fred Hutchinson Cancer Research Center, Seattle), K. Struhl (Harvard University, Boston), and W. J. Shia (Stowers Institute for Medical Research) for yeast strains and plasmids. We also thank C. Bausch, R. Camahort, T. Kusch, and other members of the J.L.W. laboratory for useful discussions and technical suggestions. S.G.P. was supported by a Postdoctoral Research Fellowship from the Natural Sciences and Engineering Research Council of Canada. J. Gutiérrez was supported by the Pew Latin American Fellows Program in the Biomedical Sciences. This work was supported by a grant from the National Institute of General Medical Sciences (to J.L.W.).
Author contributions: B.L. and C.S. designed research; B.L., D.L., J. Gutiérrez, S.G.P., and C.S. performed research; B.L., S.G.P., J.C., C.S., J. Gerton, and J.L.W. analyzed data; D.L., J.C., C.S., and J. Gerton contributed new reagents/analytic tools; and B.L., S.G.P., J. Gerton, and J.L.W. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: pol II, polymerase II; IP, immunoprecipitation; ChIP, chromatin IP; TAP, tandem affinity purification; IGR, intergenic region.
References
- 1.Workman, J. L. & Kingston, R. E. (1998) Annu. Rev. Biochem. 67, 545–579. [DOI] [PubMed] [Google Scholar]
- 2.West, M. H. & Bonner, W. M. (1980) Biochemistry 19, 3238–3245. [DOI] [PubMed] [Google Scholar]
- 3.Kamakaka, R. T. & Biggins, S. (2005) Genes Dev. 19, 295–310. [DOI] [PubMed] [Google Scholar]
- 4.Santisteban, M. S., Kalashnikova, T. & Smith, M. M. (2000) Cell 103, 411–422. [DOI] [PubMed] [Google Scholar]
- 5.Dhillon, N. & Kamakaka, R. T. (2000) Mol. Cell 6, 769–780. [DOI] [PubMed] [Google Scholar]
- 6.Meneghini, M. D., Wu, M. & Madhani, H. D. (2003) Cell 112, 725–736. [DOI] [PubMed] [Google Scholar]
- 7.Faast, R., Thonglairoam, V., Schulz, T. C., Beall, J., Wells, J. R., Taylor, H., Matthaei, K., Rathjen, P. D., Tremethick, D. J. & Lyons, I. (2001) Curr. Biol. 11, 1183–1187. [DOI] [PubMed] [Google Scholar]
- 8.Clarkson, M. J., Wells, J. R., Gibson, F., Saint, R. & Tremethick, D. J. (1999) Nature 399, 694–697. [DOI] [PubMed] [Google Scholar]
- 9.van Daal, A. & Elgin, S. C. (1992) Mol. Biol. Cell 3, 593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu, X., Li, B. & Gorovsky, M. A. (1996) Mol. Cell. Biol. 16, 4305–4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rangasamy, D., Berven, L., Ridgway, P. & Tremethick, D. J. (2003) EMBO J. 22, 1599–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krogan, N. J., Keogh, M. C., Datta, N., Sawa, C., Ryan, O. W., Ding, H., Haw, R. A., Pootoolal, J., Tong, A., Canadien, V., et al. (2003) Mol. Cell 12, 1565–1576. [DOI] [PubMed] [Google Scholar]
- 13.Fan, J. Y., Rangasamy, D., Luger, K. & Tremethick, D. J. (2004) Mol. Cell 16, 655–661. [DOI] [PubMed] [Google Scholar]
- 14.Rangasamy, D., Greaves, I. & Tremethick, D. J. (2004) Nat. Struct. Mol. Biol. 11, 650–655. [DOI] [PubMed] [Google Scholar]
- 15.Zhang, H., Richardson, D. O., Roberts, D. N., Utley, R., Erdjument-Bromage, H., Tempst, P., Cote, J. & Cairns, B. R. (2004) Mol. Cell. Biol. 24, 9424–9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kobor, M. S., Venkatasubrahmanyam, S., Meneghini, M. D., Gin, J. W., Jennings, J. L., Link, A. J., Madhani, H. D. & Rine, J. (2004) PLoS Biol. 2, E131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizuguchi, G., Shen, X., Landry, J., Wu, W. H., Sen, S. & Wu, C. (2004) Science 303, 343–348. [DOI] [PubMed] [Google Scholar]
- 18.Suto, R. K., Clarkson, M. J., Tremethick, D. J. & Luger, K. (2000) Nat. Struct. Biol. 7, 1121–1124. [DOI] [PubMed] [Google Scholar]
- 19.Fan, J. Y., Gordon, F., Luger, K., Hansen, J. C. & Tremethick, D. J. (2002) Nat. Struct. Biol. 9, 172–176. [DOI] [PubMed] [Google Scholar]
- 20.Park, Y. J., Dyer, P. N., Tremethick, D. J. & Luger, K. (2004) J. Biol. Chem. 279, 24274–24282. [DOI] [PubMed] [Google Scholar]
- 21.Juan, L. J., Utley, R. T., Adams, C. C., Vettese-Dadey, M. & Workman, J. L. (1994) EMBO J. 13, 6031–6040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, B. & Reese, J. C. (2001) J. Biol. Chem. 276, 33788–33797. [DOI] [PubMed] [Google Scholar]
- 23.Li, B., Howe, L., Anderson, S., Yates, J. R., 3rd, & Workman, J. L. (2003) J. Biol. Chem. 278, 8897–8903. [DOI] [PubMed] [Google Scholar]
- 24.Luger, K., Mader, A. W., Richmond, R. K., Sargent, D. F. & Richmond, T. J. (1997) Nature 389, 251–260. [DOI] [PubMed] [Google Scholar]
- 25.Owen-Hughes, T., Utley, R. T., Steger, D. J., West, J. M., John, S., Cote, J., Havas, K. M. & Workman, J. L. (1999) Methods Mol. Biol. 119, 319–331. [DOI] [PubMed] [Google Scholar]
- 26.Iyer, V. R., Horak, C. E., Scafe, C. S., Botstein, D., Snyder, M. & Brown, P. O. (2001) Nature 409, 533–538. [DOI] [PubMed] [Google Scholar]
- 27.Bohlander, S. K., Espinosa, R., 3rd, Le Beau, M. M., Rowley, J. D. & Diaz, M. O. (1992) Genomics 13, 1322–1324. [DOI] [PubMed] [Google Scholar]
- 28.Leach, T. J., Mazzeo, M., Chotkowski, H. L., Madigan, J. P., Wotring, M. G. & Glaser, R. L. (2000) J. Biol. Chem. 275, 23267–23272. [DOI] [PubMed] [Google Scholar]
- 29.Lee, C. K., Shibata, Y., Rao, B., Strahl, B. D. & Lieb, J. D. (2004) Nat. Genet. 36, 900–905. [DOI] [PubMed] [Google Scholar]
- 30.Robert, F., Pokholok, D. K., Hannett, N. M., Rinaldi, N. J., Chandy, M., Rolfe, A., Workman, J. L., Gifford, D. K. & Young, R. A. (2004) Mol. Cell 16, 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bernstein, B. E., Liu, C. L., Humphrey, E. L., Perlstein, E. O. & Schreiber, S. L. (2004) Genome Biol. 5, R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hampsey, M. & Reinberg, D. (2003) Cell 113, 429–432. [DOI] [PubMed] [Google Scholar]
- 33.Schubeler, D., MacAlpine, D. M., Scalzo, D., Wirbelauer, C., Kooperberg, C., van Leeuwen, F., Gottschling, D. E., O'Neill, L. P., Turner, B. M., Delrow, J., et al. (2004) Genes Dev. 18, 1263–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holstege, F. C., Jennings, E. G., Wyrick, J. J., Lee, T. I., Hengartner, C. J., Green, M. R., Golub, T. R., Lander, E. S. & Young, R. A. (1998) Cell 95, 717–728. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.