Abstract
Survival of living cells and organisms is largely based on highly reliable function of their regulatory networks. However, the elements of biological networks, e.g., regulatory genes in genetic networks or neurons in the nervous system, are far from being reliable dynamical elements. How can networks of unreliable elements perform reliably? We here address this question in networks of autonomous noisy elements with fluctuating timing and study the conditions for an overall system behavior being reproducible in the presence of such noise. We find a clear distinction between reliable and unreliable dynamical attractors. In the reliable case, synchrony is sustained in the network, whereas in the unreliable scenario, fluctuating timing of single elements can gradually desynchronize the system, leading to nonreproducible behavior. The likelihood of reliable dynamical attractors strongly depends on the underlying topology of a network. Comparing with the observed architectures of gene regulation networks, we find that those 3-node subgraphs that allow for reliable dynamics are also those that are more abundant in nature, suggesting that specific topologies of regulatory networks may provide a selective advantage in evolution through their resistance against noise.
Keywords: genetic networks, biological computation, robustness, stability, computer model
The processes of life in cells and organisms rely on highly reproducible information processing. It has been a long-standing question how reliability in biological circuits is accomplished even though these involve elements with nonreproducible, noisy dynamics (1). In nerve cells, for example, firing of spikes is not fully determined by synaptic input (2). Similarly, in gene regulation, protein concentrations evolve in a quite irreproducible manner under given promoter levels (3–6). In the steady state of a system, such fluctuations can easily be dampened, even solely on the basis of properties of the single elements. In genetic transcription, for example, autoregulation of single genes successfully attenuates noise (7). In dynamical situations with rising and falling levels of activation, however, intrinsic noise of the interacting elements causes nonlocal effects, e.g., through fluctuations of switching delays (3, 8). In larger systems of interacting elements, such fluctuating delays can cause stability problems because, e.g., timing and coordination between the elements may fail. Such system failures may be incurable on the single-element level (e.g., single gene level) because, eventually, the entire circuit can be involved. In this work we study this problem in detail. We ask under which conditions a complex network of fluctuating dynamical elements can be stabilized by a suitable circuit design, maintaining an overall reproducible system dynamics. Our study is motivated by the extraordinary reliability of gene regulation networks as observed in living organisms and as exemplified by the dramatic consequences of rare system failures, e.g., at the origin of complex diseases.
The question of when a circuit is robust and the implications of desired reliability for circuit design are well known in electrical engineering. There, reliable design of electronic circuits is achieved by specific circuit architectures and has become a field of its own since the early days of electrical switching circuit design (9). In the study of biological circuits, the question of reliable circuit design, although being equally important, has not yet gained as much attention, mostly because of lack of detailed knowledge of the circuitry so far. However, during recent years, biochemical interactions involved in the information processing within or between cells of an organism have been characterized systematically, up to an emerging systemwide picture. Such networks have been obtained for gene regulation (10–13), including regulatory networks of entire cells as of yeast Saccharomyces cerevisiae (14, 15), as well as for signal transduction (16) and neuronal information processing (17). Such systemwide perspectives allow one to take a closer look at biological circuit design. Do biological networks show structural signatures that point to reliable design, similarly to the specific structures used in designed circuits?
Architectural features of biological signaling networks have been analyzed recently, drawing a surprisingly coherent picture for networks of different origins. Gene regulation networks, for example, when reduced to their graph representation (i.e., drawn as nodes and links), show strong mutual similarities despite their different origin and even when compared with networks of different function as hard-wired neural networks (18). When comparing each biological network with a randomized version of itself (19), some subgraphs are seen to be relatively abundant whereas others are strongly suppressed (20). This pattern is not shared by other, mostly nonsignaling networks, such as the World Wide Web, social acquaintance networks, and the graphs of word adjacency in various languages. Although this observation points at possible universal features in the wiring of signaling networks, its origin is a major open question at present.
We ask here whether architectural features of biological networks are related to the ability of reliable information processing. For this purpose we study reproducibility of dynamics in model networks of information-processing units in the presence of noise, here implemented as fluctuating response times of the nodes. Starting with the simplest system of two mutually coupled nodes, we observe that there are two distinct classes of dynamical circuit behavior. In the class of reliable dynamics the nodes regain synchronization after random perturbations, whereas for the class of unreliable dynamics the system does not self-synchronize. This distinction appears in a formulation as differential equations for continuous variables (as, e.g., protein concentrations), as well as in models with discrete binary state variables, both exhibiting the same synchronization properties. Turning to networks with three nodes, we find that the occurrence of reliable dynamics strongly depends on the underlying topology. One observes that reliable dynamics is more likely to appear on those triads that have been found as building blocks (motifs) in real biological networks. Further insight into the relationship between topology and reliability is gained by the analysis of cyclic behavior (attractors). Dynamics are reliable only if all switching events are connected by a single causal chain. An instructive example is isolated feedback loops, where the fraction of initial conditions with reliable dynamics is obtained easily. In the following, we start by studying the reliability of a signaling circuit consisting of two coupled nodes.
Feedback Loops of Two Nodes
Let us first formalize “reliability” of network dynamics in the presence of noise, using the simplest setting of two mutually interacting nodes. We define a criterion telling us when the deterministic dynamics of a network of dynamical nodes in the noiseless case is exactly reproducible in the presence of fluctuating switching times. Note that this requirement (which we call reliability in the course of this work) goes beyond the common definition of dynamical “robustness” or “stability” in discrete dynamical networks. The latter as, e.g., studied for Boolean networks recently, simply are synonymous to the property that errors induced by state perturbations do not percolate through the system (21–24). Our previously undescribed reliability criterion, in contrast, focuses on the standard mode of operation of the network, i.e., in the absence of discrete state errors, and allows for testing the network dynamics with infinitesimally small perturbations.
Two scenarios can be distinguished in a system with two nodes. (i) Nodes influence each other with the opposite sign. For instance node 2 represses node 1 while node 1 activates node 2. (ii) Both nodes have the same coupling, i.e. either both couplings are activating or both are repressing. Starting with scenario (i), let us describe the dynamics of the system by rate equations for normalized concentrations (resp. firing rates) u, v ∈ [0, 1] of the two nodes
 |
[1] |
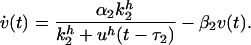 |
[2] |
The time constants αi and βi are the rates of production and degradation of the respective messenger substances. In the neural system they represent the time constants of the varying firing rate. The nonlinear production term with Hill exponent h describes collective chemical effects or a nonlinear neural response function, respectively. Note that the rate equations involve explicit delay times τ1 and τ2 for signal transmission. By randomly varying these delay variables in time, an uncertainty in response times of the dynamical elements is implemented, as motivated by the omnipresence of noise in biological circuits. To be definite, we define the initial functions of these delay differential equations by trivially extending the constant value of a fixed initial condition for each node into a sufficiently long time interval into the past. This is a natural extension, similar to gene regulation circuits in the silent state, then being activated by a constant stream of newly arriving chemical messengers.
Delay differential equations of the type 1 and 2 have been studied over the past two decades, mostly in the context of neural networks and with constant delays. The aspect of dynamical stability in the presence of noise has been a central question, and this two-neuron model system has been used to approach it in a simple and analytically accessible way (25) (for a list of references on the stability of two-neuron systems, see reference 2 in ref. 26). With delay as a potential source of oscillations, a well studied stability criterion has been the case of absence of oscillations (25), similar to stable fixed points in artificial neural networks with symmetric weights. In networks with asymmetric weights, long transients occur easily (26).
In the context of gene regulation networks, however, the criterion of reliable operation is fundamentally different from the traditional stability criterion of stable fixed points or absence of oscillations in the presence of delays. In fact, oscillations are often part of the regular function of a genetic network (27). The central question to be asked here is: How is the exact time course of transients and attractors sustained in networks of biological and, therefore, noisy and unreliable elements? To mimic this situation, here we study circuits with variable delays. The resulting potential for rearrangements in the time ordering of switching events is the major threat for dynamical reliability of the system.
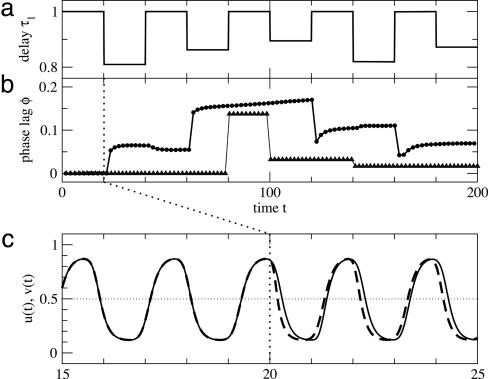
Fig. 1 shows the above system under randomly varying transmission delay τ1. The variables u(t) and v(t) oscillate with a time lag that depends on the current transmission delay τ1. Note that the dynamics remains qualitatively the same, independently of delay τ1, v(t) always lagging behind u(t) by ≈¼ of an oscillation period. Further note that the current phase lag does not depend on the history of the system. The system retains synchronization irrespective of the fluctuations in the transmission delay τ1.
Fig. 1.
Dynamics of the feedback loop with antisymmetric coupling. (a) Fluctuations of transmission delay τ1.(b) Evolution of phase lag between u(t) and v(t) for the continuous case (filled circles) and the discrete approximation (open triangles). Compare Eqs. 1, 2 and 3, 4, respectively. (c) The phase lag is defined as the time difference between subsequent passages of variables u (dashed curve) and v (solid curve) across the value ½ from below, as indicated by the black double arrows. Parameters in Eqs. 1 and 2 have values τ2 = 0.5, α1 = α2 = β1 = β2 = 10, k1 = k2 = 101/2, h = 2. Broad variations of these parameters give qualitatively the same results.
The sigmoid response curves allow a further simplification via approximating them by step functions. Considering the limit of fast production and degradation αi, βi → ∞ in Eqs. 1 and 2 we obtain the simplified dynamics
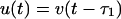 |
[3] |
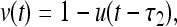 |
[4] |
with binary-state variables u, v ∈ {0, 1}. The initial functions are chosen as in the continuous example.
When turning to discrete maps we have to keep in mind that in certain parameter regimes even Boolean difference equations may exhibit extraordinarily complex dynamics as a result of the delays (28, 29). In this study, however, we consider systems that are far away from such parameter conditions. Mapping our continuous equations onto simple discrete maps is meaningful here because the nodes always work in a stable, nonchaotic mode. This fact is due to the low-pass filter characteristics and the delay of the original nodes as defined by Eqs. 1 and 2. Thus, while dynamical systems with delay are often intuitively associated with chaotic system behavior, in our present system delay is a stabilizing factor, as also is observed in different contexts of coupled maps (30). Fig. 1 shows that the approximate description by means of discrete functions has the same qualitative behavior as the original continuous-rate equations.
Let us now turn to case (ii) above, with two mutually coupled nodes with the same type of coupling in both directions. Here we take both couplings to be inhibitory. Again we describe the system in terms of rate equations
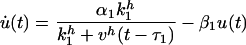 |
[5] |
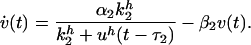 |
[6] |
The approximation by binary-state variables now reads
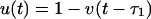 |
[7] |
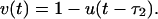 |
[8] |
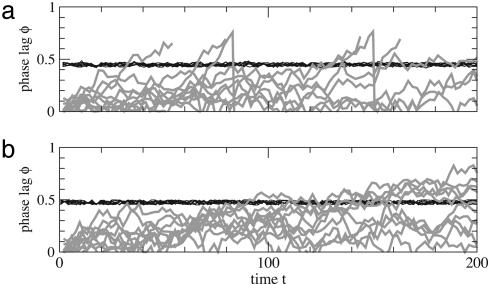
The dynamics for the continuous and the binary system are shown in Fig. 2. Initially, the variables u(t) and v(t) are perfectly synchronized, due to identical initial values. Changing the transmission delay τ1 causes a nonzero phase lag that is retained even when the transmission delay is reset to the original value. In fact, varying τ1 inside a small interval causes accumulation of phase lag between u(t) and v(t). The system does not self-synchronize and may therefore be driven out of phase by slight fluctuations in the transmission delays. Consequently, system (i) with antisymmetric interaction and system (ii) with symmetric interactions differ with respect to reproducibility (see Fig. 3). With transmission delays varying randomly with time and across several runs, system (i) shows reproducible behavior. The phase lag practically remains constant, as the oscillation with a given phase lag is the stable (attractive) mode. In contrast, the behavior of system (ii) varies across runs. Synchronized oscillation is a marginally stable mode of this system such that the fluctuations drive the system away from this mode. We call these systems nonreproducible (or unreliable).
Fig. 2.
Dynamics of the feedback loop with symmetric coupling (Eqs. 5 and 6) under fluctuations of the transmission delay τ1. We keep τ2 = 1.0 constant; all other parameters and plotting details for a, b, and c are the same as in Fig. 1 a, b, and c, respectively.
Fig. 3.
Systems with and without stable synchrony. (a) Phase lag as a function of time for systems of two mutually coupled nodes with continuous state variables under fluctuating transmission delays. The system with antisymmetric coupling (Eqs. 1 and 2) remains synchronous with a phase lag close to 0.5 (black curves). In the system with symmetric coupling (Eqs. 5 and 6) the phase lag between the oscillating nodes is not stable against fluctuations (gray curves). (b) Same as in a for systems with discrete (Boolean) state variables with antisymmetric coupling (Eqs. 3 and 4) and symmetric coupling (Eqs. 7 and 8).
Three Nodes
In the following, we shall see that the clear distinction between reproducible dynamics with intrinsically stable synchronicity and nonreproducible dynamics sensitive to fluctuations extends beyond simple oscillations in systems of two nodes. Let us study dynamics on 3-node circuits as shown in Fig. 4 Top. We now need to define the dynamics of nodes with more than one input. Consider a node i that is directly influenced by nodes j and k. Restricting ourselves to the binary approximation of states in the following, node i switches according to
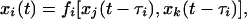 |
[9] |
where we define τi as the (time-dependent) transmission delay of node i. The Boolean function fi maps the four pairs of binary states (xj, xk) to the set {0, 1}. We choose fi from the set of canalizing functions (31), i.e., we do not use the exclusive-or function and its negation. From the 14 canalizing functions we further exclude those that are constant with respect to one or both of the inputs. We are then left with eight Boolean functions of two inputs (these are OR, AND, and their variants generated by negation of one or both inputs). For nodes with one input, we only allow the nonconstant Boolean functions Identity (output = input) and Negation (output ≠ input).
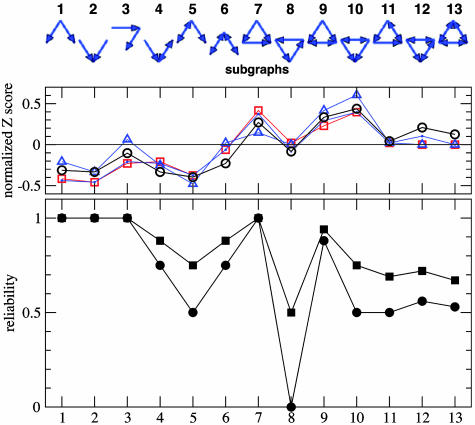
Fig. 4.
Abundance and dynamical reliability of subgraphs. (Top) All directed connected graphs of three nodes. (Middle) Experimental network profiles based on the normalized Z-score for various real networks: Signal transduction in mammalian cells (squares), genetic transcription for development in the fruit fly (dots), and the sea-urchin (triangles), and synaptic connections in Caenorhabditis elegans (circles). See ref. 18 for details. (Bottom) Graph reliability (squares) and strict graph reliability (circles) as defined in the text.
As “graph reliability” we define the probability of obtaining reliable dynamics when preparing a random initial condition and a random assignment of Boolean functions. The “strict graph reliability” is the probability that all initial conditions yield reliable dynamics for a random assignment of Boolean functions. The small system size allows us to obtain the exact values of these quantities by full enumeration of all combinations of initial conditions and function assignments. See Methods for details on the simulation procedure.
Viewing the results in Fig. 4 Bottom, we first note that feed-forward wiring (subgraphs 1, 2, 3, and 7) always yields reliable dynamics: From any initial condition the system reaches a fixed point after a short time. Such motifs are frequently seen in genetic networks (12). For example the feed-forward loop, 7, forms a central part of the flagella motor network of Escherichia coli (32). The least reproducible dynamics is obtained for the pure feedback loop (subgraph 8) with strict reliability zero. For an explanation see the below section on Feedback Loops of Arbitrary Length. This motif is rarely seen in biological networks and is best known from the stable oscillatory mode of its artificial implementation, the so-called “repressilator” (33). Interestingly, the feedback loop with an additional link in the “opposite” direction (subgraph 11) has a larger strict reliability ½. Now let us compare the dynamical reliability of the subgraphs with their relative abundance in some examples of biological signaling networks shown in Fig. 4 Middle. The relative abundance of a specific subgraph is here quantified by the so-called Z-score, comparing the occurrences of this subgraph in the real network Nreali with its average occurrence 〈Nrandi 〉 in an ensemble of randomized instances of this same network. The randomized network is obtained, e.g., by swapping a link between unconnected pairs of connected links: This process conserves the network at the single-node level (i.e., numbers of in- and out-links of each node), but shuffles the higher-order connectivity patterns (19). The Z-score Zi of each subgraph i is calculated as
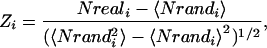 |
[10] |
and the resulting vector Zi normalized to length one (18). As shown in Fig. 4, our profile for dynamical reliability of the subgraphs is similar to the set of complex signaling networks. Let us compare this set in more detail with the dynamical predictions of reliability. Consider first subgraphs 7–13, which contain a closed triad. Among these, subgraph 7 (the feed-forward loop) and subgraph 9 (two bidirectionally coupled nodes receiving from a common third node) have particularly large reliability. Both subgraphs are also observed to be highly abundant in biological signaling networks. Furthermore, subgraph 12 has larger reliability than subgraphs 11 and 13. This superiority is also reflected by abundance measure in the networks. The large abundance of subgraph 10, however, cannot be predicted from the dynamical reliability. For the remaining subgraphs 1–6, wiring diagrams without closed triads, there is a clear correlation between the empirical Z-score and our measure of dynamical reliability.
The presence or absence of a given subgraph cannot be fully explained by the reliability measure presented here. First, the Z-score tends to increase with the number of connections in the subgraph. The bias towards densely (but not fully) connected motifs is a consequence of the networks' modular structure with functional clusters of nodes (34). This property of the networks is not directly related to robust dynamics and, therefore, is not reflected in our reliability measure. Second, we have neglected the network environment of the motifs, which may greatly change the dynamics. For instance, the feed-forward subgraph 3 alone yields perfectly reliable dynamics. The 4-node feedback loop 1 → 2 → 3 → 4 → 1, however, does not give reliable dynamics (as shown below), even though subgraph 3 is its only motif. Third, we have defined reliability of a subgraph by averaging over all assignments of plausible functions. In reality, however, the nodes' functions may be correlated with the wiring diagram. For instance, subgraph 10 gives perfectly reliable dynamics if one assumes antisymmetric influence (one promoter, one repressor) between the bidirectionally linked nodes. Despite these limitations of the present analysis, the correlation between abundance and reliability of subgraphs in Fig. 4 is remarkable. In the following we gain more detailed insight into the mechanism leading to reliability by considering attractors in feedback loops.
Attractors and Causality
Even though in biological systems transmission delays fluctuate, it is instructive to regard constant transmission delays as a reference case. Setting τi = 1 for all nodes i in Eq. 9, we recover the time-discrete synchronous update mode often employed in Boolean network models. In this idealized picture, all signal transmissions require exactly the same time, and nodes flip synchronously as if driven by a central clock. The deterministic dynamics eventually reaches a periodic attractor, an indefinitely repeated sequence of network states as illustrated in Fig. 5. Reliability under fluctuating transmission delays becomes an issue when noise perturbs the stability of this perfectly synchronized mode. When slightly perturbed by one retarded signal transmission, does the system autonomously reestablish synchronicity? Or does the system “remember” the perturbed timing? In the latter case, just as in Fig. 2, a series of perturbations may add up to drive the system away from the predictable periodic behavior by destroying the causal structure of the attractor. Let us call a “flipping event” a pair (i, t), given that node i changes state at time t. For the attractor in Fig. 5 we draw arrows from each flipping event to all flipping events it causes to happen in the next time step. In the resulting plot, Fig. 6a, there is a single closed chain of flipping events. Retardation of one event by a time tret simply retards all subsequent events by the same amount of time. Event (i, t) in the unperturbed scenario becomes event (i, t + tret), but the sequence of states encountered by the system remains the same. This built-in compensation of fluctuations renders the dynamics reliable.
Fig. 5.
Attractor of a Boolean network. (Left) A Boolean network with N = 3 nodes with possible states 0 and 1. Each node has a lookup table to determine its state given the input from other nodes. Here the two light nodes simply copy their single input, while the dark node performs the Boolean function NOR on its inputs. (Right) In the deterministic case of constant transmission delays, the system always reaches a reproducible periodic sequence of states. Filled nodes are in state 1 (on); open nodes in state 0 (off).
Fig. 6.
Causal structure of attractors. (a) The attractor shown in Fig. 5 has one causal chain of flipping events triggering each other. (b) A different attractor (on the same wiring diagram) with two independent causal chains of flipping events. The attractor in b is obtained after replacing the NOR in Fig. 5 with the Boolean function that gives 1 if and only if it receives 0 from the top node and 1 from the left node.
An example of an unreliable dynamical attractor is shown in Fig. 6b. In this case there are two separate chains of flipping events, one connecting the on-events and the other connecting the off-events. Retarding an event in one of the chains does not influence the timing of the events in the other chain. By repeatedly retarded on-events, the time span that a node spends in the on-state is gradually reduced and eventually reaches zero. Then the system is caught in a fixed point and does not follow the attractor any longer. In general, the dynamics is reliable if and only if the attractor contains exactly one causal chain.
Feedback Loops of Arbitrary Length
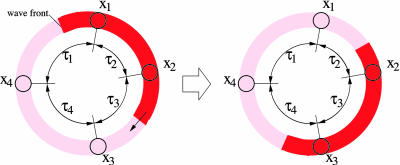
The relation between topology and reproducibility is particularly obvious in isolated feedback loops. Such loops are rare in biological contexts [short oscillating loops occur in the gene networks controlling the cell cycle (34)], however, are tractable and therefore useful in theoretical models. Consider N ≥ 2 nodes connected in a directed cycle, i.e., each node receives input only from its (clockwise) predecessor. If node i – 1 changes state (“flips”) at time t, node i will change state at time t + τi, node i + 1 will change state at time t + τi + τi+1 and so forth. The dynamics can be interpreted as “wave fronts” of flipping events traveling at constant speed on a ring. The nodes are located on this ring at distances given by the transmission delays τi, as illustrated in Fig. 7. For constant transmission delays, the dynamics is periodic (with period τ1 + ··· + τN). However, when a transmission delay τi fluctuates, consecutive passages of wave fronts from node i – 1 to node i take different times. Eventually wave fronts encounter and annihilate: Flipping from 0 to 1 and back to 0 at the same time results in no flipping at all. Annihilations of wave fronts happen stochastically because they are driven by the random fluctuations of transmission delays.
Fig. 7.
The wave front picture of dynamics in a feedback loop with N = 4 nodes all performing the Boolean function Identity.
Consequently, the dynamics is reproducible only if wave fronts cannot annihilate. This condition is fulfilled in the cases of a single wave front or no wave fronts at all. In the latter case the state of the system is a fixed point. When all nodes perform the function Identity, as in the example of Fig. 7, the number of wave fronts is even. Two or more wave fronts can annihilate, eventually leading to irreproducible dynamics. The only reproducible dynamics in this case is a system that stays on a fixed point, corresponding to zero wave fronts. However, if one of the nodes performs the function Negation then this node acts as a resting wave front, because states on the two sides are always different. The total number of wave fronts (including the resting one) is still even, but now the number of traveling wave fronts is odd. Initial conditions exist such that there is a single traveling wave front, giving reproducible dynamics. The two cases generalize easily. For an even number of inhibitory couplings (i.e., an even number of nodes performing Inversion) the dynamics is reliable if and only if one of the two fixed points is chosen as initial condition. Then the fraction of initial conditions with reproducible dynamics is
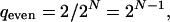 |
[11] |
for a feedback loop of N nodes. Analogously we find for the feedback loop with an odd number of inhibitory couplings
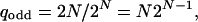 |
[12] |
where 2N initial conditions generate a singular wave front. Note that the only feedback loop that yields reproducible dynamics for all initial conditions has N = 2 nodes, one performing Inversion and the other Identity. This is the system studied as case (i) given by Eqs. 1 and 2.
Summary and Conclusions
Biological information processing systems as circuits of intrinsically noisy elements are constrained by the need for reproducible output. Effects of fluctuations on the system level can be suppressed through a suitable circuit design. This finding is a main result of our study of the influence of network topology on the reliability of information processing in networks of switches with fluctuating response times. In the reliable scenario, the elements cooperatively suppress fluctuations and tend to synchronize their operations. In the unreliable scenario, in contrast, networks desynchronize and show irreproducible behavior when response times fluctuate.
The occurrence of the two dynamical classes is strongly biased by the topology. Whether or not the system shows reliable dynamics can to a large degree be deduced from the unlabeled wiring diagram without information on the type of couplings and functions of switches.
When comparing our findings with empirical networks of genetic transcription, signal transduction, and the nervous system, we observe that the statistics of their local wiring structure is closely related to our reliability measure. Reliable triads tend to occur significantly more frequently in natural networks than in randomized versions of the networks, whereas unreliable triads are typically suppressed.
Our study suggests that biological signaling networks (as gene or neural networks) are shaped by the selective advantage of the ability to robust signal processing. This finding adds to the earlier evidence that the dynamical attractor of the yeast cell cycle is already robustly designed against switching errors as well as mutations of single genes (35). We suggest here that a second mechanism secures the standard operation mode against noise, even before any switching errors occur.
There are numerous ways to extend this work. Quite obviously, in our dynamical study of 3-motifs we have neglected the network context by assuming all external input to be constant. Further work could go beyond this limitation by looking at larger networks. Another interesting outlook is a study of the dynamics on an empirical network (23) and to compare its reliability with rewired counterparts. We expect that dynamical studies on biological network topologies will teach us about origin and function of these systems, even while we are still lacking full knowledge of network properties and dynamics (36). Reliable dynamics is a key property of biological signaling networks.
Methods
Differential equations are integrated by first-order Euler method using a time increment Δt = 10–5. For systems with Boolean variables (Eqs. 3, 4, and 7–9) integration is performed with continuous time t (exact up to machine precision). In the simulations in Three Nodes, transmission delays τi are varied as follows: Whenever node i changes state, a new target delay time τ*i is drawn from the homogeneous distribution on the interval [0.9; 1]. Then the delay τi approaches the target value τ*i according to 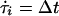 sgn (τ*i – τi) (otherwise, if τi were set to a new value abruptly, temporal order of signal transmission would not be conserved).
sgn (τ*i – τi) (otherwise, if τi were set to a new value abruptly, temporal order of signal transmission would not be conserved).
Criterion for reliability: For a given dynamical scenario (choice of Boolean functions and initial condition) 100 independent simulation runs of duration T = 1,000 are performed, and the sequence of sustained states is recorded. A sustained state is a state (x1,..., xN) that is assumed by the system at least for a time span of tsus = ½. The given dynamical scenario on the given subgraph is called reproducible if the series of sustained states of all 100 runs are identical. Varying the number of runs and their duration by one order of magnitude does not change the set of reliable scenarios.
Acknowledgments
We thank Ron Milo for providing material from ref. 18 and Yigal Nochomovitz as well as three anonymous referees for valuable comments. This work was supported by the Deutsche Forschungsgemeinschaft DFG through the Interdisciplinary Center for Bioinformatics at the University of Leipzig where most of this work was done.
Author contributions: S.B. designed research; and K.K. and S.B. performed research and wrote the paper.
Conflict of interest statement: No conflicts declared.
References
- 1.Rao, C. V., Wolf, D. M. & Arkin, A. P. (2002) Nature 420, 231–237. [DOI] [PubMed] [Google Scholar]
- 2.Allen, C. & Stevens, C. F. (1994) Proc. Natl. Acad. Sci. USA 91, 10380–10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McAdams, H. H. & Arkin, A. (1997) Proc. Natl. Acad. Sci. USA 94, 814–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elowitz, M. B., Levine, A. J., Siggia, E. D. & Swain, P. S. (2002) Science 297, 1183–1186. [DOI] [PubMed] [Google Scholar]
- 5.Rosenfeld, N., Young, J. W., Alon, U., Swain, P. S. & Elowitz, M. B. (2005) Science 307, 1962–1965. [DOI] [PubMed] [Google Scholar]
- 6.Pedraza, J. M. & van Oudenaarden, A. (2005) Science 307, 1965–1969. [DOI] [PubMed] [Google Scholar]
- 7.Becskei, A. & Serrano, L. (2000) Nature 405, 590–593. [DOI] [PubMed] [Google Scholar]
- 8.Beierholm, U., Nielsen, C. D., Ryge, J., Alstrom, P. & Kiehn, O. (2001) J. Neurophysiol. 86, 1858–1868. [DOI] [PubMed] [Google Scholar]
- 9.Keister, W., Ritchie, A. E. & Washburn, S. H. (1951) The Design of Switching Circuits, The Bell Telephone Laboratories Series (Van Nostrand, New York).
- 10.Thieffry, D., Huerta, A. M., Pérez-Rueda, E. & Collado-Vides, J. (1998) BioEssays 20, 433–440. [DOI] [PubMed] [Google Scholar]
- 11.Davidson, E. H., Rast, J. P., Oliveri, P., Ransick, A., Calestani, C., Yuh, C. H., Minokawa, T., Amore, G., Hinman, V., Arenas-Mena, C., et al. (2002) Science 295, 1669–1678. [DOI] [PubMed] [Google Scholar]
- 12.Shen-Orr, S., Milo, R., Mangan, S. & Alon, U. (2002) Nat. Genet. 31, 64–68. [DOI] [PubMed] [Google Scholar]
- 13.Levine, M. & Davidson, E. H. (2005) Proc. Natl. Acad. Sci. USA 102, 4936–4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee, T. I., Rinaldi, N. J., Robert, F., Odom, D. T., Bar-Joseph, Z., Gerber, G. K., Hannett, N. M., Harbison, C. T., Thompson, C. M., Simon, I., et al. (2002) Science 298, 799–804. [DOI] [PubMed] [Google Scholar]
- 15.Tong, A. H. Y., Lesage, G., Bader, G. D., Ding, H., Xu, H., Xin, X., Young, J., Berriz, G. F., Brost, R. L., Chang, M., et al. (2004) Science 303, 808–813. [DOI] [PubMed] [Google Scholar]
- 16.Galperin, M. Y. (2004) Environ. Microbiol. 6, 552–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White, J. G., Southgate, E., Thompson, J. N. & Brenner, S. (1986) Philos. Trans. R. Soc. London Ser. B 314, 1–340. [DOI] [PubMed] [Google Scholar]
- 18.Milo, R., Itzkovitz, S., Kashtan, N., Levitt, R., Shen-Orr, S., Ayzenshtat, I., Sheffer, M. & Alon, R. (2004) Science 303, 1538–1542. [DOI] [PubMed] [Google Scholar]
- 19.Maslov, S. & Sneppen, K. (2002) Science 296, 910–913. [DOI] [PubMed] [Google Scholar]
- 20.Milo, R., Shen-Orr, S., Itzkovitz, S., Kashtan, N., Chklovskii, D. & Alon, U. (2002) Science 298, 824–827. [DOI] [PubMed] [Google Scholar]
- 21.Aldana, M. & Cluzel, P. (2003) Proc. Natl. Acad. Sci. USA 100, 8710–8714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shmulevich, I., Lähdesmäki, H., Dougherty, E. R., Astola, J. & Zhang, W. (2003) Proc. Natl. Acad. Sci. USA 100, 10734–10739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kauffman, S., Peterson, C., Samuelsson, B. & Troein, C. (2003) Proc. Natl. Acad. Sci. USA 100, 14796–14799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kauffman, S., Peterson, C., Samuelsson, B. & Troein, C. (2004) Proc. Natl. Acad. Sci. USA 101, 17102–17107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marcus, C. M. & Westervelt, R. M. (1989) Phys. Rev. A 39, 347–359. [DOI] [PubMed] [Google Scholar]
- 26.Pakdaman, K., Grotta-Ragazzo, C. & Malta, C. P. (1998) Phys. Rev. E 58, 3623–3627. [Google Scholar]
- 27.Hirata, H., Yoshiura, S., Ohtsuka, T., Bessho, Y., Harada, T., Yoshikawa, K. & Kageyama, R. (2002) Science 298, 840–843. [DOI] [PubMed] [Google Scholar]
- 28.Dee, D. & Ghil, M. (1984) SIAM J. Appl. Math. 44, 111–126. [Google Scholar]
- 29.Ghil, M. & Mullhaupt, A. (1985) J. Stat. Phys. 41, 125–173. [Google Scholar]
- 30.Atay, F. M., Jost, J. & Wende, A. (2004) Phys. Rev. Lett. 92, 144101. [DOI] [PubMed] [Google Scholar]
- 31.Kauffman, S. A. (1984) Physica D 10, 145–156. [Google Scholar]
- 32.Kalir, S., Mangan, S. & Alon, U. (2005) Mol. Syst. Biol. 4100010, E1–E6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elowitz, M. B. & Leibler, S. (2000) Nature 403, 335–338. [DOI] [PubMed] [Google Scholar]
- 34.Milo, R., Itzkovitz, S., Kashtan, N., Levitt, R. & Alon, R. (2004) Science 305, 1107d. [Google Scholar]
- 35.Li, F., Long, T., Lu, Y., Ouyang, Q. & Tang, C. (2004) Proc. Natl. Acad. Sci. USA 101, 4781–4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bornholdt, S. (2005) Science 310, 449–451. [DOI] [PubMed] [Google Scholar]