Abstract
The biochemistry of early stages of hematopoietic differentiation is difficult to study because only relatively small numbers of precursor cells are available. The murine EML cell line is a multipotential cell line that can be used to model some of these steps. We found that the lineage– EML precursor cells can be separated into two populations based on cell surface markers including CD34. Both populations contain similar levels of stem cell factor (SCF) receptor (c-Kit) but only the CD34+ population shows a growth response when treated with SCF. Conversely, the CD34– population will grow in the presence of the cytokine IL-3. The human β-globin locus control region hypersensitive site 2 plays different roles on β-globin transcription in the CD34+ and CD34– populations. The two populations are present in about equal amounts in culture, and the CD34+ population rapidly regenerates the mixed population when grown in the presence of SCF. We suggest that this system may mimic a normal developmental transition in hematopoiesis.
Keywords: c-Kit, CD34, EML cells, hypersensitive site 2
The early stages in the differentiation of hematopoietic stem cells are difficult to study in normal cells because of the paucity of such precursors relative to more mature cells. As a surrogate several murine cell lines have been developed that can be differentiated into more than one lineage in vitro (1–3). EML is one such line, developed from murine bone marrow cells transfected with a vector expressing a dominant negative form of the retinoic acid receptor (1). EML cells are indefinitely propagatable in medium containing stem cell factor (SCF) and can be differentiated into erythroid, myeloid, and perhaps lymphoid cells in vitro. EML cells also can be recloned without losing their multipotential character, and similar cell lines can be repeatedly derived from murine marrow.
We have examined the lineage-negative cells (Lin– cells) present in a population of EML cells growing exponentially in SCF and found they can be separated into two subpopulations on the basis of surface CD34 expression. The two populations are present in about equal amounts, and growth of the CD34+ population in SCF rapidly regenerates a mixed population containing about equal amounts of CD34+ and CD34– cells. Although the two populations resemble each other in morphology, levels of SCF receptor expression, and overall pattern of mRNA expression, they differ in growth characteristics and patterns of receptor phosphorylation in response to cytokines. Contrary roles of locus control region (LCR) hypersensitive site 2 (HS2) on β-globin promoter in the two populations were observed. We suggest that these two populations represent models for an early step in the differentiation of normal hematopoietic precursors.
Materials and Methods
Cell Culture. EML C1 cells were the kind gift of Schickwann Tsai (University of Utah, Salt Lake City) and were cultured as described (1). The cells were induced to differentiate into erythroid, myeloid, or lymphoid cells by the protocols described in ref. 1. For myeloid differentiation, most of the cells died after the cultures were switched from the combination of SCF, IL-3, and all-trans retinoic acid (10 μM) medium into granulocyte–macrophage colony-stimulating factor medium on day 3. The dead cells were removed by using the dead cells removal kit (Miltenyi Biotech, Auburn, CA), and RNA was extracted from the live cells. For lymphoid differentiation, the differentiated EML cells were separated from the stromal cell W20 feeder cells (gift of Schickwann Tsai) at the time of harvest by FACS using size scatter. The EML cells were clearly separated from W20 cells because of their smaller side scatter.
RNA Extraction and Microarray Analysis. RNA from the different cell samples was extracted with an RNeasy kit (Qiagen, Valencia, CA). RNA quality was evaluated by measuring the ratio of 28S to 18S rRNA on agarose gel electrophoresis and determining the ratio of optical absorbance at 260 and 280 nm. Samples were rejected if the A280/A260 was <1.8, the 18S/28S rRNA was >0.6, or the samples showed any traces of degradation or DNA contamination on gel electrophoresis. RNA expression analysis was performed on the mouse MG-U74 or A430 chip sets by using standard techniques for probe preparation and the Affymetrix (Santa Clara, CA) mas 5.0 normalization procedure.
Lentviral Reporter Assays. The human β-globin promoter (sequence: 5′-GGTACGGCTGTCATCACTTAGACCTCACCCTGTGGAGCCACACCCTAGGGTTGGCCAATCTACTCCCAGGAGCAGGGAGGGCAGGAGCCAGGGCTGGGCATAAAAGTCAGGGCAGAGCCATCTATTGCTTACATTTGCTTCTGACACAAC-3′) or β-globin promoter with a upstream insert of the enhancer regions of the β-globin LCR HS2 (sequence: 5′-CAGTTTTTCCTTAGTTCCTGTTACATTTCTGTGTGTCTCCATTAGTGACCTCCCATAGTCCAAGCATGAGCAGTTCTGGCCAGGCCCCTGTCGGGGTCAGTGCCCCACCCCCGCCTTCTGGTTCTGTGCAACCTTCTAAGCAAACCTTCTGGCTCAAGCACAGCAATGCTGAGTCATGATGAGTCATGCTGAGGCTTAGGGTGTGTGCCCAGATGTTCTCAGCCTAGAGTGATGACTCCTATCTGGGTCCCCAGCAGGATGCTTACAGGGCAGATGGCAAAAAAAAGGAGAAGCTGACCACCTGACTAAAACTCCACCTCAAACGGCATCATAAAGAAAATGGATGCCTG-3′) was cloned upstream of the destabilized eGFP gene (d2-eGFP, half-life of 2 h; Clontech). These cassettes were then cloned into a self-inactivating lentiviral vector plasmid (gift of Irving L. Weissman's laboratory at Stanford University, Stanford, CA). Lentiviruses were produced as described (4, 5). Briefly, 293T cells were transfected with the transfer vector plasmids, the vesicular stomatitis virus glycoprotein envelope-encoding plasmid pMD.G, and the packaging plasmid CMV R8.74. Viral supernatant was collected 24 h later and stored at 4°C. Fresh media were added to the plates, and virus-containing medium was collected 24 h later again. The final supernatants were pooled together and concentrated by ultracentrifugation. EML cells (1 × 105) were transduced with at least 1 × 106 of the reporter lentiviruses and cultured in SCF medium (Iscove's modified Dulbecco medium supplemented with 15% BHK conditional medium and 15% horse serum). GFP expression and cell surface markers were analyzed by FACS 2 days after transduction of the cells.
Separations of Lin–CD34+ and Lin–CD34– EML Cells. EML cells were collected by centrifugation and washed twice with FACS buffer (PBS supplemented with 0.5% BSA and 2 mM EDTA) by centrifugation. Isolation of the Lin– subpopulation EML cells with a lineage depletion kit (Miltenyi Biotec) was performed according to the manufacturer's protocol. The Lin–-enriched fraction was labeled with CD34 antibody, lineage marker antibody mixture (CD45R, Ter119, CD11b, Gr.1, and 7/4) (Becton Dickinson Immunocytometry Systems) and Sca-1 (eBioscience, San Diego, CA) or isotypic control antibodies (from the corresponding vendor) following the instructions of the manufacturers. Lin–CD34+ and Lin–CD34– EML cell populations were separated by using a Moflo high flow cytometer (Cytomation, Fort Collins, CO).
Cell Proliferation and Cell Apoptosis Analyses. The separated cells were subcultured in the different media at the same cell concentration (1 × 105 cells per ml). Dead cells were detected by trypan blue staining. Apoptotic cells were detected by using a vibrant apoptosis assay kit (Molecular Probes).
Real-Time Quantitative PCR and PCR. RNA samples were the same as those that had been used for microarray analysis, and cDNA was reverse-transcribed by using a first-strand cDNA synthesis kit for RT-PCR(AMV) (Roche Applied Science) as described in the manufacturer's protocol. The real-time PCRs were carried out from cDNA samples by using the iQ SYBR green supermix (Bio-Rad), as described in the manufacturer's protocol, and the following sequence of DNA primers: CD34, forward ACCACAGACTTCCCCAACTG and reverse TCGGAGCAGAAGATGATGTG; c-Kit, forward GGGCTAGCCAGAGACATCAG and reverse AGGAGAAGAGCTCCCAGAGG; Sca-1, forward CCCCTACCCTGATGGAGTCT and reverse TTCCTGGCAACAGGAAGTCA; CSF2rb1, forward CCACTCTCTGCCTGATCTCC and reverse CCCACACTGCACATCCATAG; EPO R, forward CCCAAGTTTGAGAGCAAAGC and reverse TGCAGGCTACATGACTTTCG; GATA1, forward CCCAAGAAGCGAATGATTGT and reverse TTCCTCGTCTGGATTCCATC; GATA3, forward CTGGAGGAGGAACGCTAATG and reverse GTTGAAGGAGCTGCTCTTGG; and GAPDH, forward AACTTTGGCATTGTGGAAGG and reverse ACACATTGGGGGTAGGAACA. GAPDH was used to normalize all real-time PCR fractions for comparison. The sequence of destabilized GFP (dGFP) was amplified from genomic DNA extracted from Lin–CD34+GFP– and Lin–CD34+GFP+ EML cells transduced with an HS2-β-globin promoter-dGFP cassette by standard PCR using the following DNA primers: forward TATATCATGGCCGACAAGCA and reverse CTGGGTGCTCAGGTAGTGGT.
Immunoprecipitation and Immunoblotting Analyses. The sorted Lin–CD34+ and Lin–CD34– EML cells were incubated in SCF medium for 2 h and then washed with PBS twice. The cells subsequently were starved (cultured in Iscove's modified Dulbecco medium with 0.5% BSA without serum or SCF at 1 × 106 per ml) for 4 h in the incubator. Starved cells were treated with 100 ng/ml purified SCF or 5 ng/ml purified IL-3 (PeproTech, Rocky Hill, NJ) or the combination of SCF and IL-3 for 10 min. Cells were pelleted, washed with ice-cold PBS, and lysed at 107/ml with ice-cold lysis buffer as described (6). The lysates were cleared by centrifugation for 10 min at 18,000 × g. c-Kit and IL-3 receptor (IL-3R)/β-chain were immunopreciptated from the cleared lysates with the relevant antibodies [antibodies recognizing c-Kit (SC1494) and IL-3/IL-5/granulocyte–macrophage colony-stimulating factor receptor β(SC-678) from Santa Cruz Biotechnology] at 1:50 dilution for 3 h on ice. After addition of protein G-Sepharose the suspension was incubated by rotating for 1 h. After three washes in lysis buffer, the beads were boiled in SDS/PAGE sample buffer for 5 min and analyzed by Western blotting. Samples were resolved with SDS/PAGE, transferred to Immobilion-P, and immunoblotted with the relevant antibodies (antibodies specific for pTyr 719-Kit, 3391 and phosphor-tyrosine P-Tyr-100, 9411 from Cell Signaling Technology, Beverly, MA), according to the antibody manufacturer's instruction.
Results
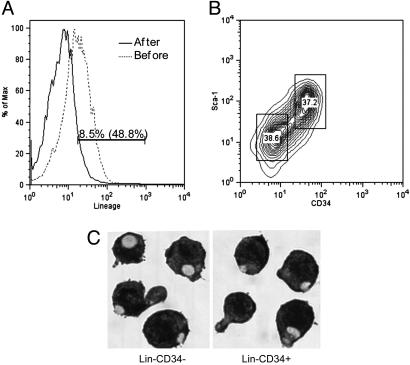
As described in the literature, our culture of EML cells grew with an apparent doubling time of 24 h, when cultured in SCF medium. Because this cell line can be differentiated into more than one lineage we were interested in looking for heterogeneity in the precursor populations. We therefore used magnetic beads coated with a mixture of antibodies directed against lineage markers to prepare by negative selection a population of Lin– EML cells from an exponentially growing culture. We then analyzed this preparation by FACS, using antibodies directed against CD34 and Sca-1 markers of immature hematopoietic cells. As shown in Fig. 1, in each of several experiments we observed that the cells could be separated into two slightly overlapping populations of approximately equal size. One population was CD34– and Sca-1low and the other was CD34+ and Sca-1high.
Fig. 1.
Isolation of Lin–CD34+ and Lin–CD34– EML cells. (A) Expression of the lineage markers on EML cells before lineage depletion (dotted line) or after lineage depletion (solid line) by magnetic cell sorting. The cells were stained with phycoerythrin (PE)-cy7-lineage mixture antibodies and analyzed by FACS. (B) Lin–-enriched EML cells were stained with PE-cy5-lineage mixture antibodies, PE-conjugated CD34 antibody, and allophycocyanin-conjugated Sca-1 antibody. Subsequently, the Lin+ cells were gated out and Lin– EML cells were sorted into CD34+ and CD34– EML cell populations by Moflo high flow cell cytometry. (C) Lin–CD34– and Lin–CD34+ EML cells cytospun onto slides were stained with Wright-Giemsa by standard protocols. (Magnification: ×400.)
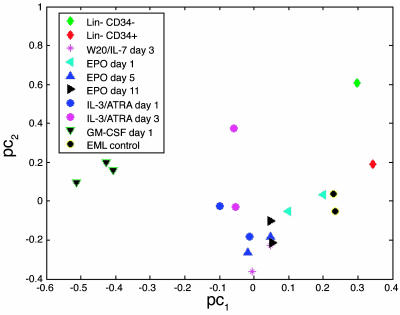
We have previously shown that, after a binormalization procedure, principal component analysis of mRNA expression data derived from Affymetrix oligonucleotide arrays clearly separates multiple lineages of mature hematopoietic cells into nonoverlapping clusters (7, 8). Protocols have been presented for differentiating EML cells toward erythroid cells, myeloid cells, or cells resembling lymphocytes by manipulation of the growth conditions and cytokine milieu of these cells. We prepared EML cells differentiated toward each of these cell types and compared the RNA analysis from these cells with parental EML cells and Lin–CD34+ and Lin–CD34– cells, separated by preparative FACS analysis. As shown in Fig. 2, the mRNA expression patterns in the two subpopulations of the Lin– cells mapped close to one another and near the mixed population of Lin– cells when compared with the various partially differentiated populations.
Fig. 2.
Principal component analysis of transcription factor expression by Lin–CD34+, Lin–CD34–, parental EML, and different differentiated EML cells. Principal component analysis allows us to present the distribution of the samples in the multidimensional transcription factor expression space in a 2D graph. A preliminary step of the analysis is a binormalization procedure that simultaneously centers the transcription factor profile of each sample and expression intensities across all samples of each transcription factor around zero. The principal components are linear combinations of the normalized transcription factor expression profiles. The first two leading principal components capture most of the variation of the data. Therefore, the data can be displayed (with a minor loss of information) in a 2D graph by projection of the 19 samples onto a subspace spanned by the two leading principal components. The axes legends pc1 and pc2 stand for the first two principal components. The distances between the samples in this 2D space is an approximation of the dissimilarities between their corresponding transcription factor expression profiles. The cell types are represented by the symbols indicated in Inset.
Examination of the mRNA patterns of the two subpopulations of cells showed that both expressed erythroid cell markers such as mRNA for the globin early and light chains and GATA1 (9, 10). However, these were expressed at a significantly higher level in the CD34– cell population as were Pira and Pirb, Ig-like surface molecules associated with myeloid cells, and mast cell proteases. This constellation of mRNAs from more than one mature lineage would be consistent with the CD34– cells being roughly comparable to the chronic myelogenous leukemia common myeloerythroid precursor described by Weissman and colleagues (11–13). In particular, the relative elevation of mRNAs for a number of erythroid genes suggested that either a subpopulation of the CD34– cells was differentiated into purely erythroid precursors or these cells represented a mixed differentiation state with prominent erythroid potential (Table 1). Conversely, the CD34+ cells expressed higher levels of the early myeloid marker myeloperoxidase and a few genes, such as BCL3 (14), vav (15), and CD7 (16) that have been associated with lymphocyte development or function, and might reflect a potential for differentiation into nonmyeloid cells.
Table 1. Gene expression change.
| Folds of gene expression change (Lin-CD34+/Lin-CD34-)
|
||
|---|---|---|
| Gene | Microarray | Real-time PCR |
| CD34 | 7.97 | 8.26 |
| c-Kit | 0.75 | 0.92 |
| Sca-1 | 1.69 | 1.61 |
| CSF2rb1 | 0.25 | 0.23 |
| EPO R | 0.33 | 0.1 |
| Gata1 | 0.27 | 0.23 |
| Gata3 | 3.15 | 7.56 |
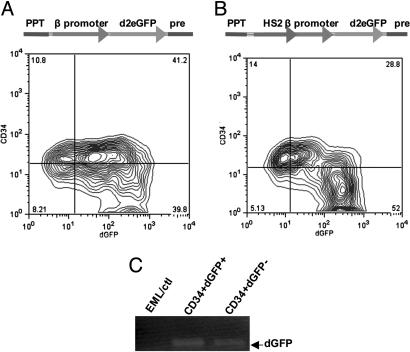
Because the CD34+ cells expressed significant amounts of mRNA for globins, it was possible that they represented a mixed population with some cells differentiated along the erythroid lineage and others from different lineages but without having matured to the point where they expressed detectable amounts of lineage-specific surface markers. An additional possibility was that cells fluctuated between a state in which they produced erythroid genes and other states in which these genes were suppressed. To test this possibility, we prepared self-inactivating lentiviral vectors expressing an unstable GFP driven by the β-globin promoter. Two constructs were prepared that were identical except that one construct contained a mini-LCR (HS2) inserted upstream of the β-globin promoter (17). Cells were transduced with either construct and analyzed by FACS to compare GFP expression in the CD34+ and CD34– populations.
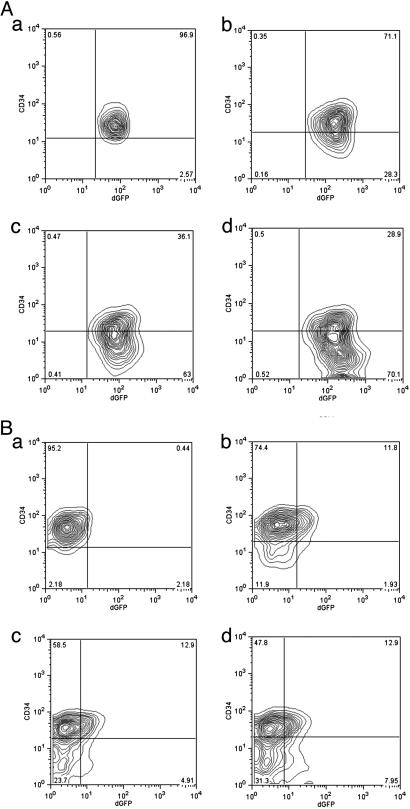
In the absence of the LCR enhancer region GFP was strongly expressed in most of the cells in the culture, and there was no distinction in the levels of expression of GFP between the two cell types (Fig. 3A). However, somewhat unexpectedly, as shown in Fig. 3B, with constructs containing the LCR, the expression of GFP in the CD34+ cells was significantly reduced compared with either the CD34– cells or the CD34+ cells carrying a construct without the LCR. Although some of the cells not expressing GFP may have evaded infection by the lentivirus vector, PCR of viral sequences showed that the vector sequences were present in the GFP– cells at levels similar to those in the GFP+ cells (Fig. 3C). Also, cultivation of the CD34high, GFPlow cells in SCF regenerated CD34low, GFPhigh cells (Fig. 4A). These results raise the possibility that the LCR elements can act to dampen the levels of globin expression in immature cells and augment it as erythroid cells mature. Further, with the exception of a few CD34+ cells that expressed GFP strongly, most of the CD34+ cells fell into a single cluster well separated from the CD34– cells on the basis of GFP expression. Culture of the CD34+ GFP-expressing cells in SCF rapidly generated a population of largely CD34– GFP-expressing cells (Fig. 4B), suggesting that the GFP-CD34+ cells may have partially differentiated toward CD34– cells but still retained CD34 protein.
Fig. 3.
The expression of dGFP driven by the β-globin promoter alone or with an upstream insert of the HS2 enhancer. (A) The map of the integrated β promoter-dGFP (Upper) and the expression of dGFP in Lin–CD34+ or Lin–CD34– EML cells infected with a lentiviral reporter containing the β-promoter-dGFP cassette (Lower). PPT, polypurine tract. (B) The map of the integrated HS2-β promoter-dGFP (Upper) and the expression of dGFP in Lin–CD34+ or Lin–CD34– EML cells infected with a lentiviral reporter containing the HS2-β-promoter-dGFP cassette (Lower). (C) Seventy five nanograms of genomic DNA from parental EML cells (no lentivirus infection) or Lin–CD34+dGFP+ and Lin–CD34+dGFP– EML cells (infected with a lentiviral reporter containing the cassette of HS2-β-promoter-dGFP) was amplified with the primers for the dGFP gene by PCR. This process showed that there was a similar amount of lentiviral DNA in each of the two cell types, even though GFP expression was lower in the CD34+ cells.
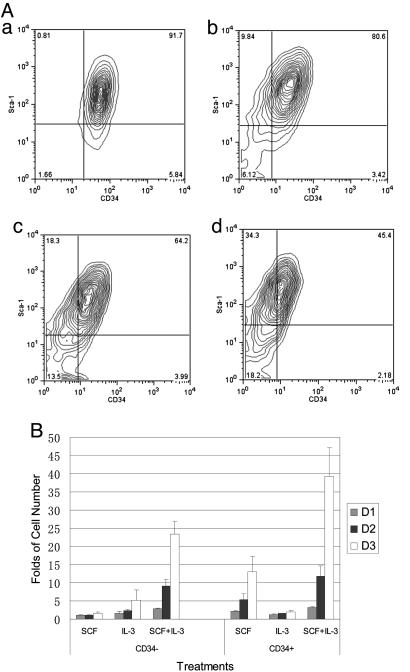
Fig. 4.
The differentiation of Lin–CD34+dGFPlow and Lin–CD34+dGFP– EML cells transfected with HS2–2b-dGFP lentivirus. (A) The expression of dGFP in CD34+ or CD34– EML cells differentiated from Lin–CD34+dGFPlow EML cells. (a) The purification of the sorted Lin–CD34+dGFPlow. (b–d) The sorted Lin–CD34+dGFPlow cells were cultured in SCF medium for 1 day (b), 2 days (c), and 3 days (d). (B) The expression of dGFP in CD34+ or CD34– EML cells differentiated from Lin–CD34+dGFP– EML cells. (a) The purification of the sorted Lin–CD34+dGFP–. (b–d) The sorted Lin–CD34+dGFP– cells were cultured in SCF medium for 1 day (b), 2 days (c), and 3 days (d).
Certain receptors were elevated in the CD34– cells compared with the CD34+ cells. This finding prompted us to examine the growth requirements of separated populations of CD34+ and CD34–, Lin– EML cells. Growth of CD34+ cells in SCF rapidly regenerated an ≈50–50 mixture of CD34+ and CD34– cells (Fig. 5A). The CD34– cells in the same medium slowly began to accumulate some CD34+ cells (data no shown), but the time scale is such that they could have come from contaminating CD34+ cells in the preparation.
Fig. 5.
The differentiation and proliferation of Lin–CD34– or Lin–CD34+EML cells in response to different cytokines. (A) The expression of cell surface markers CD34 and Sca-1 on Lin–CD34+ EML cells in SCF medium at different times, as analyzed by FACS. (a) Purification check after the sorting. (b–d) Lin–CD34+ cells were subcultured in SCF medium after 1 day (b), 2 days (c), and 4 days (d). (B) The growth curve of the sorted Lin–CD34– and Lin–CD34+ EML cells incubated in Iscove's modified Dulbecco medium supplemented with 15% horse serum in the presence of SCF (100 ng/ml), IL-3 (5 ng/ml), or a combination of SCF (100 ng/ml) and IL-3 (5 ng/ml) for different times. The live cells were counted by using trypan blue staining. The data represent three independent experiments.
The CD34– cells did not increase in cell number when cultivated with SCF within 3 days after the FACS. They grew somewhat slowly in IL-3 and rapidly when the medium was supplemented with both IL-3 and SCF (Fig. 5B). In contrast, the CD34+ cells grew in SCF alone although they grew more rapidly with the combination of SCF and IL-3 (Fig. 5B). The apoptosis analysis showed that there were ≈15% more apoptotic cells in CD34– cells cultured in SCF medium compared with CD34+ cells (data no shown).
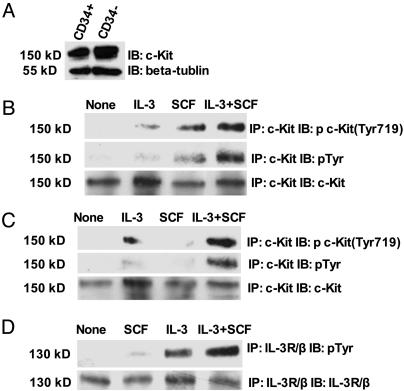
By Affymetrix cDNA analysis, real-time PCR (Table 1) and Western blotting (Fig. 6A) both the CD34+ and CD34– cell fractions had similar levels of expression of the SCF receptor c-Kit. However, the CD34– cells had several-fold more mRNA for the common IL-3R/β chain than did the CD34+ cells (Table 1). To investigate why the CD34– cells did not grow in SCF, we immunoprecipitated the c-Kit receptor or the IL-3R/β chain from cells grown under various conditions, and then checked for the presence of phosphotyrosine in either receptor. As expected the c-Kit receptor rapidly became tyrosine-phosphorylated after addition of SCF to the CD34+ cells (Fig. 6B). However, even in the presence of the same amount of receptor and SCF, the c-Kit receptor in the CD34– cells did not become tyrosine-phosphorylated (Fig. 6C). Curiously, addition of IL-3 instead of SCF produced some level of increased phosphotyrosine in the c-Kit from the CD34– cells (Fig. 6C). Addition of SCF and IL-3 together synergistically produced marked phosphorylation of c-Kit in CD34+ and CD34– cells (Fig. 6 B and C). In CD34– cells, stimulated with IL-3 for 10 min, the IL-3R/β chain became tyrosine-phosphorylated. Also significant phosphorylation of IL-3R/β chain could be detected after SCF treatment. Furthermore, stimulation with IL-3 together with SCF markedly enhanced the phosphorylation of IL-3R/β chain (Fig. 6D). These results suggest that transphosphorylation or some other form of transregulation between c-Kit and IL-3R/β chain occurs in CD34+ and CD34– cells.
Fig. 6.
The phosphorylation of c-Kit or IL-3R/β in Lin–CD34– and Lin–CD34+EML cells in response to different cytokines. (A) Total cell lysates of Lin–CD34+ and Lin–CD34– EML cells were separated by SDS/PAGE and transferred to an Immobion-P membrane. The membrane was probed with anti-c-Kit antibody, stripped, and reprobed with anti-β-tubulin antibody. IB, immunoblotting. (B and C) Lin–CD34+ (B) and Lin–CD34– (C) EML cells were starved and treated with no stimulation, SCF (100 ng/ml), IL-3 (5 ng/ml), or the combination IL-3 (5 ng/ml) and SCF (100 ng/ml) for 10 min. Cells were harvested, and c-Kit was immunopreciptated (IP) with c-Kit antibody (M14). The immunopreciptates were analyzed by SDS/PAGE, followed by immunoblotting with the indicated antibodies. After being hybridized with antiphosphotyrosine Y719-specific c-Kit antibody, the same membrane was stripped and reprobed with the general antiphosphotyrosine antibody PY100 and anti-c-Kit antibody. (D) Lin–CD34– EML cells were starved and treated with no stimulation, SCF (100 ng/ml), IL-3 (5 ng/ml), or the combination IL-3 (5 ng/ml) and SCF (100 ng/ml) for 10 min. Cells were harvested, and IL-3R/β was immunopreciptated with anti-IL-3R/β antibody (K17). The immunopreciptates was analyzed by SDS/PAGE, followed by immunoblotting with antiphosphotyrosine antibody PY100. The membrane was then striped and reprobed with IL-3R/β antibody.
Discussion
EML cells are described as a multipotent cell line, with indications that they may differentiate into megakaryocyte- or lymphocyte-like cells. There is definite evidence for their capacity to differentiate along the erythroid lineage or give rise to cells that can develop into mature neutrophils. The mRNA analyses presented here are consistent with this differentiation potential, although there was pronounced erythroid bias as represented by expression of globins, GATA1 (but also GATA2), EKLF, erythropoietin receptor, and several other genes involved in red cell cytoskeleton formation or heme synthesis (18–20). Because the EML cell line can be repeatedly cloned and has been rederived from normal marrow, it has been suggested that it reflects the features of normal multipotential hematopoietic precursor cells. Nevertheless, the block to granulocytic differentiation induced by the dominant negative retinoic acid receptor raises the caution that these cells may not reflect in its entirety any single stage of normal development.
The heterogeneity in normal cultures of EML cells propagated for relatively long periods of time indicate that there must be both ongoing self-renewal of precursor cells and differentiation of progeny in these cultures. This potential is strikingly demonstrated by the observation that the CD34+Lin– cell population can, within a few days of exponential growth, re-establish a mixed culture in which there are about equal numbers of CD34+ and CD34–Lin– cells. The CD34– cells cannot proliferate in media containing only SCF, suggesting that they are derived from the CD34+ cells. However, the population as a whole is growing rapidly so that overall each CD34+ cell appears to have an equal probability of generating two CD34+ cells or at least one CD34– cell. In several models of multizoate stem cell division, a niche containing other types of cells provides an environment that modulates the outcome of stem cell divisions, sometimes leaving one stem cell attached to a site in the niche, whereas the other progeny leaves the niche and differentiates (21–25). However, with EML cells in solution there is no anatomical niche as the cells grow in suspension with no matrix. This observation suggests that the choice is regulated by either a tightly controlled stochastic mechanism or a determined pattern of selective differentiation such as occurs in dividing Schizosaccharomyces pombe cells (26–28).
A construct expressing unstable GFP driven by the β-globin promoter produced strong GFP signals in essentially all cells whether CD34+ or CD34–. However, when the enhancer regions of the β-globin LCR HS2 was inserted upstream from the β-globin promoter, there was a marked down-regulation of GFP expression in the CD34+ but not the CD34– cells. Both CD34+ and CD34– cells express GATA2, an early hematopoietic transcription factor, and GATA1, a factor associated with mature erythropoiesis and megakaryocytopoiesis (10, 29, 30), and both express mRNA for a number of erythroid genes. However, the levels of GATA1 and globin mRNA are higher in the CD34– cells. This difference could have been caused by heterogeneity in the populations or alternatively could reflect a more erythroid character of the entire CD34– population. FACS analysis of GFP expression on a cell-by-cell basis favored the latter interpretation. Conversely, the FACS analysis suggested that there might be some heterogeneity in the CD34+ cell fraction, with the existence of a minor population of these cells that are more erythroid in character than the major population.
The globin HS2 enhancer has been shown in a variety of studies to act as an enhancer of transcription in erythroid cells (17, 30–32). The present studies raise the possibility that these same sequences may act to limit the expression of linked promoters in early hematopoietic precursors, and it will be of interest to extend these results and determine which factors bind the LCR in the CD34+ cells. After the present experiments were completed, reports (33, 34) appeared suggesting that the globin LCR may act as a negative regulator of gene expression under some circumstances, and our present observations are consistent with that proposal.
The failure of the CD34– population of cells to grow in SCF medium was somewhat surprising as they express levels of the SCF receptor (c-Kit) that are virtually the same as the levels in the SCF-responsive CD34+ cells. In contrast, the CD34– cells grew to moderate extent in IL-3 alone, whereas the CD34+ cells did not. This growth may be related to an increased level of mRNA for the IL-3R/common β chain in the CD34– cells.
The shift from SCF-dependent growth to a requirement for other cytokines such as IL-3 is a plausible early differentiation event, permitting the body to separately regulate the proliferation of different stages of precursor cells (35–37). The present studies show that this shift is related to a failure of SCF to cause detectable c-Kit tyrosine phosphorylation in the CD34– cells. This lack of c-Kit phosphorylation could, in principle, be caused by either failure to activate the receptor or a phosphatase activity that rapidly dephosphorylated the tyrosines on the receptor (38). However, curiously, IL-3 treatment without SCF caused some phosphorylation of c-Kit in the CD34– cells, so that phosphatase inhibition is less likely. The synergistic growth effect of IL-3 together with SCF correlated with a very prominent phosphorylation of c-Kit and IL-3R/β chain in the CD34– cells treated with the cytokine combination. This finding suggests the transphosphorylation between c-Kit and IL-3R occurred in CD34+ and CD34– cells. Our results correlate with the results for c-Kit and erythropoietin receptor where transphosphorylation has been observed and suggested to contribute to synergy (39). The full explanation for these phenomena awaits further research. However, it is interesting that a physical association between IL-3R and SCF receptor has been noted in other cell types (6), and the present results would be consistent with a functional interaction between at least some subunits of the receptors in the EML cells.
In summary, we present evidence that EML cells cultured in SCF are a mixed culture that includes a subpopulation that is replicating and gives rise to a second SCF nonresponsive population to maintain a fairly precise ratio of cell populations in the culture. The change from the CD34+ replicating population to the nonreplicating cells is associated with a specific loss of response of the receptor c-Kit to SCF together with the acquisition of a growth response to IL-3. This system provides an opportunity to study bulk cultures of precursor cells that undergo self-renewal and differentiation in a quantitatively precise way in the absence of any stem cell niche.
Acknowledgments
We thank Dr. Millind C. Majajan, Dr. Chen Liao, Dr. Yashiro Nakayama, Brian Reed, and Dr. Yin Lu for technical assistance; the Hematology Seminar Group at the Yale University School of Medicine for stimulating discussion; Dr. Schickwann Tsai for EML and W20 cell lines; and Dr. Irving L. Weissman for the lentiviral vectors. This work was supported by National Institutes of Health Grant 1R01 AG23111-02 and National Heart, Lung, and Blood Institute Contract N01-HV-28186.
Author contributions: Z.-j.Y. and S.M.W. designed research; Z.-j.Y. and Z.L. performed research; Z.-j.Y. and Y.K. analyzed data; and Z.-j.Y. and S.M.W. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: SCF, stem cell factor; LCR, locus control region; HS2, hypersensitive site 2; Lin cells, lineage cells; dGFP, destabilized GFP; IL-3R, IL-3 receptor.
References
- 1.Tsai, S., Bartelmez, S., Sitnicka, E. & Collins, S. (1994) Genes Dev. 8, 2831–2841. [DOI] [PubMed] [Google Scholar]
- 2.Pinto do, O. P., Kolterud, A. & Carlsson, L. (1998) EMBO J. 17, 5744–5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu, W. M., Hawley, T. S., Hawley, R. G. & Qu, C. K. (2002) Blood 100, 3828–3831. [DOI] [PubMed] [Google Scholar]
- 4.Dull, T., Zufferey, R., Kelly, M., Mandel, R. J., Nguyen, M., Trono, D. & Naldini, L. (1998) J. Virol. 72, 8463–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reya, T., Duncan, A. W., Ailles, L., Domen, J., Scherer, D. C., Willert, K., Hintz, L., Nusse, R. & Weissman, I. L. (2003) Nature 423, 409–414. [DOI] [PubMed] [Google Scholar]
- 6.Lennartsson, J., Shivakrupa, R. & Linnekin, D. (2004) J. Biol. Chem. 279, 44544–44553. [DOI] [PubMed] [Google Scholar]
- 7.Kluger, Y., Lian, Z., Zhang, X., Newburger, P. E. & Weissman, S. M. (2004) BioEssays 26, 1276–1287. [DOI] [PubMed] [Google Scholar]
- 8.Kluger, Y., Tuck, D. P., Chang, J. T., Nakayama, Y., Poddar, R., Kohya, N., Lian, Z., Ben Nasr, A., Halaban, H. R., Krause, D. S., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 6508–6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raich, N., Clegg, C. H., Grofti, J., Romeo, P. H. & Stamatoyannopoulos, G. (1995) EMBO J. 14, 801–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weber, G. J., Choe, S. E., Dooley, K. A., Paffett-Lugassy, N. N., Zhou, Y. & Zon, L. I. (2005) Blood 106, 521–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Passegue, E., Wagner, E. F. & Weissman, I. L. (2004) Cell 119, 431–443. [DOI] [PubMed] [Google Scholar]
- 12.Shizuru, J. A., Negrin, R. S. & Weissman, I. L. (2005) Annu. Rev. Med. 56, 509–538. [DOI] [PubMed] [Google Scholar]
- 13.Jamieson, C. H., Ailles, L. E., Dylla, S. J., Muijtjens, M., Jones, C., Zehnder, J. L., Gotlib, J., Li, K., Manz, M. G., Keating, A., Sawyers, C. L. & Weissman, I. L. (2004) N. Engl. J. Med. 351, 657–667. [DOI] [PubMed] [Google Scholar]
- 14.Chen, D. & Zhang, G. (2001) Exp. Hematol. 29, 971–980. [DOI] [PubMed] [Google Scholar]
- 15.Caloca, M. J., Zugaza, J. L., Matallanas, D., Crespo, P. & Bustelo, X. R. (2003) EMBO J. 22, 3326–3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin-Henao, G. A., Quiroga, R., Sureda, A. & Garcia, J. (1999) Am. J. Hematol. 61, 178–186. [DOI] [PubMed] [Google Scholar]
- 17.Kong, S., Bohl, D., Li, C. & Tuan, D. (1997) Mol. Cell. Biol. 17, 3955–3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Terskikh, A. V., Miyamoto, T., Chang, C., Diatchenko, L. & Weissman, I. L. (2003) Blood 102, 94–101. [DOI] [PubMed] [Google Scholar]
- 19.Letting, D. L., Chen, Y. Y., Rakowski, C., Reedy, S. & Blobel, G. A. (2004) Proc. Natl. Acad. Sci. USA 101, 476–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horak, C. E., Mahajan, M. C., Luscombe, N. M., Gerstein, M., Weissman, S. M. & Snyder, M. (2002) Proc. Natl. Acad. Sci. USA 99, 2924–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rattis, F. M., Voermans, C. & Reya, T. (2004) Curr. Opin. Hematol. 11, 88–94. [DOI] [PubMed] [Google Scholar]
- 22.Reynaud, D., Ravet, E., Titeux, M., Mazurier, F., Renia, L., Dubart-Kupperschmitt, A., Romeo, P. H. & Pflumio, F. (2005) Blood 106, 2318–2328. [DOI] [PubMed] [Google Scholar]
- 23.Takano, H., Ema, H., Sudo, K. & Nakauchi, H. (2004) J. Exp. Med. 199, 295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore, K. A. & Lemischka, I. R. (2004) Cell 118, 139–140. [DOI] [PubMed] [Google Scholar]
- 25.Zhu, J. & Emerson, S. G. (2004) BioEssays 26, 595–599. [DOI] [PubMed] [Google Scholar]
- 26.Petersen, J. & Hagan, I. M. (2005) Nature 435, 507–512. [DOI] [PubMed] [Google Scholar]
- 27.Oliva, A., Rosebrock, A., Ferrezuelo, F., Pyne, S., Chen, H., Skiena, S., Futcher, B. & Leatherwood, J. (2005) PloS. Biol. 3, e225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daga, R. R. & Chang, F. (2005) Proc. Natl. Acad. Sci. USA 102, 8228–8232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ling, K. W., Ottersbach, K., van Hamburg, J. P., Oziemlak, A., Tsai, F. Y., Orkin, S. H., Ploemacher, R., Hendriks, R. W. & Dzierzak, E. (2004) J. Exp. Med. 200, 871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuasa, H., Oike, Y., Iwama, A., Nishikata, I., Sugiyama, D., Perkins, A., Mucenski, M. L., Suda, T. & Morishita, K. (2005) EMBO J. 24, 1976–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cavallesco, R. & Tuan, D. (1997) Blood Cells Mol. Dis. 23, 8–26. [DOI] [PubMed] [Google Scholar]
- 32.Morley, B. J., Abbott, C. A., Sharpe, J. A., Lida, J., Chan-Thomas, P. S. & Wood, W. G. (1992) Mol. Cell. Biol. 12, 2057–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng, Y. Q., Warin, R., Li, T., Olivier, E., Besse, A., Lobell, A., Fu, H., Lin, C. M., Aladjem, M. I. & Bouhassira, E. E. (2005) Mol. Cell. Biol. 25, 3864–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, J., Liu, H., Lin, C. M., Aladjem, M. I. & Epner, E. M. (2005) J. Biol. Chem. 280, 23340–23348. [DOI] [PubMed] [Google Scholar]
- 35.Wandzioch, E., Edling, C. E., Palmer, R. H., Carlsson, L. & Hallberg, B. (2004) Blood 104, 51–57. [DOI] [PubMed] [Google Scholar]
- 36.Stein, M. I., Zhu, J. & Emerson, S. G. (2004) Exp. Hematol. 32, 1129–1136. [DOI] [PubMed] [Google Scholar]
- 37.Era, T. (2002) Int. J. Hematol. 76, 35–43. [DOI] [PubMed] [Google Scholar]
- 38.Lennartsson, J., Jelacic, T., Linnekin, D. & Shivakrupa, R. (2005) Stem Cells 23, 16–43. [DOI] [PubMed] [Google Scholar]
- 39.Wu, H., Klingmuller, U., Besmer, P. & Lodish, H. F. (1995) Nature 377, 242–246. [DOI] [PubMed] [Google Scholar]