Abstract
The Arc repressor of bacteriophage P22 is a dimeric member of the ribbon–helix–helix family of transcription factors. Residues 9–14 of each wild-type Arc subunit pair to form two antiparallel β-strands and have the alternating pattern of polar and nonpolar residues expected for a β-ribbon with one solvent-exposed face and one face that forms part of the hydrophobic core. Simultaneously switching Asn-11 to Leu and Leu-12 to Asn changes the local binary sequence pattern to that of an amphipathic helix. Previous studies have shown that this double mutation results in replacement of the wild-type β-ribbon by two right-handed 310-helices. Moreover, an Arc variant bearing just the Asn-11 → Leu mutation has an ambiguous binary pattern and can form either the ribbon or the helical structures, which interchange rapidly. Here, we study Arc mutants in which position 11 is occupied by Gly, Ala, Val, Ile, Leu, Met, Phe, or Tyr. These mutants adopt the wild-type β-ribbon structure in a sequence context that stabilizes this fold, but they assume the alternative helical structure in a sequence background in which the wild-type fold is precluded by negative design. In an otherwise wild-type sequence background, the detailed chemical properties of the position 11 side chain dictate which of the two competing conformational folds is preferred.
Keywords: protein folding, secondary structure, structural evolution
Understanding how amino acid sequence determines a protein fold is critical for predicting structure, for protein design, and for understanding folding and misfolding. The binary pattern of polar (P) and hydrophobic (H) residues in a sequence appears to be one of the most basic features that determine the fold of a protein (1, 2). Indeed, the importance of binary pattern has been impressively demonstrated by the isolation of stable α-helical or β-ribbon proteins from designed libraries (3). Some binary patterns, however, do not uniquely distinguish between competing folds or structural states. For example, protein misfolding in prion disease can result when one hydrophobic residue is mutated to another (4–6). In general, it seems clear that the molecular details of side-chain packing, hydrogen-bonding, electrostatics, etc., will be important determinants of the precise native conformation of a protein and of the stability of this structure to denaturation.
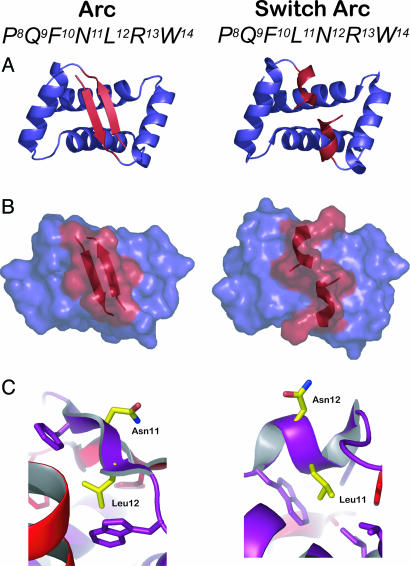
In the Arc repressor homodimer of phage P22, interchange of adjacent polar (Asn-11) and hydrophobic (Leu-12) residues causes a dramatic change in local structure (7, 8). As shown in Fig. 1A, the surrounding N-terminal sequence folds as an antiparallel two-stranded β-ribbon in the wild-type dimer but forms two right-handed 310-helices in the NL11/LN12 mutant dimer, which is also called “switch” Arc (7–10). Asn-11 is on the protein surface of wild-type Arc, and Leu-12 is buried in the core (Fig. 1C Left). In switch Arc, by contrast, Leu-11 forms part of the hydrophobic core, and Asn-12 is exposed to solvent (Fig. 1C Right). Residues 9–14 have an alternating PHPHPH β-ribbon pattern in wild-type Arc and a helical PHHPPH pattern in the switch Arc protein. Arc bearing just the NL11 mutation has an ambiguous binary pattern (PHHHPH) and exists in a dynamic equilibrium between the wild-type β-ribbon fold and the switch helical fold (11). It is plausible that the binary-pattern change caused by the NL11/LN12 mutations is largely responsible for the observed changes in secondary and tertiary structure. However, the extent to which the detailed properties of the position 11 side chain dictate the structural preference of Arc for the competing wild-type and switch conformations is unknown.
Fig. 1.
Structures from the wild-type Arc dimer (AB dimer of Protein Data Bank ID code 1PAR) are shown on the Left; structures from the switch Arc dimer (Protein Data Bank ID code 1QTG) are shown on the Right.(A) Backbone cartoon representations. The region shown in red forms an antiparallel β-ribbon in wild-type Arc and two 310 helices in switch Arc. (B) Surface representations using the same color scheme as in A. (C) In wild-type Arc, Asn-11 is on the protein surface, and Leu-12 is part of the protein core. In switch Arc, Leu-11 is part of the core, and Asn-12 is on the protein surface.
Here, we determine the properties of Arc mutants containing Gly-11, Ala-11, Val-11, Ile-11, Leu-11, Met-11, Phe-11, or Tyr-11 in three different sequence contexts. When variants contained the LN12 substitution, which effectively precludes the β-ribbon fold of Arc, each position 11 mutant adopted a “switch-like” fold, demonstrating that the precise details of core packing by the position 11 side chain are relatively unimportant determinants of this conformation. By contrast, the position 11 variants appeared to adopt the wild-type fold in the context of Leu-12 and the PL8 mutation, which stabilizes the β-ribbon conformation (12). In an otherwise wild-type sequence background, however, the identity of residue 11 played a significant role in determining the relative proportions of the competing wild-type and switch protein folds. Together, these results demonstrate that the detailed properties of the position 11 side chain become critical determinants of fold specificity only when the remaining portions of the Arc sequence contribute roughly equally to stabilization of the two competing protein folds.
Materials and Methods
Protein Expression and Purification. Mutations were constructed in the arc-st11 gene of plasmid pET800 or pSA700 by cassette mutagenesis or Stratagene QuikChange site-directed mutagenesis. The st11 tag encodes the C-terminal sequence H6KNQHE (13), which allows affinity purification and improves expression by reducing degradation. Mutants in pET800 or pSA700 backgrounds were overexpressed in Escherichia coli strains X90(λDE3) and X90, respectively. Strains used for protein purification were freshly transformed and plated on LB agar containing 100 μg/ml ampicillin at 37°C. A single colony was picked into LB broth plus 100 μg/ml ampicillin, grown for 10–14 h at 37°C, and diluted 1:100 into the same medium. When the OD600 reached 0.6 to 1.0, the culture was induced with 0.1 mg/ml isopropyl-β-d-thiogalactoside (IPTG) and grown for an additional 3.5 h. Cells were harvested by centrifugation, the supernatant was discarded, and the cell pellet was frozen at –80°C before protein purification.
Mutant proteins were purified by nickel-affinity and anion-exchange chromatography by using SP-Sephadex (13). Cells from 1 liter of cell culture were resuspended in 20 ml of buffer A (0.01 M Tris/0.1 M NaH2PO4/6 M guanidine hydrochloride, pH 8.0), stirred at 4°C for 1 h to allow lysis, and centrifuged at 12,000 × g for 30 min. Imidazole was added to the supernatant to a final concentration of 10 mM, and this material was passed over a 2- to 3-ml Ni2+-nitrilotriacetic acid (NTA) column that had been preequilibrated with buffer A plus 10 mM imidazole. Two 50-ml washes with buffer A plus 10 mM imidazole were performed, and bound protein was eluted from the column by using 15 ml of buffer F (0.2 M acetic acid/6 M guanidine hydrochloride). The eluant was dialyzed against two 6-liter changes of buffer B (0.01 M Tris, pH 7.5/0.2 mM EDTA) for a minimum of 8 h. The protein sample was then loaded onto a 2- to 3-ml SP-Sephadex column preequilibrated with buffer B plus 100 mM KCl. This column was washed with 100 ml of buffer B plus 100 mM KCl and eluted with buffer B plus 2 M KCl. Fractions containing purified protein were dialyzed against two 6-liter changes of storage buffer (50 mM Tris, pH 7.5/250 mM KCl/0.2 mM EDTA) for a minimum of 8 h. For 15N-labeling of mutants, proteins were overexpressed from pET800 in E. coli strain BL21(λDE3)-pLysS by using M9T minimal medium containing 15NH4Cl (0.8 g/liter) as the sole nitrogen source. The 15N-labeled mutants were purified as described above, but after purification they were dialyzed against three 6-liter changes of distilled H2O for a minimum of 8 h before being lyophilized.
Circular Dichroism (CD) and Fluorescence. CD spectra were taken in an Aviv Associates (Lakewood, NJ) model 60DS instrument. Near-UV CD spectra from 260 nm to 300 nm (step size 0.5 nm; averaging time 3 s) were the average of five scans determined at 100 μM protein concentration in storage buffer at 15°C in a 1-cm pathlength cuvette and were corrected by subtracting a buffer blank. Fluorescence emission spectra from 300 to 420 nM were determined by using a Photon Technology International QM-2000–4SE instrument (excitation wavelength 280 nm). Spectra were the average of three scans (step size 1 nm; averaging time 1 s) determined by using 100 μM protein in storage buffer at 15°C in a 1-cm pathlength cuvette. Basis spectra for the β-ribbon conformation (PL8 background) and switch conformation (LN12 background) of each position 11 variant were used to calculate the conformational preference in an otherwise wild-type sequence context as described (11).
NMR. Lyophilized 15N-labeled protein was resuspended at a concentration of ≈4 mM in a buffer containing 20 mM NaPO4 (pH 5.0), 10% D2O, and 1 mM 3-(trimethylsilyl)-propionic acid (TMSP) as the internal chemical-shift standard. The pH was adjusted to 4.9 by using HCl, but no correction for the effect of D2O on pH was made (14). NMR spectra at 30°C were acquired by using a Bruker DMX500 spectrometer. The 1H and 15N chemical shifts are relative to TMSP (15). Heteronuclear single quantum correlation (HSQC) spectra were acquired with standard Bruker pulse sequences (16), and pulsed-field gradients were used for coherence selection and solvent suppression (17). Presaturation was used for solvent suppression for the wild-type sample. For the mutants, data were collected as the average of four scans of 2,048 data points for each of 256 t1 transients. For switch Arc, the data were collected with eight scans of 2,048 data points for each of 64 t1 transients. Raw data were apodized with shifted sine bell functions and zero filled in both dimensions before Fourier transformation by using Bruker xwin-nmr software.
Denaturation Assays. For thermal-denaturation studies, CD ellipticity at 222 nm was monitored from 4°C to 100°C, with a step size of 2°C, an averaging time of 30 s, and an equilibration time of 1 min. The protein concentration was 10 μM in storage buffer. For urea denaturation, ellipticity at 230 nm was monitored at 25°C for samples containing from 0 to 9 M urea. At each urea concentration, the ellipticity of 5 μM protein in storage buffer was averaged over 60 data points taken at 1 s intervals after 1 min of sample mixing. Denaturation curves were fitted to equations for a concerted reaction in which the native dimer denatures and dissociates to two unfolded monomers (18–20). For fitting of urea denaturation data, the m value was fixed at –1.3 kcal/mol·M.
Results
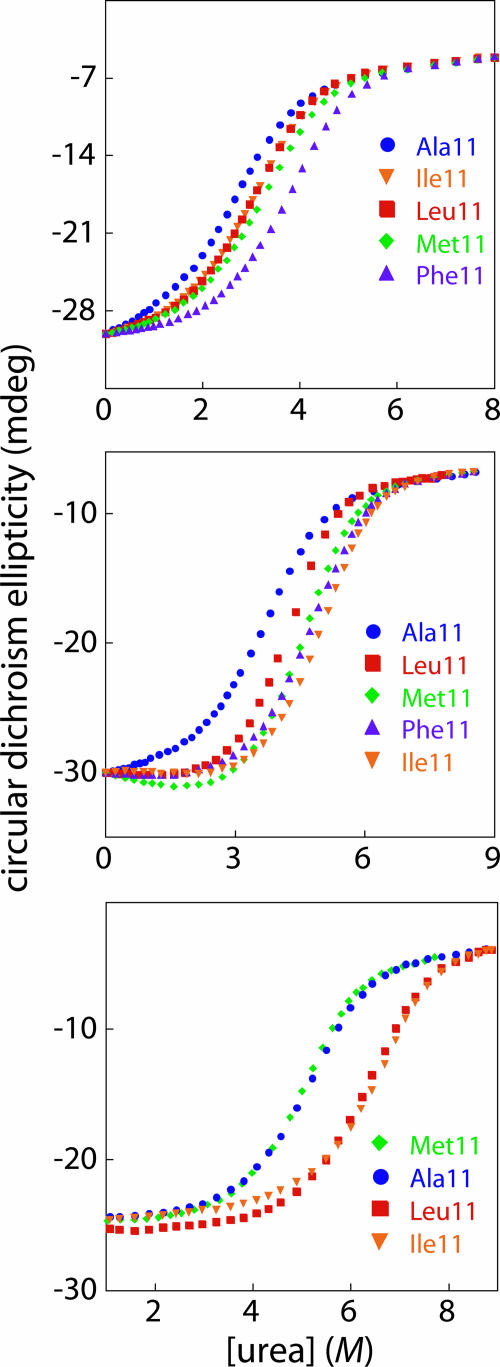
To explore the effects of different nonpolar side chains at position 11, we constructed and purified Arc mutants containing Gly, Ala, Val, Ile, Leu, Met, Phe, or Tyr at this position in an otherwise wild-type sequence context, in proteins containing one of the switch mutations (Leu-12 → Asn), and in proteins containing the Pro-8 → Leu mutation. The LN12 mutation destabilizes the wild-type Arc fold by replacing a hydrophobic-core residue with a polar residue (7–10), whereas the PL8 mutation stabilizes the wild-type fold of Arc by introducing two additional hydrogen bonds into the β-ribbon (12). In each of the three sequence backgrounds, the position 11 mutants showed cooperative thermal melts with Tms ranging from 46°C to 80°C (Table 1). For the mutants tested, increasing concentrations of urea also resulted in cooperative denaturation/dissociation (Fig. 2) with free energies of unfolding ranging from 10.6 to 15.8 kcal/mol (Table 1). Hence, the sequence changes at position 11 are compatible with stable folding in the LN12 background, the PL8 background, and in an otherwise wild-type sequence background.
Table 1. Stabilities of Arc position 11 mutants to thermal or urea denaturation in three sequence backgrounds.
| Sequence background
| ||||||
|---|---|---|---|---|---|---|
| LN12
|
WT
|
PL8
|
||||
| Residue 11 | Tm, °C | ΔGu, kcal/mol | Tm, °C | ΔGu, kcal/mol | Tm, °C | ΔGu, kcal/mol |
| Gly | 46 | ND | 56 | ND | 70 | ND |
| Ala | 51 | 10.6 | 63 | 11.9 | 72 | 13.7 |
| Val | 52 | ND | 71 | ND | ND | ND |
| Leu | 57 | 11.0 | 67 | 12.5 | 76 | 15.2 |
| Ile | 56 | 11.1 | 71 | 13.5 | 80 | 15.8 |
| Met | 55 | 11.3 | 67 | 12.9 | 73 | 13.6 |
| Phe | 59 | 12.0 | 68 | 13.2 | ND | ND |
| Tyr | 56 | ND | 66 | ND | 76 | ND |
Tms were determined at 10 μM protein concentrations in 50 mM Tris (pH 7.5), 250 mM KCl, and 0.2 mM EDTA. Under these conditions, Tm is 58°C for WT Arc and 72°C for Arc PL8 (12, 13). Stabilities to urea denaturation at 25°C were determined by using 5 μM protein in the same buffer. Under these conditions, ΔGu is 10.9 kcal/mol for WT Arc and 13.6 kcal/mol for Arc PL8 (12, 13). ND, not determined.
Fig. 2.
Urea denaturation (25°C, pH 7.5, 5 μM protein) was monitored by changes in CD ellipticity at 230 nm for selected position 11 Arc mutants in the LN12 sequence background (Top), in an otherwise wild-type sequence background (Middle), and in the PL8 sequence background (Bottom). Free energies of Arc dissociation/denaturation in the absence of urea (ΔGu) were calculated from these data and are listed in Table 1.
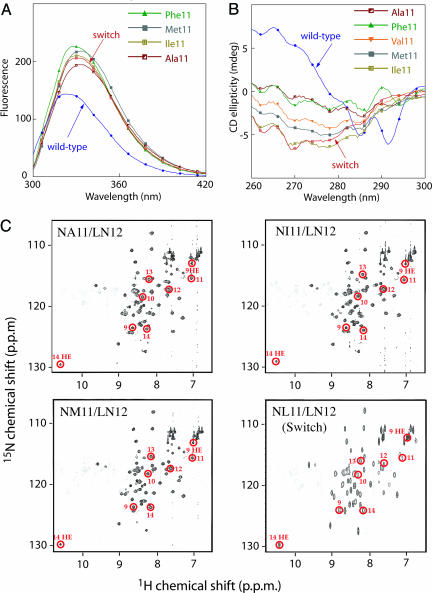
In the LN12 Background, Position 11 Mutants Adopt Switch-Like Structures. In switch Arc (NL11/LN12), the side chain of Asn-12 is on the protein surface, whereas the side chain of Leu-11 forms part of the hydrophobic core but is also partially solvent exposed (7, 8). Spectroscopic and NMR studies provided evidence that other nonpolar residues at position 11 also resulted in a structure similar to switch Arc when the position 12 residue was Asn (Fig. 3). For example, in the LN12 background, the position 11 mutants had fluorescence spectra more similar to switch Arc than to wild-type Arc both in intensity and in terms the spectral red shift (Fig. 3A). Moreover, none of the near-UV CD spectra of the position 11 mutants in the LN12 background showed the distinct negative minima at 285 and 292 nm that are characteristic of the wild-type conformation but not of switch Arc (Fig. 3B). Finally, HSQC NMR spectra of the NA11/LN12, NI11/LN12, and NM11/LN12 mutants displayed a number of resonances (circled in Fig. 3C) that are characteristic of the 310-helical structure of switch Arc (7, 8). Importantly, amide nitrogen and proton resonances for residues 9–14 appear at very different positions in the HSQC spectrum of wild-type Arc (9).
Fig. 3.
Spectral properties of selected position 11 mutants in the LN12 sequence background. (A) Fluorescence spectra of four position 11/LN12 mutants, wild-type Arc (blue), and NL11/LN12 switch Arc (red). (B) Near-UV CD spectra of five position 11/LN12 mutants, wild-type Arc (blue), and switch Arc (red). (C) HSQC NMR spectra. The circled resonances, which have been assigned to residues 9–14 in the NL11/LN12 switch Arc spectrum (8, 9), have similar chemical shifts in the spectra of the NA11/LN12, NI11/LN12, and NM11/LN12 mutants, indicating that these residues adopt a switch-like 310-helical fold in all of these proteins.
The experiments described above suggest that the switch conformation can tolerate a wide variety of nonpolar side chains at position 11, which forms part of the hydrophobic core when the N-terminal residues of Arc fold as 310-helices (7, 8). Consistent with this model, mutants with LN12 and small side chains at position 11 (Gly, Ala, and Val) were less stable than those with larger hydrophobic side chains (Leu, Ile, Met, Phe, and Tyr) both in temperature unfolding and urea denaturation studies (Table 1). We assume that the protein core is better packed in the mutants with larger hydrophobic side chains at position 11. It is also worth noting that the position 11 side chain in the switch conformation packs against Trp-14. Because the details of the burial and packing of Trp-14 in the protein core play a significant factor in determining the fluorescence and near-UV CD spectra of variants that assume a switch-like conformation, it is not surprising that the spectra of most position 11/LN12 mutants are somewhat different from the spectra of switch Arc (NL11/LN12).
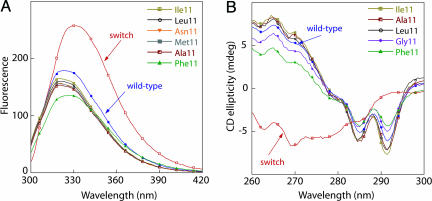
In the PL8 Background, Mutants Have Structures Like Wild-Type Arc. Crystallographic and biophysical studies have previously shown that the PL8 mutation results in formation of an additional hydrogen bond at each end of the wild-type β-ribbon, creating a hyperstable mutant with a structure essentially identical to wild-type Arc (12). We found that mutants with Gly-11, Ala-11, Leu-11, Ile-11, Met-11, Phe-11, or Tyr-11 in addition to the PL8 substitution had fluorescence spectra (Fig. 4A) and near-UV CD spectra (Fig. 4B) that were significantly more similar to those of wild-type Arc than to those of switch Arc, suggesting that these variants also adopt a β-ribbon structure similar to that of wild-type Arc. The NV11/PL8 mutant was not characterized.
Fig. 4.
Fluorescence (A) and near-UV CD (B) spectra of selected position 11 mutants in the PL8 sequence background. In both A and B, the corresponding spectra of wild-type Arc (blue) and NL11/LN12 switch Arc (red) are shown for comparison.
The spectroscopic results suggest that position 11 side chains ranging in size from Gly to Tyr can be accommodated into the β-ribbon structure of the Arc PL8 dimer. In this structural context, the two position 11 side chains would be on the protein surface but positioned closely enough to pack against each other. Such packing probably explains why some of the PL8 variants with larger nonpolar side chains at position 11 (Ile, Leu, and Tyr) are more stable than those with smaller side chains (Ala and Gly) at this position (Table 1). Indeed, the Tms of mutants with Leu-11, Ile-11, and Tyr-11 were even higher than the Tm of the PL8 protein with Asn-11, the wild-type side chain.
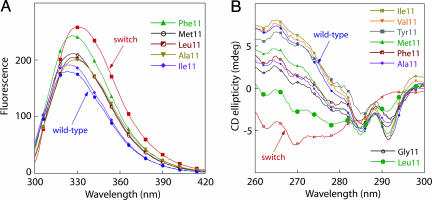
Position 11 Mutants Adopt both Folds in a “Wild-Type” Sequence Context. Arc bearing the NL11 mutation can adopt either the wild-type structure or the switch structure in a dynamic equilibrium that depends on solution conditions (8, 11). In an otherwise wild-type sequence background, variants with the NI11 and NV11 mutations had near-UV and/or fluorescence spectra that resembled those of wild-type Arc, whereas those with the NM11, NF11, NA11, and NG11 substitutions had spectra that were intermediate between those of wild-type Arc and switch Arc (Fig. 5). These results suggested that the first group of mutants was largely in the wild-type structure, whereas the latter group equilibrated between the wild-type and switch conformations. By using the near-UV CD spectra in either the LN12 or PL8 backgrounds as basis spectra for the switch and wild-type conformations of each position 11 mutant, structural preferences for each NX11 variant were calculated. Based on this analysis, the proportion of wild-type structure was >95% for NI11 and NV11, 75% for NF11, 65% for NM11, 60% for NY11, and 30% for NA11, NG11, and NL11 under the conditions of the assay (15°C, storage buffer). Hence, the identity of the position 11 side chain appears to have a large influence on the conformational preferences of these variants, with β-branched residues strongly favoring the wild-type fold.
Fig. 5.
Fluorescence (A) and near-UV CD (B) spectra of selected position 11 Arc mutants in an otherwise wild-type sequence background. In both A and B, the spectra of wild-type Arc (blue) and NL11/LN12 switch Arc (red) are shown for comparison.
The relative equilibrium populations of the switch and wild-type conformations of Arc NL11 are influenced by solvent and temperature, with the switch fold favored at 0°C in 250 mM potassium thiocyanate (KSCN) and the wild-type fold favored at room temperature in 10% ethanol (11). Similar effects were observed for several of the other position 11 mutants. Specifically, the far-UV CD spectra of the NG11, NM11, NF11, and NY11 mutants were shifted to positions expected for an increased population of the switch structure at 0°C in KSCN and to positions consistent with an increased population of the wild-type structure at 15°C in 10% ethanol (data not shown). These results are consistent with the supposition that this set of position 11 mutants are able to adopt both alternative structures, with the equilibrium between these structures being strongly influenced by the chemical identity of the mutant side chain as well as by temperature and solvent composition.
Discussion
The identity of the position 11 side chain is an important determinant of the surrounding secondary and tertiary structure in Arc repressor. In the wild-type dimer, position 11 is Asn, and residues 9–14 in one subunit form a β-strand that pairs with the corresponding β-strand in the other subunit to create an antiparallel β-ribbon. When Asn-11 is changed to Leu, two alternative conformations are possible: either the wild-type β-ribbon or the 310 helices of the switch Arc conformation (11). Remarkably, the β-ribbon and switch conformations interconvert at rates that can exceed 1,000 sec–1 in the native NL11 protein (11). The Asn-11 → Leu mutation can be viewed as an evolutionary bridge between the two competing folds, because Leu-11 is able to serve as a hydrophobic core residue in the helical structure or as a surface position in the β-ribbon structure. The studies presented here show that the switch conformation also becomes accessible when Asn-11 is replaced with nonpolar residues other than Leu. The evolutionary bridge connecting the wild-type and switch folds is therefore reasonably wide and contains a fair number of essentially structurally degenerate sequences. Importantly, however, we find that the equilibrium between the two competing structures is highly dependent on the identity and detailed structure of the position 11 side chain. Indeed, >95% of the molecules of the Arc variant containing Ile-11 assumed the wild-type conformation, whereas ≈70% of the molecules of the Leu-11 variant adopted the switch conformation under the same conditions. Hence, moving a single methyl group from the β-carbon to the γ-carbon of the position 11 side chain results in a dramatic change in structural preference. The net balance between favorable and unfavorable interactions made by each position 11 side chain in the β-ribbon and 310-helical conformations ultimately determines structural preference. Thus, Ile-11 must make better interactions on the surface of the β-ribbon than in the core of the switch structure, whereas the opposite must be true of Leu-11.
Residues 9–14 have a β-strand pattern (P9H10P11H12P13H14) in wild-type Arc and a helical pattern (P9H10H11P12P13H14) in the double-mutant switch Arc protein. In otherwise wild-type Arc, mutating Asn-11 to a nonpolar residue results in an ambiguous pattern (P9H10H11H12P13H14), allowing either the β-ribbon fold with the H11 side chain on the surface or the 310-helical fold with the H12 side chain on the surface. The presence of a polar Asn at position 11 in the Arc sequence can be viewed as a strong negative design element that basically precludes the helical conformation, because of the extreme energetic cost of burying a polar residue in the protein core. Similarly, a polar Asn at position 12 effectively blocks access to the β-ribbon conformation. Indeed, in the LN12 sequence background, we found that a stable switch-like conformation was supported by Gly-11, Ala-11, Val-11, Ile-11, Met-11, Phe-11, and Tyr-11 and by the previously described Leu-11 (7, 8). In this sequence context, the detailed properties of the position 11 side chain are clearly far less important than the negative design constraints introduced by the presence of the Asn-12 side chain.
Most native protein structures appear to be stabilized by a combination of positive and negative design features. The PL8 mutation can be viewed as a positive design element that favors the wild-type β-ribbon conformation of Arc. In wild-type Arc, the β-ribbon is stabilized by six interstrand hydrogen bonds (9, 10). In the PL8 mutant, by contrast, the newly introduced main-chain amide hydrogen of each Leu-8 forms a hydrogen bond with the carbonyl oxygen of Trp-14 in the other subunit, increasing the total number of β-ribbon hydrogen bonds to eight and the stability of the protein by 2.5 kcal/mol (12). In the PL8 sequence background, we found that all of the position 11 mutants had spectral properties consistent with the β-ribbon fold. Although a protein like PL8/NL11 should still be able to form the switch conformation, the transition from the wild-type to the switch fold would now require breaking the additional hydrogen bonds and thus be far less energetically favorable. For example, under standard solution conditions, the switch conformation of the NL11 protein is only ≈0.5 kcal/mol more stable than the wild-type conformation, resulting in a slight excess of molecules in the switch conformation. In PL8/NL11, by contrast, the β-ribbon conformation should be 2 kcal/mol more stable than the competing switch conformation (assuming that Leu-8 neither stabilizes nor destabilizes the 310-helical fold directly), and thus 97% of the molecules would be expected to assume the wild-type β-ribbon fold.
As might have been expected, neither the hydrophobicities (21), molecular weights, nor secondary-structure propensities (22) of the mutant position 11 side chains were well correlated with stability in all three of the sequence contexts explored. In the LN12 background, for example, the Tms of the mutant variants showed reasonable correlations with hydrophobicity (R = 0.93) and molecular weight (R = 0.88) but poor correlations with β-sheet and α-helical propensities (Rs of 0.41 and 0.36). In the PL8 background, however, the observed correlation coefficients were all relatively poor: 0.55 (hydrophobicity), 0.44 (molecular weight), 0.31 (β-sheet propensity), and 0.26 (α-helical propensity). Finally, the correlation between the fraction of molecules in the β-ribbon conformation and position 11 properties was reasonable for β-sheet propensity (R = 0.90) but relatively poor for hydrophobicity (R = 0.58), molecular weight (R = 0.40), and α-helical propensity (R = 0.15) in the otherwise wild-type Arc background.
The β-ribbon of wild-type Arc is a key determinant of its function as a repressor of transcription. Indeed, the side chains at positions 9, 11, and 13 contact bases in the major groove of operator DNA and are essential for proper DNA recognition (10, 23). Interestingly, at the position corresponding to Asn-11 of Arc, many paralogs have Gly, Val, Met, or Leu. These evolutionarily related proteins all function as transcription factors and thus almost certainly assume the β-ribbon fold. How is the inactive switch fold avoided or destabilized? One possibility is that other sequence positions in these proteins favor the β-ribbon fold in a positive fashion, much like the PL8 mutation in Arc. Alternatively, binding to specific or nonspecific DNA in the cell may stabilize the active β-ribbon conformation by mass action (11). Finally, the fraction of “active” protein might be regulated by environmental conditions and/or by binding to small ligands.
Acknowledgments
We thank J. McKnight for assistance and use of equipment in collecting NMR spectra. This work was supported by National Institutes of Health Grant AI-15706.
Author contributions: T.A.A. and M.H.J.C. designed research; T.A.A. performed research; T.A.A. and R.T.S. analyzed data; M.H.J.C. contributed new reagents/analytic tools; and T.A.A., M.H.J.C., and R.T.S. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: P, polar residue; H, hydrophobic residue; CD, circular dichroism; HSQC, heteronuclear single quantum correlation.
References
- 1.Dill, K. A., Bromberg, S., Yue, K., Fiebig, K. M., Yee, D. P., Thomas, P. D. & Chan, H. S. (1995) Protein Sci. 4, 561–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cordes, M. H., Davidson, A. R. & Sauer, R. T. (1996) Curr. Opin. Struct. Biol. 6, 3–10. [DOI] [PubMed] [Google Scholar]
- 3.Hecht, M. H., Das, A., Go, A., Bradley, L. K. & Wei, Y. (2004) Protein Sci. 13, 1711–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen, F. E. (1999) J. Mol. Biol. 293, 313–320. [DOI] [PubMed] [Google Scholar]
- 5.Carrell, R. W. & Gooptu, B. (1998) Curr. Opin. Struct. Biol. 8, 799–809. [DOI] [PubMed] [Google Scholar]
- 6.Prusiner, S. B. (1994) Philos. Trans. R. Soc. London B 343, 447–463. [DOI] [PubMed] [Google Scholar]
- 7.Cordes, M. H., Walsh, N. P., McKnight, C. J. & Sauer, R. T. (1999) Science 284, 325–328. [DOI] [PubMed] [Google Scholar]
- 8.Cordes, M. H., Walsh, N. P., McKnight, C. J. & Sauer, R. T. (2003) J. Mol. Biol. 326, 899–909. [DOI] [PubMed] [Google Scholar]
- 9.Breg, J. N., van Opheusden, J. H., Burgering, M. J., Boelens, R. & Kaptein, R. (1990) Nature 346, 586–589. [DOI] [PubMed] [Google Scholar]
- 10.Raumann, B. E., Rould, M. A., Pabo, C. O. & Sauer, R. T. (1994) Nature 367, 754–757. [DOI] [PubMed] [Google Scholar]
- 11.Cordes, M. H., Burton, R. E., Walsh, N. P., McKnight, C. J. & Sauer, R. T. (2000) Nat. Struct. Biol. 7, 1129–1132. [DOI] [PubMed] [Google Scholar]
- 12.Schildbach, J. F., Milla, M. E., Jeffrey, P. D., Raumann, B. E. & Sauer, R. T. (1995) Biochemistry 34, 1405–1412. [DOI] [PubMed] [Google Scholar]
- 13.Milla, M. E., Brown, B. M. & Sauer, R. T. (1993) Protein Sci. 2, 2198–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bundi, A. & Wuthrich, K. (1979) Biopolymers 18, 285–297. [Google Scholar]
- 15.Wishart, D. S., Bigam, C. G., Yao, J., Abildgaard, F., Dyson, H. J., Oldfield, E., Markley, J. L. & Sykes, B. D. (1995) J. Biomol. NMR 6, 135–140. [DOI] [PubMed] [Google Scholar]
- 16.Bodenhausen, G. & Ruben, D. J. (1980) Chem. Phys. Lett. 69, 185–189. [Google Scholar]
- 17.Davis, A. L., Keeler, J., Laue, E. D. & Moskau, D. (1992) J. Magn. Reson. 98, 207–216. [Google Scholar]
- 18.Bowie, J. U. & Sauer, R. T. (1989) Biochemistry 28, 7139–7143. [DOI] [PubMed] [Google Scholar]
- 19.Milla, M. E., Brown, B. M. & Sauer, R. T. (1994) Nat. Struct. Biol. 1, 518–523. [DOI] [PubMed] [Google Scholar]
- 20.Milla, M. E. & Sauer, R. T. (1995) Biochemistry 34, 3344–3351. [DOI] [PubMed] [Google Scholar]
- 21.Fauchere, J. L., Charton, M., Kier, L. B., Verloop, A. & Pliska, V. (1988) Int. J. Pept. Protein Res. 32, 269–278. [DOI] [PubMed] [Google Scholar]
- 22.Levitt, M. (1978) Biochemistry 17, 4277–4285. [DOI] [PubMed] [Google Scholar]
- 23.Brown, B. M., Milla, M. E., Smith, T. L. & Sauer, R. T. (1994) Nat. Struct. Biol. 1, 164–168. [DOI] [PubMed] [Google Scholar]