Abstract
Activation of the Wnt/β-catenin pathway promotes the development of several cancers and is an attractive target for chemopreventive and chemotherapeutic agents. Nonsteroidal antiinflammatory drugs (NSAIDs) have been reported to antagonize β-catenin function, but their mechanism of action is not known. We demonstrate here that interference with β-catenin function by NSAIDs does not correlate with cyclooxygenase (COX) inhibition. Instead, NSAID inhibition of β-catenin requires the high level expression of peroxisome proliferator-activated receptor γ (PPAR-γ) and its co-receptor retinoid-X-receptor α (RXR-α). Immunoprecipitation experiments show that β-catenin interacts with RXR-α and PPAR-γ in some malignant cells. Repression of β-catenin-dependent transcription by NSAIDs is thus indirect and depends on the coexpression of other nuclear receptors.
Keywords: peroxisome proliferator-activated receptor γ, retinoid X receptor α
The long-term ingestion of various nonsteroidal antiinflammatory drugs (NSAIDs) is associated with a reduced incidence of colorectal cancer, and probably of prostate and breast cancer (1–6). However, the general inhibitors of cyclooxygenases (COX) have side effects that preclude their use in the general population (7–9). The development of the more specific COX-2 inhibitors was an effort to overcome this problem. Unfortunately, these drugs may increase the risk of cardiovascular diseases (10–14). Accordingly, there is a major therapeutic need for agents that retain the chemopreventive actions of NSAIDs, but that are devoid of their potential toxicities.
Activation of the multifunctional protein β-catenin has been shown to play a critical role in the development of colorectal cancer and has been implicated in prostate and breast cancer as well (15–21). Wnt ligand interaction with a seven-transmembrane receptor of the frizzled family induces a signaling cascade wherein molecular complexes are recruited close to the membrane. The disheveled (Dsh) protein alters axin interactions, which then results in the release of β-catenin from a cytoplasmic complex (22–24). Unphosphorylated β-catenin migrates to the nucleus (25–27). There, it associates with transcription factors of the T cell factor (TCF) and lymphocyte-enhancing factor (LEF) family, as well as with various transcriptional cofactors, to form multiprotein complexes that regulate genes important for proliferation, differentiation, and apoptosis (21, 28).
Different NSAIDs have been demonstrated to inhibit the activity of β-catenin-dependent reporter genes in malignant cell lines, and to induce β-catenin degradation (29–34). Moreover, the colonic polyps of patients treated with NSAIDs have reduced nuclear accumulation of β-catenin (29). The biochemical and clinical data imply that NSAIDs inhibit β-catenin activity or its stability (31, 32). However, the biochemical basis for these effects is uncertain, insofar as NSAIDs have not been shown to interact directly with β-catenin.
At concentrations far higher than those required to inhibit COX-1 and COX-2, NSAIDs can weakly interact with the nuclear hormone receptor peroxisome proliferator-activated receptor-γ (PPAR-γ) (35). The binding could be related to the chemopreventive actions of the drugs, because high-affinity synthetic ligands of PPAR-γ, such as troglitazone, can prevent the development of colon, prostate, and breast cancer in some animal models (36–38). Recently, the NSAID-like molecule R-etodolac was also reported to bind the PPAR-γ coactivator retinoid X receptor α (RXR-α) (39). However, it is not clear how the functions of β-catenin, RXR-α, and PPAR-γ are connected. Indeed, some transgenic mice that express constitutively active PPAR-γ have accelerated mammary gland tumors, compared with control animals from the same strains (40).
Activating ligands for different nuclear hormone receptors, including retinoid, androgen, and vitamin D receptors, have been reported to inhibit β-catenin-dependent transcription, possibly by competition for a common pool of cofactors (41–44). The phenomenon has been referred to as transrepression. Because NSAIDs can bind PPAR-γ and RXR-α, we hypothesized that their effects on β-catenin function might also be attributable to transrepression. The current experiments indicate that NSAIDs efficiently inhibit β-catenin function only in cells that coexpress both PPAR-γ and RXR-α. These results have implications for the development and selective use of NSAID-like drugs as cancer chemopreventive and chemotherapeutic agents.
Materials and Methods
R-etodolac was prepared by fractional crystallization from pharmaceutical grade tablets of racemic etodolac as described (45). The enantiomeric purity was >97% as assayed by HPLC on a chiral column (AGP, ChromTech, Hagersten, Sweden). The PPAR-α activator WY14,643 was purchased from ChemSyn Laboratory (Lenexa, KS). Unless otherwise indicated, other reagents were from Sigma.
Plasmids. The PPAR-γ reporter plasmid p(AOX)3-TK-Luc, and the expression vectors for PPAR-γ, PPAR-γEA469, and RXR-α have been described, and were kindly provided by C. Glass (University of California at San Diego) (46). The PPAR-γEA469 expression vector has a mutation in the AF2 domain that destroys its transactivation, transrepression, and coactivator interactions (47). The β-catenin expression plasmid, and the β-catenin-dependent TCF/LEF TOPflash reporter plasmid were gifts from H. Clevers (University of Utrecht, Utrecht, The Netherlands). An expression plasmid for human Dsh was purchased from Origene Technologies (Rockville, MD). S. Ho (University of California at San Diego) provided the expression plasmid for nuclear factor of activated T cells (NFAT) (48). The expression plasmids pCMXβgal and H-RasV12 have been described (49). The pAP-1 and pNFAT-Luc reporter plasmids were purchased from BD Biosciences. The UAS-TK-Luc reporter for Gal4, and expression plasmids for Gal4, Gal4-PPAR-γ binding protein (PBP), and VP16 for the mammalian two-hybrid system have been described (50, 51). The VP16-PPAR-γ expression vector was constructed by ligating PPAR-γ cDNA into the corresponding KpnI and NheI sites of the pVP16 vector.
Cell Culture and Transfections. LNCaP cells were grown in RPMI medium 1640 (Life Technologies) supplemented with 10% FBS and 100 μg/ml penicillin and 100 μg/ml streptomycin. The human embryonic kidney cell line HEK293 was maintained in DMEM supplemented with 10% FBS. The cells were transfected by using the FuGENE transfection reagent (Roche Diagnostics) according to the manufacturer's instructions, typically with 0.5 μg of reporter plasmid, 0.1–0.2 μg of control plasmid pCMXβgal, 0.1–0.2 μg of expression plasmid, and carrier DNA pBluescriptKSII for a total of 1 μg per well. After overnight incubation, the cells were washed and given fresh medium that contained 0.5% FBS supplemented with the different drugs, as indicated in the figure legends. If DMSO was used, the control cultures received the same solvent. For luciferase assays, cells were lysed in potassium phosphate buffer containing 1% Triton X-100, and light emission was detected in the presence of luciferin by using a microtiter plate luminometer (MicroBeta TriLux, Gaithersburg, MD). The luciferase values were normalized for variations in transfection efficiency by using the β-gal internal control, and are expressed either as relative luciferase units (RLU) or as fold stimulation of luciferase activity compared with the designated control cultures. All of the transfection results represent means of a minimum of three independent transfections assayed in duplicate, ± SEM.
Immunoprecipitation and Immunoblotting. Cells were washed twice with PBS and collected in 0.5 ml of lysis buffer (20 mM Tris·HCl, pH 8.0/10% glycerol/5 mM MgCl2/0.15 M KCl/0.1% Nonidet P-40/protease inhibitors). The lysates of 0.5 to 1 × 107 cells were incubated overnight at 4°C with saturating amounts of agarose beads linked to monoclonal antibodies specific for β-catenin or PPAR-γ (Santa Cruz Biotechnology). The beads were washed twice with lysis buffer and once with PBS. Bound proteins were eluted by boiling the samples in SDS sample buffer and were then resolved by 10% SDS/PAGE and transferred to nitrocellulose membranes.
After removal of medium, cells were disrupted in lysis buffer (25 mM Tris·HCl/150 mM KCl/5 mM EDTA/1% Nonidet P-40/0.5% sodium deoxycholic acid/0.1% SDS) including phosphatase and protease inhibitor cocktails. The samples were heated to 70°C for 10 min in loading buffer with or without 10 mM DTT. Each lane of an SDS/PAGE gel was loaded with 20 μg of protein. After electrophoresis, the proteins were transferred to a poly(vinylidene difluoride) (PVDF) membrane, blocked with 2% I-Block (Tropix, Bedford, MA) containing 0.05% Tween 20 in PBS, and then incubated with primary antibody. Antibodies to PPAR-γ, RXR-α, and β-catenin (all from Santa Cruz Biotechnology) were used. Horseradish peroxidase-conjugated anti-IgG was used as the secondary antibody. The membranes were developed by using a chemiluminescence system (ECL detection reagent, Amersham Pharmacia Life Science). The membranes were stripped with Re-Blot Western blot recycling kit (Chemicon International, Temecula, CA) and reprobed.
Results
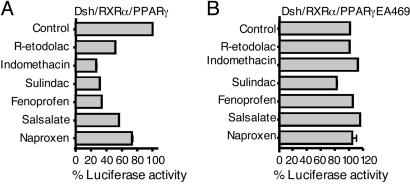
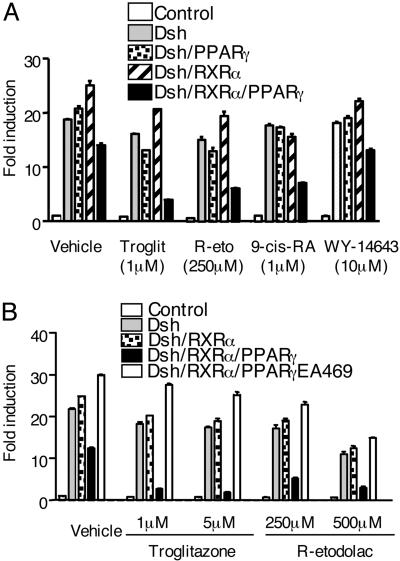
Dependence of β-Catenin Antagonism on PPAR-γ and RXR-α. Nineteen approved NSAIDs, plus R-etodolac, were tested for β-catenin antagonism in a cell-based screen, by using a reporter construct for TCF-dependent signal transmission. The HEK293 cells were transiently transfected with plasmids for (i) Dsh or β-catenin, (ii) the same plasmids, plus plasmids for RXR-α and PPAR-γ, and (iii) the TCF/LEF-dependent TOPflash reporter gene. All of the tested NSAIDs, with the exception of rofecoxib, inhibited β-catenin signaling, as assessed by TOPflash expression of active luciferase in cells that expressed high levels of wild-type PPAR-γ and RXR-α (Fig. 1 and Table 1). The NSAIDs did not efficiently inhibit the TCF/LEF-dependent reporter gene in cells transfected with the inactive PPAR-γEA469 vector (Figs. 1 and 2), despite expression of the recombinant protein in immunoblots (data not shown). The β-catenin antagonistic activity of the NSAIDs was also diminished in cells transfected with a PPAR-γ plasmid alone, compared with cells transfected with plasmids for both PPAR-γ and RXR-α (Fig. 2).
Fig. 1.
NSAIDs inhibit Dsh/PPAR-γ/RXR-α-mediated transcription. HEK293 cells were transfected with a TCF/LEF-dependent reporter gene, expression plasmids for Dsh, RXR-α, and either active PPAR-γ (A) or inactive PPAR-γEA469 (B). After overnight incubation, the cells were treated for 24 h with 250 μM R-etodolac, 125 μM indomethacin, 250 μM sulindac, 200 μM fenoprofen, 300 μM salsalate, and 250 μM naproxen or vehicle alone, after which the reporter gene activity was measured. All cells were also transfected with a β-gal reporter gene to control for transfection efficiency. The results are expressed as % control TCF/LEF-dependent reporter gene activity ± SEM (n = 3).
Table 1. Inhibition of β-catenin/PPAR-γ/RXR-α signaling by NSAIDs.
| Percent control
|
||||||
|---|---|---|---|---|---|---|
| Drugs | 20 μM | 40 μM | 80 μM | 104 μM | 208 μM | 416 μM |
| Aspirin | 96.3 ± 0.6 | 92.3 ± 0.4 | 89.1 ± 4.8 | |||
| Naproxen | 100.3 ± 7.7 | 90.7 ± 0.9 | 55.2 ± 2.4 | |||
| Ketoprofen | 79.4 ± 2.4 | 65.3 ± 2.9 | 53.1 ± 3.8 | |||
| Ketorolac | 98.7 ± 3.2 | 78.5 ± 0.3 | 47.9 ± 1.1 | |||
| Ibuprofen | 78.1 ± 2.9 | 62.8 ± 0.4 | 50.3 ± 0.3 | |||
| Piroxicam | 101.9 ± 3.4 | 94.4 ± 2.8 | 88.4 ± 3.8 | |||
| Meloxicam | 98.3 ± 0.0 | 93.8 ± 10.6 | 78.6 ± 3.6 | |||
| Salsalate | 74.4 ± 4.6 | 53.8 ± 4.9 | 43.6 ± 3.9 | |||
| Flurbiprofen | 109.8 ± 0.1 | 79.0 ± 1.3 | 41.7 ± 1.5 | |||
| Sulindac | 62.7 ± 6.4 | 41.3 ± 0.3 | 28.5 ± 3.9 | |||
| Tolmetin | 85.3 ± 0.6 | 67.7 ± 1.7 | 53.8 ± 0.3 | |||
| Fenoprofen | 75.3 ± 5.2 | 50.6 ± 0.1 | 30.3 ± 1.4 | |||
| R-etodolac | 76.2 ± 2.4 | 58.5 ± 3.1 | 35.6 ± 1.1 | |||
| Diflunisal | 75.2 ± 4.5 | 58.0 ± 0.3 | 18.9 ± 0.2 | |||
| Diclofenac | 84.1 ± 5.6 | 53.8 ± 4.6 | 9.1 ± 0.1 | |||
| Indomethacin | 41.2 ± 0.4 | 28.8 ± 1.3 | 5.2 ± 0.4 | |||
| Meclofenamic acid | 82.0 ± 1.3 | 41.4 ± 1.3 | 13.1 ± 0.2 | |||
| Nabumetone | 101.2 ± 1.4 | 52.8 ± 6.6 | 24.9 ± 1.0 | |||
| Rofecoxib | 104.5 ± 1.2 | 99.2 ± 8.4 | 96.4 ± 1.5 | |||
| Celecoxib | 95.0 ± 1.1 | 77.3 ± 4.3 | 6.2 ± 0.2 | |||
HEK293 cells were transfected with TOPflash reporter and expression plasmids for Dsh, PPAR-γ, and RXR-α. After overnight incubation, the cells were treated for 24 h with the indicated concentrations of NSAIDs. Presented are the means of the percentages of TOPflash activity relative to cells treated with the vehicle alone (DMSO treatment = 100%) ± SD. Values in bold represent the concentrations of approximately 50% of the control value. The compounds are listed in rank order of increasing cytotoxicity from top to the bottom based on an independent MTT-based assay.
Fig. 2.
Troglitazone and R-etodolac inhibit Dsh/β-catenin-mediated transcription through a process that requires PPAR-γ and RXR-α. (A) TOPflash reporter was transfected into HEK293 cells with expression plasmids for Dsh, PPAR-γ, and RXR-α, as indicated. After overnight incubation, the cells were treated for 24 h with 1 μM troglitazone, 1 μM9-cis-RA, 10 μM of the PPAR-α activator WY14,643, 250 μM R-etodolac, or vehicle alone. (B) This experiment was performed similar to A, except the inactive PPAR-γEA469 plasmid was used along with the indicated concentrations of troglitazone and R-etodolac. TOPflash reporter gene activity was measured. The results are expressed as fold induction of TCF/LEF-dependent reporter gene activity ± SEM (n = 3). All cells were also transfected with a β-gal reporter gene to control for transfection efficiency.
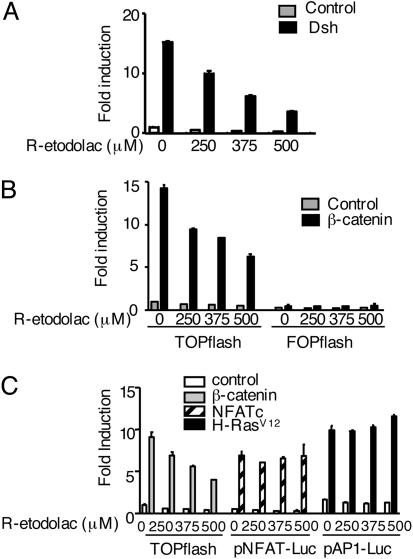
In control experiments, inhibition of β-catenin induced transcription by the PPAR-γ activator troglitazone, and the RXR-α activator 9-cis-retinoic acid (RA), also required coexpression of both nuclear receptors (Fig. 2). Inhibition of the TOPflash reporter by an NSAID was equivalent in cells transfected with either Dsh or β-catenin plasmids (Fig. 3 A and B). The inhibition of β-catenin signaling via the TCF reporter plasmid was specific, because the signals from reporter genes for NFAT and activator protein 1 (AP-1) were unaffected by the same NSAID (Fig. 3C).
Fig. 3.
NSAID inhibition of TCF/LEF-dependent transcription is specific and downstream of β-catenin. (A) The TCF/LEF-dependent reporter was transfected into HEK293 cells with expression plasmids for either Dsh (A) or β-catenin (B). Then, the cells were treated with the indicated concentrations of R-etodolac for 24 h. The fold induction values are the ratios of the normalized luciferase activities in cells transfected with both expression and reporter plasmids, compared with the activities in cells receiving the respective reporter plasmids alone. The results are the mean ± SEM of triplicate experiments. (C) Reporter plasmids for TCF/LEF, activator protein 1 (AP-1), or NFAT were transfected into HEK293 cells along with the respective expression plasmids for β-catenin, H-RasV12 and NFAT, as indicated. Transfected cells were treated with the indicated concentrations of R-etodolac for 24 h, and the fold increase in luciferase activities was determined. The results are the mean ± SEM of triplicate determinations.
Role of Cyclooxygenases (COXs). Because inhibition of β-catenin signaling by NSAIDs requires both PPAR-γ and RXR-α, the effect of these drugs may be indirect, and a consequence of COX blockade. However, the concentrations of NSAIDs required to inhibit β-catenin function were manyfold higher than the levels reported to block COX-1 or COX-2 (Table 1). Moreover, the COX-inactive R-stereoisomer of etodolac, as well as the very weak COX inhibitor salsalate, impeded β-catenin-stimulated transcription as well as conventional NSAIDs (52). Thus, COX inhibition was not necessary for β-catenin antagonism.
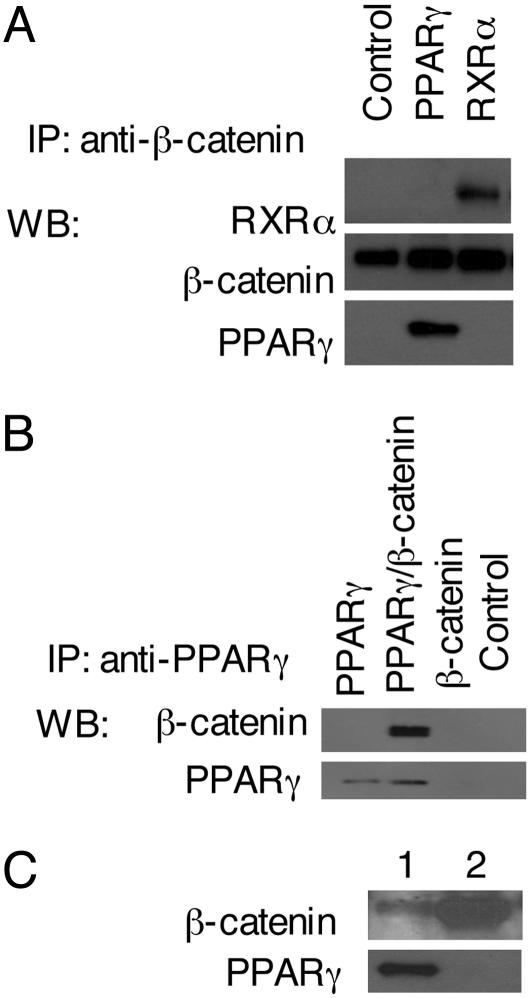
Interaction of β-Catenin with RXR-α and PPAR-γ. The overexpression of both RXR-α and PPAR-γ sensitized cells to NSAID suppression of TOPflash activity, suggesting that there may be a direct interaction between β-catenin and these proteins. Immunoprecipitation of cells overexpressing PPAR-γ and RXR-α with antibody to β-catenin pulled down all three proteins, as detected by immunoblotting (Fig. 4A). Similarly, an anti-PPAR-γ antibody pulled down β-catenin (Fig. 4B). The association was not an artifact of overexpression, because antibodies to PPAR-γ coimmunoprecipitated β-catenin in otherwise unmanipulated LNCaP prostate cancer cells (Fig. 4C).
Fig. 4.
Interaction of PPAR-γ and β-catenin. (A) Expression plasmids for Dsh, PPAR-γ, and RXR-α were cotransfected into HEK293 cells as indicated. At 48 h after transfection, cell extracts were prepared for immunoprecipitation (IP) with an anti-β-catenin monoclonal antibody. The immune complexes were analyzed by immunoblotting with anti-β-catenin, anti-PPAR-γ, and anti-RXR-α.(B) HEK293 cells were transfected with expression plasmids for PPAR-γ in the presence or absence of β-catenin. At 48 h after transfection, cell extracts were prepared for IP with the anti-PPAR-γ monoclonal antibody. The immune complexes were analyzed by immunoblotting with anti-β-catenin or anti-PPAR-γ antibodies. (C) LNCaP cells were grown at 37°C for 24–36 h. Cells were lysed, and IP was completed with anti-PPAR-γ (lane 1) and anti-β-catenin (lane 2) monoclonal antibodies. The immune complexes were analyzed by immunoblotting with anti-β-catenin or anti-PPAR-γ antibodies.
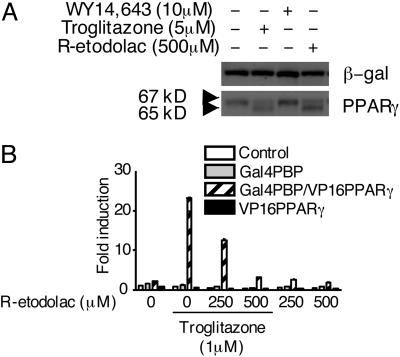
Interaction of R-Etodolac with PPAR-γ. The COX-inactive R-stereoisomer of etodolac was used to study the interactions of NSAIDs with PPAR-γ. In preliminary experiments, we were unable to measure the binding of [3H]R-etodolac to the recombinant ligand-binding domain of PPAR-γ, presumably because of its relatively low affinity (39), or potentially the requirement for interaction with RXRα. However, exposure of PPAR-γ-transfected cells to either R-etodolac or troglitazone caused the appearance in immunoblots of a new, lower molecular weight species, possibly representing a proteolytic product (Fig. 5A). Similar results were observed with other NSAIDs (data not shown). Moreover, in a mammalian two-hybrid reporter gene assay, R-etodolac inhibited the interaction of PPAR-γ with the PBP at the same concentrations that antagonized β-catenin function (Fig. 5B).
Fig. 5.
Interaction of R-etodolac with PPAR-γ. (A) HEK293 cells were transfected with expression plasmids for PPAR-γ and β-gal. After overnight incubation, the cells were treated for 24 h with 5 μM troglitazone, 10 μM WY14,643, 500 μM R-etodolac, and DMSO alone. Cell lysates were prepared and analyzed by SDS/PAGE under reducing conditions, transferred to a membrane, and probed with anti-PPAR-γ and anti-β-gal (β-gal) antibodies. (B) Inhibition by R-etodolac of the ligand-dependent interaction between PBP and PPAR-γ in a mammalian two-hybrid system. The UAS-TK-Luc reporter and expression plasmids for Gal4-PBP, VP16, VP16-PPAR-γ were transfected into HEK293 cells. After 16 h, the cells were treated with the indicated amount of R-etodolac, troglitazone, and vehicle alone for 24 h.
Discussion
Promiscuous activation of β-catenin is a principal cause of colorectal cancer. In prospective studies, the administration of NSAIDs has been demonstrated to inhibit the growth of premalignant polyps in patients with mutations in the adenomatosis polyposis coli (APC) gene, which regulates β-catenin activation and degradation (29). Hence, it is logical to assume that NSAIDs inhibit β-catenin function in some way. Indeed, reduced nuclear expression of β-catenin has been observed in some colonic polyps from patients treated with an NSAID (30–32). However, the clinical observations have been difficult to explain at a molecular level. In part, this is because β-catenin is not an enzyme that can be targeted with active site directed inhibitors, but rather is a multifunctional docking protein with roles in transcription and cell adhesion. How inhibition of COX enzymes can regulate β-catenin activity is not clear. High throughput screens for β-catenin antagonists have not yielded COX inhibitors or β-catenin-binding agents, but rather compounds that interact with its transcriptional coactivator cAMP binding protein (CBP) (53). The sulfone metabolite of the NSAID sulindac has been reported to induce β-catenin degradation as a consequence of inhibition of a cGMP-dependent phosphodiesterase (54). Other NSAIDs have also been shown to cause β-catenin proteolysis by caspases in some malignant cells (55, 56).
In a preliminary cell-based screen of known drugs for antagonism of β-catenin-dependent transcription, the NSAIDs and the PPAR-γ ligands were the two compound classes that most frequently scored positive. However, we were unable to demonstrate a direct interaction between NSAIDs and β-catenin. Rather, repression of β-catenin function by NSAIDs required the high-level expression of PPAR-γ and RXR-α. In cells transfected with a mutated PPAR-γ plasmid that lacked transactivation or transrepression function, NSAIDs did not inhibit β-catenin-induced transcription. The NSAID inhibition of β-catenin was also minimal in cells with low levels of RXR-α. Thus, antagonism of β-catenin by NSAIDs was context-dependent.
The β-catenin inhibitory activity of NSAIDs did not correlate with inhibition of COX function, because the COX inactive R-stereoisomer of the NSAID etodolac, and the weak COX inhibitor salsalate, were both active. Previously, Hedvat et al. (57) reported that R-etodolac weakly activated a PPAR-γ-dependent reporter gene in transfected cells and induced the degradation of PPAR-γ in human prostate cancer xenografts. Subsequently, Kolluri et al. (39) showed that R-etodolac bound to RXR-α inhibited its transactivation function and caused its degradation in murine prostate tumors. The current experiments demonstrate that β-catenin, RXR-α, and PPAR-γ can interact directly in cells, suggesting that a multireceptor complex is involved in NSAID inhibition of β-catenin.
Not only NSAIDs, but also the PPAR-γ ligand troglitazone, and the RXR-α ligand 9-cis-retinoic acid, antagonized β-catenin-dependent reporter gene activity (58, 59). However, antagonism of β-catenin by R-etodolac could not be attributed to PPAR-γ or RXR-α activation. This NSAID-like agent has been shown to inhibit the ability of a high-affinity RXR-α ligand to activate transcription (39). As shown here, it also blocked the troglitazone-induced association of PPAR-γ with its binding protein (Fig. 5). Moreover, the concentrations of PPAR-γ and RXR-α activators required to block β-catenin-induced transcription far exceed the concentrations that have been reported to stimulate the respective receptors. The results of these experiments are consistent with a transrepression model of β-catenin inhibition, which depends on the high-level expression of both RXR-α and PPAR-γ.
Many chemoprevention and chemotherapy studies with NSAIDs, PPAR-γ activators, and RXR agonists have been curtailed because of the toxicities associated with the primary actions of these drugs, and because of lack of efficacy in some systems. Screens for agents that bind RXR-α and/or PPAR-γ, without activating the receptors, may yield drugs capable of antagonizing β-catenin function in malignant cells with minimal side effects. However, agents of this type would be effective only in cells that highly express all three proteins. Thus, the proteonomic profile of a malignant cell may control the ability of NSAIDs, and related agents, to inhibit β-catenin-dependent growth and survival.
Acknowledgments
We thank M. Rosenbach, K. Pekny, P. Charos, L. Ronacher, R. Tawatao, and Y. Zhao for technical assistance and N. Noon for secretarial support. This work was supported in part by National Institutes of Health Grants CA81534 and CA113318 and U.S. Army Medical Research and Material Command Award W81XWH-04-1-0453.
Author contributions: D.L. performed research; H.B.C. contributed new reagents/analytic tools; M.C. and D.A.C. analyzed data; D.A.C. designed research; and M.C. and D.A.C. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: NSAID, nonsteroidal antiinflammatory drug; COX, cyclooxygenase; TCF, T cell factor; LEF, lymphoid-enhancing factor; PPAR, peroxisome proliferator-activated receptor; RXR-α, retinoid X receptor α; PBP, PPAR-binding protein; Dsh, dishevelled; RA, retinoic acid; NFAT, nuclear factor of activated T cells.
References
- 1.Collet, J. P., Sharpe, C., Belzile, E., Boivin, J. F., Hanley, J. & Abenhaim, L. (1999) Br. J. Cancer 81, 62-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harris, R. E., Chlebowski, R. T., Jackson, R. D., Frid, D. J., Ascenseo, J. L., Anderson, G., Loar, A., Rodabough, R. J., White, E. & McTiernan, A. (2003) Cancer Res. 63, 6096-6101. [PubMed] [Google Scholar]
- 3.Irani, J., Ravery, V., Pariente, J. L., Chartier-Kastler, E., Lechevallier, E., Soulie, M., Chautard, D., Coloby, P., Fontaine, E., Bladou, F., et al. (2002) J. Urol. 168, 1985-1988. [DOI] [PubMed] [Google Scholar]
- 4.Khuder, S. A. & Mutgi, A. B. (2001) Br. J. Cancer 84, 1188-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Langman, M. J., Cheng, K. K., Gilman, E. A. & Lancashire, R. J. (2000) Br. Med. J. 320, 1642-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts, R. O., Jacobson, D. J., Girman, C. J., Rhodes, T., Lieber, M. M. & Jacobsen, S. J. (2002) Mayo Clin. Proc. 77, 219-225. [DOI] [PubMed] [Google Scholar]
- 7.Allison, M. C., Howatson, A. G., Torrance, C. J., Lee, F. D. & Russell, R. I. (1992) N. Engl. J. Med. 327, 749-754. [DOI] [PubMed] [Google Scholar]
- 8.DeWitt, D. L., Meade, E. A. & Smith, W. L. (1993) Am. J. Med. 95, 40S-44S. [DOI] [PubMed] [Google Scholar]
- 9.Traversa, G., Walker, A. M., Ippolito, F. M., Caffari, B., Capurso, L., Dezi, A., Koch, M., Maggini, M., Alegiani, S. S. & Raschetti, R. (1995) Epidemiology 6, 49-54. [DOI] [PubMed] [Google Scholar]
- 10.Howard, P. A. & Delafontaine, P. (2004) J. Am. Coll. Cardiol. 43, 519-525. [DOI] [PubMed] [Google Scholar]
- 11.Mamdani, M., Juurlink, D. N., Lee, D. S., Rochon, P. A., Kopp, A., Naglie, G., Austin, P. C., Laupacis, A. & Stukel, T. A. (2004) Lancet 363, 1751-1756. [DOI] [PubMed] [Google Scholar]
- 12.Solomon, D. H., Glynn, R. J. & Avorn, J. (2002) Lancet 360, 90. [DOI] [PubMed] [Google Scholar]
- 13.Solomon, D. H., Glynn, R. J., Levin, R. & Avorn, J. (2002) Arch. Intern. Med. 162, 1099-1104. [DOI] [PubMed] [Google Scholar]
- 14.Weir, M. R., Sperling, R. S., Reicin, A. & Gertz, B. J. (2003) Am. Heart J. 146, 591-604. [DOI] [PubMed] [Google Scholar]
- 15.Chesire, D. R., Ewing, C. M., Gage, W. R. & Isaacs, W. B. (2002) Oncogene 21, 2679-2694. [DOI] [PubMed] [Google Scholar]
- 16.Chesire, D. R. & Isaacs, W. B. (2003) Endocr. Relat. Cancer 10, 537-560. [DOI] [PubMed] [Google Scholar]
- 17.Giles, R. H., van Es, J. H. & Clevers, H. (2003) Biochim. Biophys. Acta 1653, 1-24. [DOI] [PubMed] [Google Scholar]
- 18.Hovanes, K., Li, T. W., Munguia, J. E., Truong, T., Milovanovic, T., Lawrence Marsh, J., Holcombe, R. F. & Waterman, M. L. (2001) Nat. Genet. 28, 53-57. [DOI] [PubMed] [Google Scholar]
- 19.Li, H., Pamukcu, R. & Thompson, W. J. (2002) Cancer Biol. Ther. 1, 621-625. [DOI] [PubMed] [Google Scholar]
- 20.Li, Y., Hively, W. P. & Varmus, H. E. (2000) Oncogene 19, 1002-1009. [DOI] [PubMed] [Google Scholar]
- 21.Tetsu, O. & McCormick, F. (1999) Nature 398, 422-426. [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto, H., Kishida, S., Kishida, M., Ikeda, S., Takada, S. & Kikuchi, A. (1999) J. Biol. Chem. 274, 10681-10684. [DOI] [PubMed] [Google Scholar]
- 23.Yanagawa, S., van Leeuwen, F., Wodarz, A., Klingensmith, J. & Nusse, R. (1995) Genes Dev. 9, 1087-1097. [DOI] [PubMed] [Google Scholar]
- 24.Kishida, S., Yamamoto, H., Hino, S., Ikeda, S., Kishida, M. & Kikuchi, A. (1999) Mol. Cell. Biol. 19, 4414-4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coghlan, M. P., Culbert, A. A., Cross, D. A., Corcoran, S. L., Yates, J. W., Pearce, N. J., Rausch, O. L., Murphy, G. J., Carter, P. S., Roxbee Cox, L., et al. (2000) Chem. Biol. 7, 793-803. [DOI] [PubMed] [Google Scholar]
- 26.Fagotto, F., Gluck, U. & Gumbiner, B. M. (1998) Curr. Biol. 8, 181-190. [DOI] [PubMed] [Google Scholar]
- 27.Yost, C., Torres, M., Miller, J. R., Huang, E., Kimelman, D. & Moon, R. T. (1996) Genes Dev. 10, 1443-1454. [DOI] [PubMed] [Google Scholar]
- 28.He, T. C., Sparks, A. B., Rago, C., Hermeking, H., Zawel, L., da Costa, L. T., Morin, P. J., Vogelstein, B. & Kinzler, K. W. (1998) Science 281, 1509-1512. [DOI] [PubMed] [Google Scholar]
- 29.Boon, E. M., Keller, J. J., Wormhoudt, T. A., Giardiello, F. M., Offerhaus, G. J., van der Neut, R. & Pals, S. T. (2004) Br. J. Cancer 90, 224-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown, W. A., Skinner, S. A., Vogiagis, D. & O'Brien, P. E. (2001) Dig. Dis. Sci. 46, 2314-2321. [DOI] [PubMed] [Google Scholar]
- 31.Dihlmann, S., Klein, S. & von Knebel Doeberitz, M. (2003) Mol. Cancer Ther. 2, 509-516. [PubMed] [Google Scholar]
- 32.Gardner, S. H., Hawcroft, G. & Hull, M. A. (2004) Br. J. Cancer 91, 153-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hawcroft, G., D'Amico, M., Albanese, C., Markham, A. F., Pestell, R. G. & Hull, M. A. (2002) Carcinogenesis 23, 107-114. [DOI] [PubMed] [Google Scholar]
- 34.Lu, D., Zhao, Y., Tawatao, R., Cottam, H. B., Sen, M., Leoni, L. M., Kipps, T. J., Corr, M. & Carson, D. A. (2004) Proc. Natl. Acad. Sci. USA 101, 3118-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lehmann, J. M., Lenhard, J. M., Oliver, B. B., Ringold, G. M. & Kliewer, S. A. (1997) J. Biol. Chem. 272, 3406-3410. [DOI] [PubMed] [Google Scholar]
- 36.Sarraf, P., Mueller, E., Jones, D., King, F. J., DeAngelo, D. J., Partridge, J. B., Holden, S. A., Chen, L. B., Singer, S., Fletcher, C., et al. (1998) Nat. Med. 4, 1046-1052. [DOI] [PubMed] [Google Scholar]
- 37.Elstner, E., Muller, C., Koshizuka, K., Williamson, E. A., Park, D., Asou, H., Shintaku, P., Said, J. W., Heber, D. & Koeffler, H. P. (1998) Proc. Natl. Acad. Sci. USA 95, 8806-8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kubota, T., Koshizuka, K., Williamson, E. A., Asou, H., Said, J. W., Holden, S., Miyoshi, I. & Koeffler, H. P. (1998) Cancer Res. 58, 3344-3352. [PubMed] [Google Scholar]
- 39.Kolluri, S. K., Corr, M., James, S. Y., Bernasconi, M., Lu, D., Liu, W., Cottam, H. B., Leoni, L. M., Carson, D. A. & Zhang, X. K. (2005) Proc. Natl. Acad. Sci. USA 102, 2525-2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saez, E., Rosenfeld, J., Livolsi, A., Olson, P., Lombardo, E., Nelson, M., Banayo, E., Cardiff, R. D., Izpisua-Belmonte, J. C. & Evans, R. M. (2004) Genes Dev. 18, 528-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang, F., Li, X., Sharma, M., Sasaki, C. Y., Longo, D. L., Lim, B. & Sun, Z. (2002) J. Biol. Chem. 277, 11336-11344. [DOI] [PubMed] [Google Scholar]
- 42.Easwaran, V., Pishvaian, M., Salimuddin & Byers, S. (1999) Curr. Biol. 9, 1415-1418. [DOI] [PubMed] [Google Scholar]
- 43.Palmer, H. G., Gonzalez-Sancho, J. M., Espada, J., Berciano, M. T., Puig, I., Baulida, J., Quintanilla, M., Cano, A., de Herreros, A. G., Lafarga, M., et al. (2001) J. Cell Biol. 154, 369-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shah, S., Hecht, A., Pestell, R. & Byers, S. W. (2003) J. Biol. Chem. 278, 48137-48145. [DOI] [PubMed] [Google Scholar]
- 45.Becker-Scharfenkamp, U. & Blaschke, G. (1993) J. Chromatogr. 621, 199-207. [DOI] [PubMed] [Google Scholar]
- 46.DiRenzo, J., Soderstrom, M., Kurokawa, R., Ogliastro, M. H., Ricote, M., Ingrey, S., Horlein, A., Rosenfeld, M. G. & Glass, C. K. (1997) Mol. Cell. Biol. 17, 2166-2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li, M., Pascual, G. & Glass, C. K. (2000) Mol. Cell. Biol. 20, 4699-4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trama, J., Lu, Q., Hawley, R. G. & Ho, S. N. (2000) J. Immunol. 165, 4884-4894. [DOI] [PubMed] [Google Scholar]
- 49.Lu, D., Kiriyama, Y., Lee, K. Y. & Giguere, V. (2001) Cancer Res. 61, 6755-6761. [PubMed] [Google Scholar]
- 50.Forman, B. M. (2002) J. Biol. Chem. 277, 12503-12506. [DOI] [PubMed] [Google Scholar]
- 51.Tyree, C. M. & Klausing, K. (2003) Methods Mol. Med. 85, 175-183. [DOI] [PubMed] [Google Scholar]
- 52.Demerson, C. A., Humber, L. G., Abraham, N. A., Schilling, G., Martel, R. R. & Pace-Asciak, C. (1983) J. Med. Chem. 26, 1778-1780. [DOI] [PubMed] [Google Scholar]
- 53.Emami, K. H., Nguyen, C., Ma, H., Kim, D. H., Jeong, K. W., Eguchi, M., Moon, R. T., Teo, J. L., Kim, H. Y., Moon, S. H., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 12682-12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thompson, W. J., Piazza, G. A., Li, H., Liu, L., Fetter, J., Zhu, B., Sperl, G., Ahnen, D. & Pamukcu, R. (2000) Cancer Res. 60, 3338-3342. [PubMed] [Google Scholar]
- 55.Lee, E. J., Park, H. G. & Kang, H. S. (2003) Int. J. Oncol. 23, 503-508. [PubMed] [Google Scholar]
- 56.Rice, P. L., Kelloff, J., Sullivan, H., Driggers, L. J., Beard, K. S., Kuwada, S., Piazza, G. & Ahnen, D. J. (2003) Mol. Cancer Ther. 2, 885-892. [PubMed] [Google Scholar]
- 57.Hedvat, M., Jain, A., Carson, D. A., Leoni, L. M., Huang, G., Holden, S., Lu, D., Corr, M., Fox, W. & Agus, D. B. (2004) Cancer Cell 5, 565-574. [DOI] [PubMed] [Google Scholar]
- 58.Sharma, C., Pradeep, A., Wong, L., Rana, A. & Rana, B. (2004) J. Biol. Chem. 279, 35583-35594. [DOI] [PubMed] [Google Scholar]
- 59.Xiao, J. H., Ghosn, C., Hinchman, C., Forbes, C., Wang, J., Snider, N., Cordrey, A., Zhao, Y. & Chandraratna, R. A. (2003) J. Biol. Chem. 278, 29954-29962. [DOI] [PubMed] [Google Scholar]