Abstract
A study of the Thermus thermophilus chorismate mutase (TtCM) is described by using quantum mechanics (self-consistent-charge density-functional tight binding)/molecular mechanics, umbrella sampling, and the weighted histogram analysis method. The computed free energies of activation for the reactions in water and TtCM are comparable to the experimental values. The free energies for formation of near attack conformer have been determined to be 8.06 and 0.05 kcal/mol in water and TtCM, respectively. The near attack conformer stabilization contributes ≈90% to the proficiency of the enzymatic reaction compared with the reaction in water. The transition state (TS) structures and partial atom charges are much the same in the enzymatic and water reactions. The difference in the electrostatic interactions of Arg-89 with O13 in the enzyme–substrate complex and enzyme–TS complex provides the latter with but 0.55 kcal/mol of 1.92 kcal/mol total TS stabilization. Differences in electrostatic interactions between components at the active site in the enzyme–substrate complex and enzyme–TS complex are barely significant, such that TS stabilization is of minor importance and the enzymatic catalysis is through an entropic advantage.
Keywords: potential of mean force, self-consistent-charge density-functional tight binding, quantum mechanics/molecular mechanics
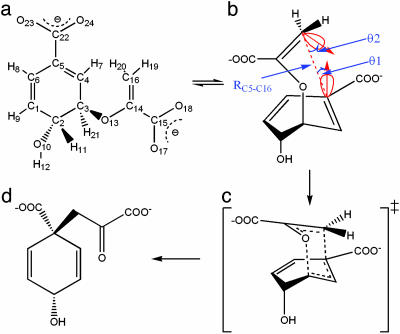
The contributions of ground-state (GS) conformations and transition-state (TS) stabilization to the proficiency of an enzymatic reaction can be more easily understood by studying a simple one-substrate enzyme reaction that involves the intramolecular rearrangement of a substrate to product without formation of a covalent intermediate (1, 2). The Claisen rearrangement of chorismate to prephenate is a well studied example (3) as shown in Fig. 1 where the reactive conformation near attack conformer (NAC) is defined.
Fig. 1.
Diagram of the chorismate → prephenate reaction. (a) Chorismate. (b) NAC of the chorismate is defined by three criteria: RC5-C16, θ1, and θ2. θ1 is the angle of the C16 approach to the C5 π -orbital. θ2 is the C16 π -orbital direction relative to the C5 atom. In the TS, θ1 = 8.2° and θ2 = 18.6°. For the NACs, the bonding angles θ1 and θ2 are allowed ±20° deviations from the TS structure. The distance RC5-C16 ≤ 3.7 Å. (c) TS. (d) Prephenate.
Our choice to research the Thermus thermophilus chorismate mutase (TtCM) relates to our current interest in the chorismate → prephenate reaction (4–6) and the questions of the temperature specificity of thermophilic enzymes (7). TtCM belongs to a rare group of the AroH-type monofunctional CMs that are principally found in Gram-positive bacteria and have been discovered as a thermophilic enzyme in the Gram-negative T. thermophilus (8, 9). The experimental free energies of activation 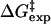 are 15.2 and 24.2 kcal/mol for the reactions in TtCM (343 K, the bacterium's optimal growth temperature) and water (298 K) (10), respectively. In this article, we describe a quantum mechanics (QM)/molecular mechanics (MM) (11) and umbrella sampling (12) study of the reactions at 343 K for TtCM and 298 K for water.
are 15.2 and 24.2 kcal/mol for the reactions in TtCM (343 K, the bacterium's optimal growth temperature) and water (298 K) (10), respectively. In this article, we describe a quantum mechanics (QM)/molecular mechanics (MM) (11) and umbrella sampling (12) study of the reactions at 343 K for TtCM and 298 K for water.
Methods
The initial coordinates of TtCM were modified from the x-ray structure of the F55S mutant of the TtCM trimer (Protein Data Bank ID code 1ODE; resolution, 1.65 Å) (8). The QM/MM simulations were carried out with charmm version 31b1 by the use of the self-consistent-charge density functional tight binding (SCCDFTB) module (13–15). For the simulations in the active site of TtCM, the chorismate was treated as the QM region. The enzyme–substrate complex (E·S) was solvated with a water cap of 25 Å radius centered at the QM region and consisted of 1,542 TIP3P water molecules (16). Stochastic boundary conditions were applied to the system (17). Umbrella sampling (18) was used to sample the conformational ensembles of the reactions. NAC definition of C5-C16 distance was chosen as the reaction coordinate in umbrella sampling. The same procedure was used for the simulations of the chorismate in water. The free energies of activation for the reactions in water [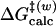 ] and TtCM [
] and TtCM [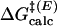 ] were determined by the potential of mean force (PMF). The free energies for NAC formation in water and enzyme were calculated from the mole fraction of the chorismate in the GS present as NAC. Detailed methods of initial coordinate preparation, QM/MM simulation setup, umbrella sampling, and calculation of free energy for NAC formation are available in Supporting Text, which is published as supporting information on the PNAS web site.
] were determined by the potential of mean force (PMF). The free energies for NAC formation in water and enzyme were calculated from the mole fraction of the chorismate in the GS present as NAC. Detailed methods of initial coordinate preparation, QM/MM simulation setup, umbrella sampling, and calculation of free energy for NAC formation are available in Supporting Text, which is published as supporting information on the PNAS web site.
Results
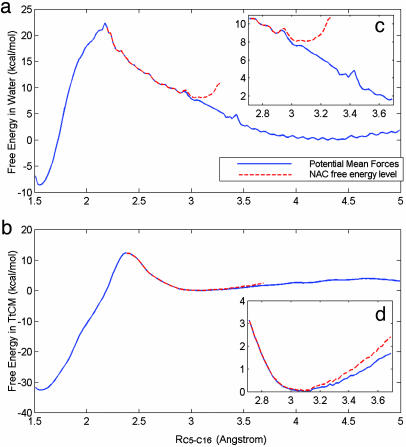
PMF Profiles Obtained from Umbrella Sampling. The free-energy profiles for the chorismate → prephenate reaction along the C5-C16 reaction coordinate in both water and the active site of TtCM, as obtained by SCCDFTB/MM and umbrella sampling, are shown in Fig. 2. The PMF profile for the reaction in water is plotted as a blue solid line, and the free-energy profile of NAC formation is plotted in a red dashed line. The activation barriers (TS) are present at reaction coordinate RC5-C16 of 2.17 and 2.37 Å in water and TtCM, respectively. The minima of the free-energy profile of NAC formation in water and TtCM are present at RC5-C16 of 3.03 and 3.09 Å. The most favorable distance of C5-C16 for NAC formation in enzyme is comparable to that in water. Thus, the substrate forms similar NAC species regardless of the environments.
Fig. 2.
Free energy profiles. (a and b) Plots of the free energy profiles of the chorismate → prephenate reactions in water (a) and TtCM (b). The blue solid lines are PMF profiles and red dashed lines are the free energy profiles of NAC formation. (c and d) Within the region of 2.7 ≤ RC5-C16 ≤ 3.7 Å, the free energy profiles were plotted in high resolution. Both PMF and NAC free energy profiles are offset by the values of free energy minima for the chorismate in GSs in PMF.
The free energies of activation in water and TtCM (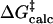 ) are 22.31 and 12.38 kcal/mol, which are in reasonable agreement with the experimental values of 24.5 and 15.4 kcal/mol, respectively (Table 1). The free energy of activation in the enzymatic reaction is 9.93 kcal/mol lower than the free energy of activation in water. The minimum of the free-energy profile of NAC formation in water [
) are 22.31 and 12.38 kcal/mol, which are in reasonable agreement with the experimental values of 24.5 and 15.4 kcal/mol, respectively (Table 1). The free energy of activation in the enzymatic reaction is 9.93 kcal/mol lower than the free energy of activation in water. The minimum of the free-energy profile of NAC formation in water [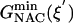 ] is 8.06 kcal/mol, which is in good agreement with previous results (8.1 kcal/mol) obtained from thermodynamic integration (4). In the enzymatic reaction, free energy for NAC formation is 0.73 kcal/mol when RC5-C16 is the distance of the NAC criteria (3.7 Å). The minimum of the free-energy profile of NAC formation in TtCM is 0.05 kcal/mol. Therefore, the GS conformations of chorismate in the active site of TtCM are essentially NACs. Using the experimental value of the free energy of reaction in enzyme (9.1 kcal/mol), the kinetic advantage of the enzymatic reaction, compared with the reaction in water, is caused 88% (8.01/9.1) by favorable NAC formation. Strajbl et al. (19), in an earlier empirical valence bond study, confirmed the importance of NAC formation in determining the proficiency of the Bacillus subtilis CM (BsCM) enzymatic reaction.
] is 8.06 kcal/mol, which is in good agreement with previous results (8.1 kcal/mol) obtained from thermodynamic integration (4). In the enzymatic reaction, free energy for NAC formation is 0.73 kcal/mol when RC5-C16 is the distance of the NAC criteria (3.7 Å). The minimum of the free-energy profile of NAC formation in TtCM is 0.05 kcal/mol. Therefore, the GS conformations of chorismate in the active site of TtCM are essentially NACs. Using the experimental value of the free energy of reaction in enzyme (9.1 kcal/mol), the kinetic advantage of the enzymatic reaction, compared with the reaction in water, is caused 88% (8.01/9.1) by favorable NAC formation. Strajbl et al. (19), in an earlier empirical valence bond study, confirmed the importance of NAC formation in determining the proficiency of the Bacillus subtilis CM (BsCM) enzymatic reaction.
Table 1. The free energies for the chorismate → prephenate reaction in both water and TtCM (kcal/mol).
| Free energies | Water | TtCM |
|---|---|---|
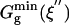 |
0.00 | 0.00 |
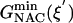 |
8.06 | 0.05 |
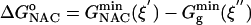 |
8.06 | 0.05 |
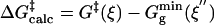 |
22.31 | 12.38 |
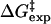 |
24.5 | 15.4 |
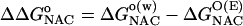 |
8.01 | |
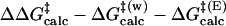 |
9.93 | |
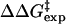 |
9.1 | |
| ΔΔGTS | 1.92 | |
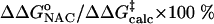 |
80.6% | |
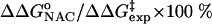 |
88.0% |
The superscripts min, o, and ‡ designate the minimum of the free energy profile, standard free energy, and free energy of the activation for the reaction, respectively. The superscripts (E) and (W) stand for the free energy change in enzyme and water, respectively. The subscripts calc and exp represent the computed and experimental values. ξ, ξ′, and ξ″ denote the positions of TS, NAC, and GS along the reaction coordinates.
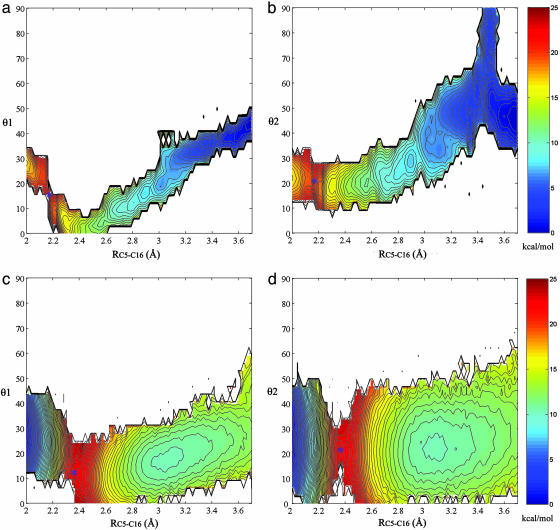
Reaction Coordinates. Because chorismate NAC is defined by three criteria, the distance RC5-C6, and the angles θ1 and θ2, 1D PMF profiles were projected into 2D PMF profiles by the weighted histogram analysis method (20, 21) along the RC5-C6 and θ1, and RC5-C6 and θ2, respectively (Fig. 3). The TS is identified from the highest saddle points in the contour plots along the reaction coordinates, labeled with blue asterisks. For the reaction in water (Fig. 3 a and b), θ1 is 14° and θ2 is 19° in the TS. For the reaction in TtCM (Fig. 3 c and d), θ1 is 12° and θ2 is 20°. Thus, θ1 and θ2 differ only by 1–2°. The TS structures in water and TtCM are very similar. With the contracting of the C5-C16 distance (starting from the GS), the reaction pathway becomes narrow. The conformations of the chorismate are all NACs when RC5-C16 < 2.83 Å in water and RC5-C16 < 2.81 Å in TtCM where the “bottleneck” shapes occur. It has also been identified from Fig. 2 that the free-energy profiles nearly overlap with the free-energy profiles of the NAC formation when RC5-C16 is < 2.83 Å in water and 2.81 Å in TtCM.
Fig. 3.
NAC conformation characterization. The RC5-C16 ranges from 2.0 to 3.7 Å, θ1 and θ2 range from 0° to 90°, and the increment of the contour line is 0.5 kcal/mol. 1D PMFs along the reaction coordinate RC5-C16 are projected to the θ1 and θ2, respectively. The TS is labeled by blue asterisks. (a and b) PMF of the reaction in water projected to θ1 (a) and θ2 (b). (c and d) PMF of the reaction in TtCM projected to θ1 (c) and θ2 (d).
Comparison of Conformations in Different Reaction Stages. The conformations of the TtCM-bound GS, NAC, TS, and product were extracted from trajectories and averaged. The interaction distances are defined as the distances between the geometric center of chorismate and that of the side chains of nearby residues (within 6 Å), which are calculated from the four averaged structures as shown in Table 2. The interaction distances do not change much in the course of the reaction. For example, the interaction distance of the chorismate and Leu-114 are 5.98, 6.00, 6.18, and 6.07 Å in the GS, NAC, TS, and product, respectively. The calculated radii of gyration for the heavy atoms of the chorismate and residues within 6 Å of the chorismate are 7.20, 7.22, 7.23, and 7.20 Å in the GS, NAC, TS, and product, respectively. Therefore, the radii of gyration also show no significant differences. The species housed in the active site were removed and the volumes of the empty cavities at various distances along the reaction coordinate were calculated by using the voidoo program (22) with a grid spacing and probe radius of 1.0 and 1.2 Å, respectively. The cavity volumes of the active site are 178 and 163 Å3 in the E·S and enzyme–TS complex (E·TS). The similarity of the cavity volumes of the active site indicates that the chorismate binds no tighter in the TS than GS. Superimposing TtCM-bound E·S and E·TS shows that the geometry of the active site structure remains unchanged (Fig. 5, which is published as supporting information on the PNAS web site).
Table 2. The geometry properties of the TtCM active site.
| Property | GS | NAC | TS | Product |
|---|---|---|---|---|
| Interaction distance, Å | ||||
| CHO—Phe-57* | 5.35 | 5.48 | 5.43 | 5.50 |
| CHO—Ala-59* | 4.94 | 4.67 | 5.22 | 4.73 |
| CHO—Arg-63* | 7.94 | 7.75 | 8.02 | 7.82 |
| CHO—Leu-72* | 6.22 | 6.21 | 6.01 | 6.28 |
| CHO—Leu-73* | 6.48 | 6.45 | 6.06 | 6.58 |
| CHO—Ser-74* | 6.24 | 6.34 | 6.22 | 6.34 |
| CHO—Arg-6 | 6.19 | 6.28 | 6.18 | 6.21 |
| CHO—Glu-77 | 6.79 | 6.80 | 6.67 | 7.03 |
| CHO—Arg-89 | 5.20 | 5.10 | 5.14 | 5.09 |
| CHO—Tyr-107 | 7.37 | 7.48 | 7.31 | 7.33 |
| CHO—Leu-114 | 5.98 | 6.00 | 6.18 | 6.07 |
| CHO—Arg-115 | 6.63 | 6.74 | 6.22 | 5.91 |
| RG, Å | 7.20 | 7.22 | 7.23 | 7.20 |
| Cavity volume, Å3 | 178.36 | 168.36 | 162.58 | 167.38 |
The interaction distances are defined as the average distances between the geometric centers of chorismate and the residue side chains that are within 6 Å of chorismate. The residues labeled with an asterisk are from the adjacent subunit. CHO denotes chorismate. RG stands for the radius of gyration of the heavy atoms of residues within 6 Å of chorismate. The cavity volume of the active site is surrounded by the residues in the vicinity of the chorismate.
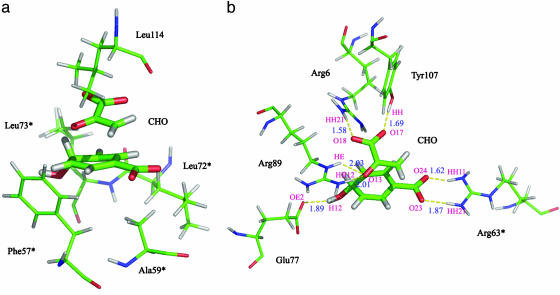
TS Structure Analysis. The hydrophobic and electrostatic interactions of TS at the active site are shown in Fig. 4. As previously mentioned, the active site changes marginally when the chorismate goes from GS to the TS. Thus, the interactions of substrate and residues in E·TS and E·S are generally alike. The most important interactions are the hydrogen bonds between Arg-6 and Arg-63* and the two carboxylates. The residues labeled with * are from the adjacent subunit. These interactions hold the substrate in a di-axial conformation required for the reaction and remain in position to bind the TS (Fig. 4b).
Fig. 4.
The structure of the TS. Only chorismate and the nearby residues (within 6 Å) are shown: residues involved in hydrophobic interactions (a) and electrostatic interactions (b). The residues marked by * are from the adjacent subunit. The hydrogen bonds between TS and residues are plotted as yellow dashed lines along with their distance.
Located beneath the TS are the nonpolar residues of the active site: Phe-57*, Ala-59*, Leu-72*, and Leu-73* (Fig. 4a), which form a basin to support the substrate. Another nonpolar residue, Leu-114, hangs over the TS, protruding from the other subunit. These two sections of the residues constrain the TS by hydrophobic interactions. The interaction distances between these nonpolar residues and the TS are all ≈6 Å, as shown in Table 2. This finding indicates the TS to be in the center of the active site and with no preference to be closer to any of the nonpolar residues. The Phe-57*, Ala-59*, Leu-72*, Leu-73*, and Leu-114 nonpolar amino acids function as “bookends” to prevent the sideways motion of the C16 and C5 atoms in the chorismate from drifting away from each other.
Charge Distributions for E·S and E·TS. The Mulliken charge distributions of the structures, averaged from the trajectories of the GS, NAC, and TS in water and TtCM, were obtained by using single-point energy calculations at the SCCDFTB/MM level (Table 3, which is published as supporting information on the PNAS web site). We have found that the charge distributions do not change much from the GS to TS. The total charges of the six atoms, C3, C4, C5, O13, C14, and C16, which are involved in the pericyclic reaction, show remarkable similarity (–0.54, –0.52, and –0.53 for the GS, NAC, and TS, respectively, in water; –0.50, –0.51, and –0.56, respectively, in TtCM). The charge distributions of the chorismate atoms involved in hydrogen bonding with the residues vary marginally from the E·S to E·TS in the thermophilic enzyme, such as H12 (0.39–0.40), O13 (–0.43 to –0.50), O17 (–0.89 to –0.83), O18 (–0.72 to –0.80), O23 (–0.76 to –0.76), and O24 (–0.88 to –0.85). Particularly, the Mulliken charge of the O13 atom, which has been considered to be critical in the enzymatic reaction (23), is –0.44, –0.43, and –0.50 in the GS, NAC, and TS, respectively, for TtCM. The electrostatic interactions between O13 and Arg-90 change only slightly from GS to TS. Comparing the TS in enzyme and water, it can be seen that the O13 charge is –0.50 in enzyme and –0.51 in water.
Discussion
In previous studies of the chorismate → prephenate reaction in the Escherichia coli CM (EcCM) using molecular dynamics, thermodynamic integration, and 2D QM/MM energy surface methods (4–6), we have shown that ≈90% of the free-energy advantage of the enzymatic reaction over that in water is caused by the ease of NAC formation in the active site of enzyme. The free energies for NAC formation in the six different systems: water, WT EcCM and E52A mutant, WT BsCM and R90Citrulline mutant, and a catalytic antibody 1F7, were calculated by using thermodynamic integration (4). A linear plot (Fig. 6, which is published as supporting information on the PNAS web site) of the free energies of NAC formation (ΔG°NAC) for these six reactions vs. the experimentally determined free energies of reactions (ΔG‡) shows slope of 1.1, which implies free energy (ΔGTS) of NAC to TS is close to being a constant and the free energies for reaction are a linear function of the free energy of NAC formation. The free energy for the NAC formation in the WT CMs was found to be very small, 0.1 kcal/mol for EcCM and 0.3 kcal/mol for BsCM at 300 K. Our results obtained by PMF are 0.05 kcal/mol for the WT TtCM at 343 K, consistent with previous studies of EcCM and BsCM at 300 K. The free energy for NAC formation is large in water, 8.06 kcal/mol obtained by PMF, and that by thermodynamic integration is 8.1 kcal/mol. The free energy for NAC formation obtained from the SCCDFTB/MM simulations with TtCM is in agreement with that of thermodynamic integration.
The total charges of the six atoms of chorismate involved in the reaction are maintained around –0.50 in the GS, NAC, and TS. The charges of atoms change only slightly from the GS to TS. This feature has previously been observed by Warshel and colleagues (19). These combined facts suggest that the TS has a marginally larger negative charge than GS, such that one should not expect a significant augment of electrostatic stabilization. Thus, small electrostatic stabilization contribution (≈10% of experimental vale of ΔΔG‡) to the proficiency of enzymatic reaction was identified.
We have previously shown that chorismate must be in one of seven defined di-axial conformations to form the reactive NAC (5). In the present study, we found that SCCDFTB/MM computations of E·S initiated with di-equatorial chorismate conformers convert to di-axial in 32 ps, which has also been demonstrated from the simulations carried out by Karplus and colleagues (24, 25) in the study of yeast CM. The reports (26, 27) that di-equatorial conformer serves as reactive conformations lead to the confusion of identifying NAC. The term NAC to recognize conformations, in which reacting atoms are at van der Waals distance regardless of the attacking angle, is tantamount to the neglect of required orbital overlaps for reaction. Contributing to the confusion is a QM/MM study of a BsCM mutant (28) in which positively charged Arg-90 is replaced by a neutral citrulline without sampling of the degrees of freedom for the protein backbone. However, it had been shown that the mutation is accompanied by rearrangements at the active site (5).
In a recent report by Hilvert and coworkers (29), a concerted, albeit asynchronous, enzyme mechanism is supported by kinetic isotope effect studies of BsCM enzymatic reaction. The secondary tritium isotope effects on thermal reaction indicate a significant cleavage of C3-O13 bond but little formation of the C5-C16 at TS. Hilvert and coworkers' density functional theory calculations at the B3LYP/6–31+G* level predict RC5-C16 of 2.66 Å and RC3-O13 of 2.13 Å in vacuo. In our SCCDFT/MM approach, RC5-C16 and RC3-O13 have been determined to be 2.55 and 2.00 Å in vacuo (6) and 2.37 and 1.94 Å in the active site of TtCM, respectively. The differences of bond breaking and making, obtained by Hilvert and coworkers (–0.55 Å in vacuo) and our studies (–0.53 Å in vacuo and –0.43 Å in TtCM) are similar. Such a concerted mechanism has also been identified by our previous results with EcCM with the 2D QM/MM energy surface method (6).
Hilvert and coworkers (29) have proposed that Arg-90 in BsCM stabilizes the ether oxygen of chorismate in asynchronous TS (29). In current simulations of TtCM, the average distances of O13 of chorismate and NH1 of Arg-89 are 3.06 and 2.83 Å for E·S and E·TS, respectively, and the charges on O13 are –0.44 and –0.50 for E·S and E·TS, respectively. The interactions of Arg-89 and chorismate were evaluated from single-point SCCDFTB/MM energy calculation, where the only side chain of the Arg-89 and chorismate were included in the calculations. A link atom was added onto the Cβ atom of the Arg-89 side chain to preserve the integrated electron and structure of the Arg-89 side chain. The average electrostatic interactions between the Arg-89 side chain and chorismate (ΔEelec) are –197.97 and –198.52 kcal/mol for E·S and E·TS, respectively. Thus, electrostatic interactions between the Arg-89 side chain and chorismate contribute 0.55 kcal/mol (ΔΔEelec) to the TS stabilization. The average van der Waals interactions between the Arg-89 side chain and chorismate (ΔEvdw) are –1.96 and –1.99 kcal/mol for E·S and E·TS, respectively, which are almost the same. The total contributions of interactions between the Arg-89 side chain and chorismate to the TS stabilization are 0.58 kcal/mol.
The role of the Arg-90 in BsCM has also been studied by the Arg-90citrulline mutant. The free energy of activation for the reaction in the BsCM Arg-90citrulline mutant increases by 5.8 kcal/mol after mutation (30). The difference in free energies for NAC formation in the active site of the mutant and WT BsCM is 3.8 kcal/mol (4.1 and 0.3 kcal/mol for the mutant and WT BsCM, respectively) (4). Thus, Arg-90 was estimated to contribute 2.0 (5.8–3.8) kcal/mol to TS stabilization in BsCM. The values of the Arg-90 contribution to the TS stabilization in BsCM obtained by the mutations are larger than the direct calculation of the interactions (0.58 kcal/mol) between Arg-89 and chorismate in TtCM, which is possibly because of the rearrangement of the active site after mutations.
Conclusion
We have obtained the PMF profiles for the chorismate → prephenate reaction both in water and a thermophilic enzyme by using the combination of SCCDFTB/MM, umbrella sampling, and the weighted histogram analysis method. The free energies of activation were determined from the PMF profile, and the free energies for NAC formation were calculated from the mole fraction of conformations in the GS present as NAC. Small numbers of the free energies for NAC formation in different CMs suggest that the GS is essentially represented by the NAC structure in the active site of the CM. The contribution of NAC formation to the lowering of the activation barrier for the reaction in the thermophilic enzyme is the same (8.01 kcal/mol at the optimum temperature, 343 K) as in the mesophilic one (8.0 and 7.8 kcal/mol for EcCM and BsCM, respectively, at 300 K). The TS structures in water and TtCM are similar. No significant changes in the electrostatic interactions and the geometry of the active site take place during the chorismate → prephenate reaction. The Mulliken charges of atoms of the GS and TS species are very similar. The electrostatic and van der Waals interactions between the ether oxygen of chorismate and Arg-90 of 0.58 kcal/mol provide minimal TS stabilization. Only a small electrostatic stabilization contribution to the proficiency of the enzymatic reaction was identified based on the charge analysis. In essence, the enzyme proficiency originates from an entropic advantage of enzyme in forming the reactive GS conformations compared with water.
Supplementary Material
Acknowledgments
We thank Dr. Xiaodong Zhang and Joe Toporowski for helpful discussion and advice and the National Partnership for Advanced Computational Infrastructure for its generous allocation of computational resources at DataStar in the University of California at San Diego Supercomputing Center. This work was supported by National Institutes of Health Grant 5R37DK9174-41.
Author contributions: T.C.B. designed research; X.Z. performed research; and X.Z. and T.C.B. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: CM, chorismate mutase; TtCM, Thermus thermophilus CM; EcCM, Escherichia coli CM; BsCM, Bacillus subtilis CM; NAC, near attack conformer; E·S, enzyme–substrate complex; E·TS, enzyme–TS complex; TS, transition state; GS, ground state; QM, quantum mechanics; MM, molecular mechanics; SCCDFTB, self-consistent-charge density-functional tight binding; PMF, potential of mean force.
References
- 1.Guilford, W. J., Copley, S. D. & Knowles, J. R. (1987) J. Am. Chem. Soc. 109, 5013–5019. [Google Scholar]
- 2.Bruice, T. C. (2002) Acc. Chem. Res. 35, 139–148. [DOI] [PubMed] [Google Scholar]
- 3.Ganem, B. (1996) Angew. Chem. Int. Ed. 35, 937–945. [Google Scholar]
- 4.Hur, S. & Bruice, T. C. (2003) Proc. Natl. Acad. Sci. USA 100, 12015–12020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hur, S. & Bruice, T. C. (2003) J. Am. Chem. Soc. 125, 5964–5972. [DOI] [PubMed] [Google Scholar]
- 6.Zhang, X. D., Zhang, X. H. & Bruice, T. C. (2005) Biochemistry 44, 10443–10448. [DOI] [PubMed] [Google Scholar]
- 7.Mazumder-Shivakumar, D. & Bruice, T. C. (2005) Biochemistry 44, 7805–7817. [DOI] [PubMed] [Google Scholar]
- 8.Helmstaedt, K., Heinrich, G., Merkl, R. & Braus, G. H. (2004) Arch. Microbiol. 181, 195–203. [DOI] [PubMed] [Google Scholar]
- 9.Tahirov, T. H., Inagaki, E., Ohshima, N., Kitao, T., Kuroishi, C., Ukita, Y., Takio, K., Kobayashi, M., Kuramitsu, S., Yokoyama, S. & Miyano, M. (2004) J. Mol. Biol. 337, 1149–1160. [DOI] [PubMed] [Google Scholar]
- 10.Mattei, P., Kast, P. & Hilvert, D. (1999) Eur. J. Biochem. 261, 25–32. [DOI] [PubMed] [Google Scholar]
- 11.Warshel, A. & Levitt, M. (1976) J. Mol. Biol. 103, 227–249. [DOI] [PubMed] [Google Scholar]
- 12.Lazaridis, T., Tobias, D. J., Brooks, C. L. & Paulaitis, M. E. (1991) J. Chem. Phys. 95, 7612–7625. [Google Scholar]
- 13.Brooks, B. R., Bruccoleri, R. E., Olafson, B. D., States, D. J., Swaminathan, S. & Karplus, M. (1983) J. Comput. Chem. 4, 187–217. [Google Scholar]
- 14.Elstner, M., Porezag, D., Jungnickel, G., Elsner, J., Haugk, M., Frauenheim, T., Suhai, S. & Seifert, G. (1998) Phys. Rev. B 58, 7260–7268. [Google Scholar]
- 15.Cui, Q. & Karplus, M. (2002) J. Am. Chem. Soc. 124, 3093–3124. [DOI] [PubMed] [Google Scholar]
- 16.Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W. & Klein, M. L. (1983) J. Chem. Phys. 79, 926–935. [Google Scholar]
- 17.Brooks, C. L. & Karplus, M. (1989) J. Mol. Biol. 208, 159–181. [DOI] [PubMed] [Google Scholar]
- 18.Beveridge, D. L. & Dicapua, F. M. (1989) Annu. Rev. Biophys. Biophys. Chem. 18, 431–492. [DOI] [PubMed] [Google Scholar]
- 19.Strajbl, M., Shurki, A., Kato, M. & Warshel, A. (2003) J. Am. Chem. Soc. 125, 10228–10237. [DOI] [PubMed] [Google Scholar]
- 20.Ferrenberg, A. M. & Swendsen, R. H. (1989) Phys. Rev. Lett. 63, 1195–1198. [DOI] [PubMed] [Google Scholar]
- 21.Kumar, S., Bouzida, D., Swendsen, R. H., Kollman, P. A. & Rosenberg, J. M. (1992) J. Comput. Chem. 13, 1011–1021. [Google Scholar]
- 22.Kleywegt, G. J. & Jones, T. A. (1994) Acta Crystallogr. D 50, 178–185. [DOI] [PubMed] [Google Scholar]
- 23.Gustin, D. J., Mattei, P., Kast, P., Wiest, O., Lee, L., Cleland, W. W. & Hilvert, D. (1999) J. Am. Chem. Soc. 121, 1756–1757. [Google Scholar]
- 24.Guo, H., Cui, Q., Lipscomb, W. N. & Karplus, M. (2001) Proc. Natl. Acad. Sci. USA 98, 9032–9037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo, H., Cui, Q., Lipscomb, W. N. & Karplus, M. (2003) Angew. Chem. Int. Ed. 42, 1508–1511. [DOI] [PubMed] [Google Scholar]
- 26.Repasky, M. P., Guimaraes, C. R. W., Chandrasekhar, J., Tirado-Rives, J. & Jorgensen, W. L. (2003) J. Am. Chem. Soc. 125, 6663–6672. [DOI] [PubMed] [Google Scholar]
- 27.Guimaraes, C. R. W., Repasky, M. P., Chandrasekhar, J., Tirado-Rives, J. & Jorgensen, W. L. (2003) J. Am. Chem. Soc. 125, 6892–6899. [DOI] [PubMed] [Google Scholar]
- 28.Guimaraes, C. R. W., Udier-Blagovic, M., Tubert-Brohman, I. & Jorgensen, W. L. (2005) J. Chem. Theory Comput. 1, 617–625. [DOI] [PubMed] [Google Scholar]
- 29.Wright, S. K., DeClue, M. S., Mandal, A., Lee, L., Wiest, O., Cleland, W. W. & Hilvert, D. (2005) J. Am. Chem. Soc. 127, 12957–12964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kienhofer, A., Kast, P. & Hilvert, D. (2003) J. Am. Chem. Soc. 125, 3206–3207. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.