Abstract
In vivo expression of human telomerase is significantly different from that of mouse telomerase. To assess the basis for this difference, a bacterial artificial chromosome clone containing the entire hTERT (human telomerase reverse transcriptase) gene was introduced in mice. In these transgenic mice, expression of the hTERT transgene was similar to that of endogenous hTERT in humans, rather than endogenous mTERT (mouse telomerase reverse transcriptase). In tissues and cells showing a striking difference in expression levels between hTERT in humans and mTERT in mice (i.e., liver, kidney, lung, uterus, and fibroblasts), expression of the hTERT transgene in transgenic mice was repressed, mimicking hTERT in humans. The transcriptional activity of the hTERT promoter was much lower than that of the mTERT promoter in mouse embryonic fibroblasts or human fibroblasts. Mutational analysis of the hTERT and mTERT promoters revealed that a nonconserved GC-box within the hTERT promoter was responsible for the human-specific repression. These results reveal that a difference in cis-regulation of transcription, rather than transacting transcription factors, is critical to species differences in tissue-specific TERT expression. Our data also suggest that the GC-box-mediated, human-specific mechanism for TERT repression is impaired in human cancers. This study represents a detailed characterization of the functional difference in a gene promoter of mice versus humans and provides not only important insight into species-specific regulation of telomerase and telomeres but also an experimental basis for generating mice humanized for telomerase enzyme and its pattern of expression.
Mouse models are widely used for understanding physiological and pathological processes in humans, including aging and cancer. However, there are some significant differences between mice and humans that are possible concerns in the use of mouse models in a variety of fields of current research. One of these differences is in the regulation of telomerase expression and telomere length (1). Telomeres, specialized structures consisting of repetitive DNA and associated proteins, function to protect chromosome ends from degradation and end-to-end fusion, thereby playing important roles in cell viability and genomic stability (2). The ribonucleoprotein enzyme telomerase synthesizes telomeric DNA repeat and can compensate telomere attrition associated with cell divisions. Most normal human somatic cells express undetectable or low levels of telomerase and undergo progressive loss of telomere length with cell divisions, eventually leading to telomere dysfunction (3). Telomere dysfunction induces permanent cell-growth arrest, termed cellular senescence, which may contribute to organismal aging in humans (4), and chromosome abnormalities, a hallmark of cancer cells (5). Activation of telomerase in human cells allows them to maintain telomere length and function, a critical step to immortalization and malignant transformation of human cells (6). In marked contrast, Mus musculus, mice used in research laboratories, have much longer telomeres and express robust levels of telomerase activity in a wider range of normal somatic tissues and cells than do humans (7–9). Telomere attrition is not a major cause of mouse-cell senescence (10), and mouse cells become immortalized and transformed more readily than human cells, at least in part, because of long telomeres and high levels of constitutive telomerase activity in mice (6, 11). These findings suggest that current mouse models for studying human cancer and aging do not always reflect the physiological and pathological status in humans (1). Indeed, knock-out mouse models of human genetic diseases associated with premature aging and cancer predisposition (e.g., Werner syndrome and Bloom syndrome) frequently do not recapitulate the human disease phenotypes; although, notably, in some instances, the disease phenotypes were manifest when mouse telomeres were shortened as a result of breeding with telomerase-null mice (12, 13). However, in this circumstance, telomerase is deficient in all tissues and cells, including those normally expressing telomerase activity in both mice and humans (e.g., germ-cell and stem-cell populations) and is, thus, discordant with the physiological status of telomerase and telomeres in humans. Thus, efforts to generate mouse models closer to humans in terms of telomere and telomerase regulation are of importance in the field of cancer and aging research.
Regulation of the telomerase reverse-transcriptase gene (TERT), which encodes the catalytic-protein subunit of telomerase enzyme, is a major determinant of telomerase activity (14). The expression level of TERT is highly correlated with telomerase activity in normal and cancer cells (15). In accordance with the differences in telomerase expression between mouse and human tissues, the expression profiles of mouse TERT (mTERT) in mouse tissues were reported to be different from those of human TERT (hTERT) in human tissues (16).
In this study, by generating transgenic mice carrying both an endogenous mTERT gene and an entire hTERT genomic transgene with considerable upstream and downstream regions and all exons and introns, we directly compare in vivo expressions of mTERT and hTERT to elucidate the molecular basis for the differential regulation. Moreover, through assays on transcriptional activities of a number of mutated mTERT and hTERT gene promoters, we identify a cis-acting DNA element that is responsible for species-specific regulation. This study provides essential data for understanding the significant difference in telomerase expression between mice and humans and for making mouse models of cancer and aging closer to humans.
Materials and Methods
Generation of Transgenic Mice. A bacterial artificial chromosome (BAC) clone containing the ≥54-kb hTERT gene (including an at least 11-kb upstream region, all of the exons and introns, and 1.2-kb downstream region) was derived from a circular yeast artificial chromosome clone that was isolated by transformation-associated recombination cloning in yeast, as described in ref. 17. The linearized BAC DNA was microinjected into C57BL/6 mouse oocytes. The screening for founder lines containing the hTERT sequence was carried out by PCR genotyping (the PCR primer sequences are available at www.cephb.fr/poltel/Map_TERT.php) by using tail DNA. Mice from two independent hTERT transgenic founder lines were bred with mTERT+/– mice that were maintained on C57BL/6 background (18) to obtain mTERT+/–hTERT+ mice. mTERT–/–-hTERT+ mice were also generated by crossing mTERT+/–hTERT+ and mTERT+/– mice. As shown in Fig. 4, which is published as supporting information on the PNAS web site, spleen cells from wild-type, mTERT–/– (18), mTERT+/–hTERT+ and mTERT–/–-hTERT+ mice were used to examine hTERT expression by RT-PCR with the following primers: 5′-GCC TGA GCT GTA CTT TGT CAA-3′ and 5′-CGC AAA CAG CTT GTT CTC CAT GTC-3′, which amplified hTERT exons 5–9 (nucleotides 2,164–2,620 in GenBank NM_003219). Detection of mTERT and mouse actin mRNA was carried out as described in refs. 18 and 19. All animals were housed at Bioqual (Rockville, MD).
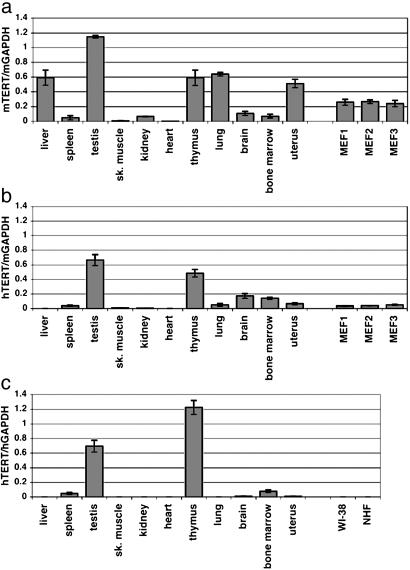
Real-Time Quantitative RT-PCR. Total RNA samples were extracted from mouse tissues and mouse embryonic fibroblasts (MEF) by using NucleoSpin kits (BD Biosciences Clontech, Palo Alto, CA). Human total RNA samples were obtained from BD Biosciences Clontech (sources of the samples are described in Supporting Materials and Methods, which is published as supporting information on the PNAS web site). One μg of total RNA was reverse-transcribed with random hexamers by using the SuperScript first-strand synthesis system for RT-PCR (Invitrogen Life Technologies). Each PCR reaction contained first-strand cDNA corresponding to 25 ng of RNA, TaqMan universal PCR master mix (Applied Biosystems), a set of primers and FAM/MGB probe for hTERT or mTERT, and a set of primers and VIC/MGB probe for human or mouse GAPDH (hGAPDH or mGAPDH, respectively) as the endogenous control (details of the primers and probes are available in Supporting Materials and Methods). Real-time detection of PCR products was performed by a PRISM 7700 sequence detector (Applied Biosystems). Reactions were in triplicate for each sample. Quantitative analysis of gene expression data was performed according to the standard-curve method in the supplier's protocol (User Bulletin #4303859B at www.appliedbiosystems.com/index.cfm) (standard curves are shown in Supporting Materials and Methods). The expression levels of mTERT or hTERT normalized with those of mGAPDH or hGAPDH (i.e., mTERT/mGAPDH, hTERT/mGAPDH, or hTERT/hGAPDH) are shown as average values ± SD in Fig. 1.
Fig. 1.
Real-time quantitative RT-PCR analysis of mTERT and hTERT expression. RNA samples from various tissues in humans and transgenic mice and from fibroblasts (MEF1–3 and two human fibroblast strains WI-38 and NHF) were examined for mTERT and hTERT mRNA levels. (a) mTERT expression normalized with mGAPDH in mTERT+/–hTERT+ transgenic mice. (b) hTERT expression normalized with mGAPDH in mTERT+/–hTERT+ transgenic mice. In a and b, tissue RNA samples (except for testis and uterus) were pooled from one male and one female 8-week-old mouse (line BAC-C10). Testis or uterus sample was from one 8-week-old male or female mouse (line BAC-C10), respectively. Tissue RNA samples similarly pooled from the other line of mTERT+/–hTERT+ mice (BAC-C2) were also examined and confirmed to give similar levels of mTERT and hTERT expression to the data shown here. MEF1, MEF2, and MEF3 are three independent preparations of fibroblasts from BAC-C10 embryos. (c) hTERT expression normalized with hGAPDH in humans. See Materials and Methods for sources of human tissue RNA samples. For human liver, another RNA sample pooled from 23 individuals was also confirmed negative for hTERT expression (data not shown).
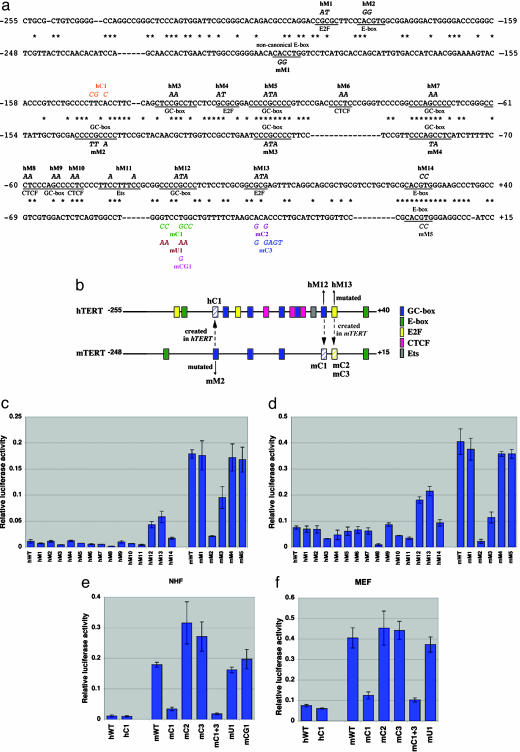
Promoter Constructs and Luciferase Assay. The 295-bp hTERT promoter cloned in pGL3-Basic vector (pBT-255, Promega) was described in ref. 20 (pBT-255). The 263-bp mTERT promoter (–248 to +15, shown in Fig. 2a) was PCR-amplified and cloned in the pGL3-Basic vector. Site-directed mutagenesis of hTERT and mTERT promoters was carried out by using the QuikChange site-directed mutagenesis kit (Stratagene). For the luciferase assay, cells were seeded on 24-well plates (8.0 × 104 to 1.5 × 105 per well, depending on cell-proliferation rate and cell size), cultured overnight, and transfected with the TERT promoter-luciferase plasmids (0.5 μg per well) or the pGL3-Control plasmid (in which the firefly luciferase reporter gene is driven by SV40 enhancer/promoter; 0.5 μg per well) by using FuGENE6 transfection reagent (Roche Diagnostics, Indianapolis). The pRL-SV40 (3 ng per well; Promega) expressing Renilla reniformis luciferase driven by SV40 enhancer/promoter was included in each transfection as a control to normalize the transcriptional activity of TERT promoter fragments. Preparation of cell lysates and measurement of luciferase activity were performed by using the Dual Luciferase reporter assay system (Promega). In Fig. 2 c–f, the normalized activity of pGL3-Control was defined as 1.0 (not shown in the figures), and the activity of each TERT promoter fragment was expressed as a relative value. All of the data (mean ± SD) were from at least three independent experiments.
Fig. 2.
Identification of a cis-element responsible for the differential transcriptional activity of hTERT versus mTERT promoter. (a) Sequence alignment of hTERT (above) and mTERT (below) promoters (24). Asterisks indicate identical nucleotides. Putative transcription-factor-binding sites are underlined and labeled along the sequences. Mutations destroying those sites are shown in italics (hM1–hM14 in hTERT and mM1–mM5 in mTERT). Mutations creating potential binding sites (hC1 in hTERT and mC1–mC3 in mTERT) and control mutations for mC1 (mU1 and mCG1) are shown in colored italics. A double mutant carrying both mC1 and mC3 mutations (mC1 + 3) was also made. (b) Schematic representation of transcription-factor-binding sites (colored rectangles) in hTERT and mTERT promoters. Also indicated are the mutations destroying (mM2, hM12, and hM13) or creating (hC1, mC1, mC2, and mC3) three candidate sites for a species-specific cis-regulatory element. The sites created in the promoter of another species are shown as hatched rectangles. (c–f) Luciferase reporter gene assays to examine the transcriptional activities of wild-type hTERT promoter (hWT) and wild-type mTERT promoter (mWT) and their derivatives with the mutations indicated in a. NHF were transfected in c and e, and MEF in d and f. In all experiments, the activity of the pGL3-Control plasmid (where SV40 enhancer/promoter drives the luciferase reporter gene) was defined as 1.0 (not shown in the figures). The activity of each promoter fragment is expressed as a relative luciferase activity (mean ± SD).
Results
Generation of hTERT BAC Transgenic Mice. After microinjecting the BAC clone containing the hTERT gene locus of at least 54 kb (17) into ≈100 C57BL/6 mouse oocytes, PCR genotyping of the resulting mice identified five founder lines containing the hTERT sequence. From two of them, the hTERT transgene was transmitted to the offspring. These offspring were found to express hTERT mRNA by RT-PCR analysis (data not shown; same primers as in Fig. 4). The hTERT-expressing offspring were bred with mTERT+/– mice (18). Two independent lines of mTERT+/–-hTERT+ mice (BAC-C2 and BAC-C10, corresponding to the two different founder lines) were analyzed in this study. Fig. 4 shows hTERT and mTERT mRNA expression in spleen lymphocytes from wild-type, mTERT–/–, mTERT+/–hTERT+, and mTERT–/–-hTERT+ mice. As expected, hTERT was expressed only in hTERT+ transgenic mice, and mTERT was expressed only in a wild-type or mTERT+/– background. In the hTERT+ transgenic mice, PCR analyses using genomic DNA confirmed the presence of the 11-kb upstream region, all 16 exon sequences, and 1.2-kb downstream region contained in the original BAC clone, and additional RT-PCR analyses suggested the expression of full-length hTERT mRNA in spleen lymphocytes (data not shown).
Cis-Regulation Determines the Differential in Vivo Expression of hTERT and mTERT. To quantitatively compare the in vivo expression of mTERT and hTERT among various organs of mTERT+/–-hTERT+ transgenic mice, quantitative real-time RT-PCR assays of mTERT mRNA (Fig. 1a) and hTERT mRNA (Fig. 1b) were performed. For comparison, hTERT mRNA expression in human organs (Fig. 1c) was also examined. As expected, in the organs where both mTERT in mice and hTERT in humans are readily detected (i.e., spleen, testis, thymus, and bone marrow), the hTERT transgene was also expressed in the transgenic mice. In organs where little or no mTERT is expressed in mice and no hTERT in humans (i.e., skeletal muscle and heart), there was similarly little or no expression of hTERT in the transgenic mice. Most informative, however, was the analysis of hTERT transgene expression in the tissues in which endogenous mTERT is expressed in mice but in which hTERT is not expressed in humans. In liver, kidney, lung, and uterus of the mTERT+/–hTERT+ transgenic mice, endogenous mTERT was expressed, but the hTERT transgene was repressed completely or expressed much less abundantly, paralleling the pattern observed for endogenous hTERT in humans. Thus, the expression profiles of the hTERT transgene in most mouse organs followed those of hTERT in humans, suggesting that cis-regulation, rather than transacting factor environments, is responsible for much of the species-specific difference in TERT expression. There was an exception: The hTERT transgene in transgenic mouse brain was expressed similarly to the endogenous mTERT, in contrast to the repression of hTERT in human brain (discussed below). The difference between hTERT and mTERT expression was also observed in MEF derived from transgenic mouse embryos, in agreement with the repression of hTERT in normal human fibroblasts (WI-38 and NHF).
We also compared the relative abundance of mTERT and hTERT mRNA in each of the organs from mTERT+/–hTERT+ mice. For this purpose, we established an RT-PCR assay in which a single pair of primers amplified both mTERT and hTERT at similar efficiencies, and the resulting PCR products were digested with an mTERT- or hTERT-specific restriction endonuclease (see Fig. 5, which is published as supporting information on the PNAS web site). In spleen, testis, thymus, brain, and bone marrow, the expression level of hTERT was comparable to that of mTERT. In liver, kidney, lung, uterus, and MEF, no or little hTERT-derived products were detected in the presence of a large quantity of mTERT-derived products. These results are consistent with the expression profiles of mTERT and hTERT shown in Fig. 1 a and b. Thus, both the quantitative comparison of mTERT or hTERT among different organs and the relative comparison of mTERT versus hTERT in each organ suggest an organ-specific, differential cis-regulation of mTERT and hTERT genes.
A Nonconserved GC-Box Within the hTERT Promoter Is Critical to the Human-Specific Repression of TERT Transcription. To assess the basis for differential cis-regulated expression of mTERT and hTERT, we analyzed the function of mouse and human promoter sequences in a transcriptional reporter assay. When the 295-bp hTERT gene promoter and the corresponding 263-bp mTERT gene promoter (Fig. 2 a and b) were examined in a luciferase reporter gene assay, the activity of mTERT promoter (mWT) was 16-fold or 5.4-fold higher than that of hTERT promoter (hWT) in NHF or MEF cells, respectively (Fig. 2 c–f), suggesting that the difference between mTERT and hTERT mRNA expression is attributable largely to the differential transcriptional activity of hTERT and mTERT promoters. These findings further suggest the critical role of a cis-acting element in determining the TERT gene-expression profiles in mice and humans.
To identify the cis-element responsible for the differential transcriptional activity (either an hTERT-specific repressive element or an mTERT-specific activating element), a number of site-directed mutations were made within conserved or nonconserved potential regulatory elements on the 295-bp hTERT promoter (hM1–hM14) and 263-bp mTERT promoter (mM1–mM5), including GC-boxes (putative Sp1 sites) (21, 22), putative E2F-binding sites (23), canonical and noncanonical E-boxes (21, 24), putative Ets-binding sites (25), and putative binding sites for a zinc-finger repressor CTCF (26) (Fig. 2a). The luciferase assay using these mutant promoter constructs in NHF (Fig. 2c) and MEF (Fig. 2d) found three nonconserved sequences as candidates for the species-specific regulatory element (Fig. 2b): Two hTERT mutants, hM12 (a GC-box) and hM13 (a putative E2F site), significantly increased promoter activity and thus appeared to abrogate repression observed with the wild-type hTERT promoter. In addition, an mTERT mutant mM2 (a GC-box) showed much weaker promoter activity than the wild-type mTERT promoter. However, a GC-box created in the hTERT promoter at the corresponding position to mM2 (hC1 in Fig. 2 a and b) did not increase transcriptional activity (hWT and hC1 in Fig. 2 e and f), inconsistent with the idea that this GC-box is an mTERT-specific activating element. Whereas the creation of a putative E2F site in the mTERT promoter at the corresponding position to hM13 (mC2 and mC3 in Fig. 2 a and b) did not reduce the mTERT promoter activity (Fig. 2 e and f), a GC-box created at the corresponding position to hM12 by itself (mC1 in Fig. 2 a and b) or in combination with the mC3 mutation (mC1 + 3) markedly reduced the mTERT promoter activity, making it similar to the hTERT promoter activity (Fig. 2 e and f). An unrelated mutation at the same position (mU1) or the creation of a CpG site, but not a GC-box (mCG1), had no effect on the promoter activity (Fig. 2 e and f). These results identified the nonconserved GC-box located at –31 to –24 in the hTERT promoter as the human-specific repressive element, at least in fibroblasts in culture. Electrophoretic mobility-shift assays showed that the Sp1/Sp3 protein complex binds to this GC-box more strongly than either Sp1 or Sp3 alone (see Fig. 6, which is published as supporting information on the PNAS web site), similar to the preferential binding of the Sp1/Sp3 complex to another GC-box within the hTERT promoter (22). It is likely that the repressive effect of the GC-box is position-dependent, because the same GC-box sequences (CCCCGCCC), located upstream, did not act as a repressive element (see hM5 and mM2 in Fig. 2 c and d).
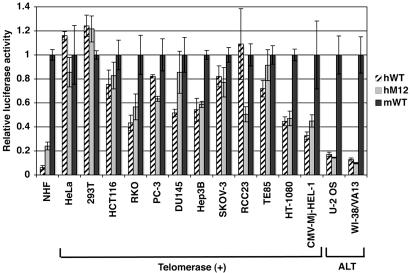
The GC-Box-Mediated Mechanism for hTERT Repression Is Impaired in Human Cancers. To examine whether the GC-box-mediated, hTERT-specific repressive mechanism is inactivated in human cancers, a variety of types of telomerase-expressing human cancer and immortalized cell lines, including those of fibroblastic origin (HT-1080 and CMV-Mj-HEL-1) that could be compared with a normal counterpart of the same cell type (NHF) were examined in the luciferase reporter gene assay using the wild-type hTERT promoter (hWT), the GC-box-mutated hTERT promoter (hM12), and the wild-type mTERT promoter (mWT) (Fig. 3). As is evident in NHF, the GC-box-mediated, hTERT-specific repression is characterized by two features in the assay: The wild-type hTERT promoter is much less active than the wild-type mTERT promoter (hWT/mWT ratio = 0.06 in NHF), and the GC-box mutation results in a significant increase in the hTERT promoter activity (hM12/hWT ratio = 3.95 in NHF). In each of the 12 telomerase-expressing cell lines examined, these two features were lost or quantitatively diminished. The hWT/mWT ratio ranged from 0.33 (CMV-Mj-HEL-1) to 1.24 (293T), and the hM12/hWT ratio ranged from 0.46 (RCC23) to 1.66 (DU145). A decrease in the hTERT promoter activity was associated with the GC-box mutation in RCC23, PC-3, and HeLa, suggesting that the GC-box may act as a positive regulatory element rather than as a repressive element in some human cancers. Interestingly, two telomerase-negative alternative lengthening of telomeres (ALT) cell lines still showed a greater repression of hWT activity (hWT/mWT ratio = 0.17 in U-2 OS and 0.12 in WI-38/VA13) than the telomerase-expressing cancer cell lines, whereas the GC-box no longer appeared to function as a repressive element in these cells (Fig. 3). Collectively, these results suggest that human-specific repression of TERT transcription is frequently abrogated in telomerase-expressing human cancer cells, but not in ALT cells, and are consistent with the notion that the telomerase component genes (i.e., hTERT and hTERC) can be repressed in ALT cells by a different mechanism than that which is active in normal cells (14, 27).
Fig. 3.
The GC-box-mediated mechanism for hTERT repression is impaired in human cancer and immortalized cells. The transcriptional activities of the wild-type hTERT promoter (hWT), the GC-box-mutated hTERT promoter (hM12) and the wild-type mTERT promoter (mWT) were examined by the luciferase reporter gene assay in the following human cell lines: HeLa, uterine cervical cancer; 293T, immortalized embryonic kidney cells expressing SV40 T-antigen; HCT116, colon cancer; RKO, colon cancer; PC-3, prostate cancer; DU145, prostate cancer; Hep3B, hepatocellular carcinoma; SKOV-3, ovarian cancer; RCC23, renal cell carcinoma; TE85, osteosarcoma; HT-1080, fibrosarcoma; CMV-Mj-HEL-1, cytomegalovirus-immortalized fibroblasts; U-2 OS, osteosarcoma; and WI-38/VA13, SV40-immortalized fibroblasts. U-2 OS and WI-38/VA13 are telomerase-negative and maintain telomeres by the alternative lengthening of telomeres (ALT) mechanism. All of the other cell lines are telomerase-positive. The promoter activities in NHF are from data in Fig. 2 and are shown for comparison. For each cell line, the activities of hWT and hM12 are expressed as the relative values to the activity of mWT (defined as 1.0). Data shown are mean ± SD of triplicate assays.
Discussion
Distinct differences exist in the in vivo expression of telomerase activity in mice versus humans (1, 7, 8). These differences are, at least in part, because of differential regulation of the TERT gene (14, 16), but the basis for the regulation of this critical gene is unknown. In this study, we generated mice carrying both the mouse and human TERT genes, each under the control of its own regulatory sequences, to study the regulation of these genes in the same cell context in vivo. Given that the regulation of the hTERT gene is cell-type-specific in humans (28) and that a transgene of large size (i.e., yeast artificial chromosome or BAC) tends to allow integration that is position-independent, copy-number-dependent, and tissue-specific in its expression (29, 30), we reasoned that introduction of the entire gene would be important for these studies. Therefore, we cloned and used a ≥54-kb genomic region containing all exons and introns and at least 11-kb upstream and 1.2-kb downstream sequences of the hTERT gene (17). Whereas the absence or instability of some human DNA sequences in conventional genomic libraries has hampered studies on functions and regulations of an entire gene locus and has left a number of gaps in the human genome sequence database, transformation-associated recombination (TAR) cloning enables the efficient, reproducible cloning and stable maintenance of large genomic regions of interest and, thus, is a powerful approach to solve these problems in genome research (31, 32). This study presents a successful application of an entire human gene isolated by TAR cloning in in vivo analysis of gene regulation. When we made transgenic mice carrying a BAC with the entire hTERT gene, we observed that the tissue-specific expression of the hTERT versus mTERT was maintained in the transgenic mice, with the possible exception of the brain (discussed below), demonstrating that the differential regulation of TERT was because of cis-acting elements of mTERT versus hTERT and enabled us to further dissect the basis for this difference.
Several cis-acting elements within the hTERT promoter have been suggested to function as repressive elements of hTERT transcription in normal human cells (14). In agreement with our previous identification of the proximal E-box (+22 to +27) as a repressive cis-element of the hTERT promoter (20), the mutation of this E-box sequence (hM14) increased the hTERT promoter activity in NHF (Fig. 2c). However, this E-box element is conserved in mice and humans (Fig. 2 a and b), and the effect of its mutation was much less than when a nonconserved GC-box was mutated (hM12). A nonconserved, potential E2F-binding site located at –13 to –9 in the hTERT promoter was a candidate for the human-specific repressive element, because its mutation hM13 abrogated the repression of hTERT promoter activity as efficiently as the GC-box mutation hM12 (Fig. 2 c and d), consistent with previous findings (23). However, creation of this E2F-binding site at the corresponding position in the mTERT promoter failed to transform its transcriptional activity into hTERT-like function (mC2 and mC3 in Fig. 2 e and f). Although one conserved (at –112 to –104) and two nonconserved (at –132 to –124 and at –31 to –24) GC-boxes in the hTERT promoter were suggested to act as repressive elements by another study (22), the nonconserved GC-box at –31 to –24 (see hM12 in Fig. 2 a–d), but not the other two (see hM3 and hM5 in Fig. 2 a–d), was found to contribute substantially to the tight repression of hTERT transcription in our in vitro reporter-gene assays. Most importantly, when this human-specific GC-box was created at the corresponding position in the mTERT promoter (Fig. 2 a and b), its transcriptional activity became repressed to levels similar to that of the hTERT promoter activity (mC1 and mC1 + 3 in Fig. 2 e and f). Based on these results, we conclude that the hTERT-specific GC-box is a cis-acting element that determines the differential expression of TERT genes in humans and mice. Further details of the GC-box- and Sp1/Sp3-mediated, hTERT-specific repression remain to be investigated. Such investigation would also be important for better understanding of hTERT activation during human carcinogenesis, because many of the human cancer cell lines studied here appeared to undergo functional inactivation of the factor or signaling pathway acting on the GC-box to repress hTERT (Fig. 3). Whereas the vast majority of studies of transcriptional regulation of a given gene in different mammalian species have emphasized conserved regulatory mechanisms, this study is a detailed characterization of the functional difference in a gene promoter of mice versus humans. It will be interesting to examine whether the chromatin status in vivo differs between mTERT and hTERT and, if so, whether and how the nonconserved GC-box at the hTERT promoter is involved in the chromatin regulation.
Ritz et al. (33) recently generated transgenic mice carrying a lacZ reporter gene driven by the 8-kb hTERT promoter (hTERTp-lacZ mice) and found that the activity of the hTERT promoter in the mouse tissues generally recapitulates the expression of hTERT in human tissues, consistent with our findings. The BAC transgenic mice generated in this study, which carried the longer promoter region, all of the exons and introns, the downstream region of the hTERT gene, and the quantitative analysis of mTERT and hTERT expression, rather than detection of a reporter gene, allowed us to compare in vivo mTERT and hTERT expression in more physiological settings. This study provides an important insight into species-specific features of telomere biology. It has been demonstrated that the expression level of TERT can affect telomere length, because a decrease in mTERT gene dosage (heterozygous knockout mice) led to reduction in telomere length in vivo (34, 35). Our findings from hTERT transgenic mice revealed the differential cis-regulation of the mouse and human TERT genes in a tissue-specific manner, suggesting a critical mechanism that determines the differential telomerase activity in many, but not all, tissues between mice and humans (8) and may also contribute to the difference in telomere-length homeostasis between the species (9). The data on the differential transcriptional activity of mTERT and hTERT promoters, attributed to an hTERT-specific repressive cis-element, provide an experimental basis for creating mouse models of cancer and aging that more closely simulate human physiology (1). Engineering of an endogenous mTERT promoter that recapitulates the properties of the hTERT promoter (i.e., creation of the hTERT-specific repressive element or replacement with the hTERT promoter) could achieve the generation of mice expressing mouse telomerase in a human-like manner. Alternatively, introduction of a human telomerase RNA component gene into mTERT–/– mice carrying the hTERT BAC (Fig. 4), which are telomerase-negative because of incompatibility between hTERT and the mouse RNA component (36) (Y.J.C., unpublished observation), could also reconstitute the human telomerase enzyme in mice.
In addition to germ line, immune system, and hematopoietic organs, some human adult organs likely contain a stem-cell-like population with a potential to express hTERT and telomerase (37). Our real-time quantitative RT-PCR assay using whole normal organs as RNA sources did not efficiently detect hTERT expression from such a stem-cell-like population in liver, skeletal muscle, kidney, heart, lung, brain, or uterus (Fig. 1c), thus allowing us to observe and analyze the differential in vivo expression of mTERT and hTERT. In combination with in situ detection of hTERT expression, the transgenic mice described in this study would also be an excellent model for dissecting in vivo hTERT regulation in adult stem-cell-like populations and the role of this regulation in human aging and carcinogenesis (38).
It is unknown why the hTERT transgene in mouse brain behaves like endogenous mTERT rather than the hTERT in human brain, in contrast to the findings in all other tissues analyzed. The hTERT promoter-driven lacZ reporter gene was also expressed in brain in an hTERTp-lacZ mouse (33). A brain-specific transacting factor(s) may exist to regulate TERT transcription. Alternatively, the brain-specific expression pattern may reflect different subpopulations of TERT-expressing cells. For example, a unique structure exists in the subventricular zone in the adult human brain (39), where telomerase-expressing neural stem cells are enriched (40). Finally, although this study identified the species-specific regulation of TERT expression as a candidate for the determinant of the difference in telomere homeostasis between mice and humans, the structural or functional divergence of mouse and human telomere-binding proteins (41) may also contribute to the species-specific telomere biology. Possible differences in the regulation and function of TERT proteins also deserve investigation. Despite these issues still to be addressed, this study provides important, mechanistic insight into the species-specific and tissue-specific regulation of telomerase and telomeres.
Supplementary Material
Acknowledgments
We thank Dr. Snorri Thorgeirsson [National Cancer Institute (NCI)] for human liver RNA and Dr. Karen Hathcock for critical reading of the manuscript. This work was supported by the Intramural Research Program of the National Institutes of Health (NIH), NCI. E.M. is supported by a Japan Society for the Promotion of Science Research Fellowship for Japanese Biomedical and Behavioral Researchers at NIH.
Author contributions: I.H., Y.J.C., L.F., S.-H.L., V.L., R.J.H., and J.C.B. designed research; I.H., Y.J.C., T.P., L.F., S.-H.L., and E.M. performed research; I.H., Y.J.C., L.F., S.-H.L., E.M., and V.L. contributed new reagents/analytic tools; I.H., Y.J.C., T.P., E.M., V.L., R.J.H., and J.C.B. analyzed data; and I.H., Y.J.C., R.J.H., and J.C.B. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: BAC, bacterial artificial chromosome; MEF, mouse embryonic fibroblasts; NHF, normal human fibroblasts; TERT, telomerase reverse transcriptase; hTERT, human TERT; mTERT, mouse TERT.
References
- 1.Rangarajan, A. & Weinberg, R. A. (2003) Nat. Rev. Cancer 3, 952–959. [DOI] [PubMed] [Google Scholar]
- 2.Blackburn, E. H. (2005) FEBS Lett. 579, 859–862. [DOI] [PubMed] [Google Scholar]
- 3.Weng, N. P. & Hodes, R. J. (2000) J. Clin. Immunol. 20, 257–267. [DOI] [PubMed] [Google Scholar]
- 4.Campisi, J. (2001) Exp. Gerontol. 36, 607–618. [DOI] [PubMed] [Google Scholar]
- 5.Hanahan, D. & Weinberg, R. A. (2000) Cell 100, 57–70. [DOI] [PubMed] [Google Scholar]
- 6.Newbold, R. F. (2002) Mutagenesis 17, 539–550. [DOI] [PubMed] [Google Scholar]
- 7.Burger, A. M., Bibby, M. C. & Double, J. A. (1997) Br. J. Cancer 75, 516–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prowse, K. R. & Greider, C. W. (1995) Proc. Natl. Acad. Sci. USA 92, 4818–4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wright, W. E. & Shay, J. W. (2000) Nat. Med. 6, 849–851. [DOI] [PubMed] [Google Scholar]
- 10.Parrinello, S., Samper, E., Krtolica, A., Goldstein, J., Melov, S. & Campisi, J. (2003) Nat. Cell Biol. 5, 741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rangarajan, A., Hong, S. J., Gifford, A. & Weinberg, R. A. (2004) Cancer Cell 6, 171–183. [DOI] [PubMed] [Google Scholar]
- 12.Du, X., Shen, J., Kugan, N., Furth, E. E., Lombard, D. B., Cheung, C., Pak, S., Luo, G., Pignolo, R. J., DePinho, R. A., et al. (2004) Mol. Cell. Biol. 24, 8437–8446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang, S., Multani, A. S., Cabrera, N. G., Naylor, M. L., Laud, P., Lombard, D., Pathak, S., Guarente, L. & DePinho, R. A. (2004) Nat. Genet. 36, 877–882. [DOI] [PubMed] [Google Scholar]
- 14.Horikawa, I. & Barrett, J. C. (2003) Carcinogenesis 24, 1167–1176. [DOI] [PubMed] [Google Scholar]
- 15.Meyerson, M. (2000) J. Clin. Oncol. 18, 2626–2634. [DOI] [PubMed] [Google Scholar]
- 16.Greenberg, R. A., Allsopp, R. C., Chin, L., Morin, G. B. & DePinho, R. A. (1998) Oncogene 16, 1723–1730. [DOI] [PubMed] [Google Scholar]
- 17.Leem, S. H., Londono-Vallejo, J. A., Kim, J. H., Bui, H., Tubacher, E., Solomon, G., Park, J. E., Horikawa, I., Kouprina, N., Barrett, J. C. & Larionov, V. (2002) Oncogene 21, 769–777. [DOI] [PubMed] [Google Scholar]
- 18.Chiang, Y. J., Hemann, M. T., Hathcock, K. S., Tessarollo, L., Feigenbaum, L., Hahn, W. C. & Hodes, R. J. (2004) Mol. Cell. Biol. 24, 7024–7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiang, Y. J., Kole, H. K., Brown, K., Naramura, M., Fukuhara, S., Hu, R. J., Jang, I. K., Gutkind, J. S., Shevach, E. & Gu, H. (2000) Nature 403, 216–220. [DOI] [PubMed] [Google Scholar]
- 20.Horikawa, I., Cable, P. L., Mazur, S. J., Appella, E., Afshari, C. A. & Barrett, J. C. (2002) Mol. Biol. Cell 13, 2585–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nozawa, K., Maehara, K. & Isobe, K. (2001) J. Biol. Chem. 276, 22016–22023. [DOI] [PubMed] [Google Scholar]
- 22.Won, J., Yim, J. & Kim, T. K. (2002) J. Biol. Chem. 277, 38230–38238. [DOI] [PubMed] [Google Scholar]
- 23.Won, J., Chang, S., Oh, S. & Kim, T. K. (2004) Proc. Natl. Acad. Sci. USA 101, 11328–11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenberg, R. A., O'Hagan, R. C., Deng, H., Xiao, Q., Hann, S. R., Adams, R. R., Lichtsteiner, S., Chin, L., Morin, G. B. & DePinho, R. A. (1999) Oncogene 18, 1219–1226. [DOI] [PubMed] [Google Scholar]
- 25.Goueli, B. S. & Janknecht, R. (2004) Mol. Cell. Biol. 24, 25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lobanenkov, V. V., Nicolas, R. H., Adler, V. V., Paterson, H., Klenova, E. M., Polotskaja, A. V. & Goodwin, G. H. (1990) Oncogene 5, 1743–1753. [PubMed] [Google Scholar]
- 27.Hoare, S. F., Bryce, L. A., Wisman, G. B., Burns, S., Going, J. J., van der Zee, A. G. & Keith, W. N. (2001) Cancer Res. 61, 27–32. [PubMed] [Google Scholar]
- 28.Ulaner, G. A., Hu, J. F., Vu, T. H., Giudice, L. C. & Hoffman, A. R. (1998) Cancer Res. 58, 4168–4172. [PubMed] [Google Scholar]
- 29.Niemann, H. & Kues, W. A. (2000) Anim. Reprod. Sci. 60–61, 277–293. [DOI] [PubMed] [Google Scholar]
- 30.Rival-Gervier, S., Viglietta, C., Maeder, C., Attal, J. & Houdebine, L. M. (2002) Mol. Reprod. Dev. 63, 161–167. [DOI] [PubMed] [Google Scholar]
- 31.Leem, S. H., Kouprina, N., Grimwood, J., Kim, J. H., Mullokandov, M., Yoon, Y. H., Chae, J. Y., Morgan, J., Lucas, S., Richardson, P., et al. (2004) Genome Res. 14, 239–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kouprina, N., Leem, S. H., Solomon, G., Ly, A., Koriabine, M., Otstot, J., Pak, E., Dutra, A., Zhao, S., Barrett, J. C. & Larionov, V. (2003) EMBO Rep. 4, 257–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ritz, J. M., Kuhle, O., Riethdorf, S., Sipos, B., Deppert, W., Englert, C. & Gunes, C. (2005) Cancer Res. 65, 1187–1196. [DOI] [PubMed] [Google Scholar]
- 34.Erdmann, N., Liu, Y. & Harrington, L. (2004) Proc. Natl. Acad. Sci. USA 101, 6080–6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hathcock, K. S., Jeffrey Chiang, Y. & Hodes, R. J. (2005) Immunol. Rev. 205, 104–113. [DOI] [PubMed] [Google Scholar]
- 36.Chen, J. L. & Greider, C. W. (2003) EMBO J. 22, 304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Urbanek, K., Torella, D., Sheikh, F., De Angelis, A., Nurzynska, D., Silvestri, F., Beltrami, C. A., Bussani, R., Beltrami, A. P., Quaini, F., et al. (2005) Proc. Natl. Acad. Sci. USA 102, 8692–8697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bell, D. R. & Van Zant, G. (2004) Oncogene 23, 7290–7296. [DOI] [PubMed] [Google Scholar]
- 39.Sanai, N., Tramontin, A. D., Quinones-Hinojosa, A., Barbaro, N. M., Gupta, N., Kunwar, S., Lawton, M. T., McDermott, M. W., Parsa, A. T., Manuel-Garcia Verdugo, J., et al. (2004) Nature 427, 740–744. [DOI] [PubMed] [Google Scholar]
- 40.Caporaso, G. L., Lim, D. A., Alvarez-Buylla, A. & Chao, M. V. (2003) Mol. Cell. Neurosci. 23, 693–702. [DOI] [PubMed] [Google Scholar]
- 41.Kim, S. H., Parrinello, S., Kim, J. & Campisi, J. (2003) Genomics 81, 422–432. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.