Abstract
Freshwater planarians have prodigious regenerative abilities that enable them to form complete organisms from tiny body fragments. This plasticity is also exhibited by the planarian germ cell lineage. Unlike many model organisms in which germ cells are specified by localized determinants, planarian germ cells appear to be specified epigenetically, arising postembryonically from stem cells. The planarian Schmidtea mediterranea is well suited for investigating the mechanisms underlying epigenetic germ cell specification. Two strains of S. mediterranea exist: a hermaphroditic strain that reproduces sexually and an asexual strain that reproduces by means of transverse fission. To date, expressed sequence tags (ESTs) have been generated only from the asexual strain. To develop molecular reagents for studying epigenetic germ cell specification, we have sequenced 27,161 ESTs from two developmental stages of the hermaphroditic strain of S. mediterranea; this collection of ESTs represents ≈10,000 unique transcripts. blast analysis of the assembled ESTs showed that 66% share similarity to sequences in public databases. We annotated the assembled ESTs using Gene Ontology terms as well as conserved protein domains and organized them in a relational database. To validate experimentally the Gene Ontology annotations, we used whole-mount in situ hybridization to examine the expression patterns of transcripts assigned to the biological process “reproduction.” Of the 53 genes in this category, 87% were expressed in the reproductive organs. In addition to its utility for studying germ cell development, this EST collection will be an important resource for annotating the planarian genome and studying this animal's amazing regenerative abilities.
Keywords: Gene Ontology, germ cells, planarians, Platyhelminthes
Germ cells represent the predecessors of the next generation and are required for the survival of sexually reproducing species. Despite the importance of understanding how germ cells are formed and how totipotency is established and maintained, the mechanisms that govern these processes remain unclear. Two distinct modes of germ cell specification are typically observed in animals: preformation and epigenesis (1, 2). In many of the best-studied model organisms (including Drosophila, Caenorhabditis elegans, Xenopus, and zebrafish), germ cells are specified early in embryogenesis by maternally supplied, cytoplasmic determinants. However, germ cell determination in many other organisms (e.g., mammals, urodele amphibians, and many basal metazoans) proceeds epigenetically, requiring inductive interactions (3, 4).
Planarian flatworms (freshwater members of the phylum Platyhelminthes) are well known for their remarkable regenerative ability, a capacity that is conferred by a population of pluripotent stem cells (neoblasts) maintained throughout life (5-7). Sexually reproducing planarians do not specify germ cells early in embryogenesis; rather, germ cells appear to be formed epigenetically, derived from neoblasts in specific regions of the adult (5, 8-11). Sexual planarians are cross-fertilizing hermaphrodites: they lay egg capsules containing many developing embryos that hatch after several weeks (12, 13). These “hatchlings” lack reproductive organs, which develop when the planarians have attained a larger size. Sexual development in planarian hermaphrodites is seasonal (8, 14), and the gonads and copulatory apparatus are formed de novo in the appropriate regions of the worm. These structures are generated in a defined order. First, the ovaries form in a region behind the cephalic ganglia; next the testes are generated dorsolaterally; and then the oviducts and vitelline glands develop, followed by the copulatory apparatus (8, 11). Reproductive maturity is achieved when the copulatory apparatus and the external opening leading to it (gonopore) are fully formed (12).
Intriguingly, the planarian germ line exhibits developmental plasticity similar to that observed in the animal's somatic tissues. T. H. Morgan (15) showed that a planarian head fragment, completely devoid of any germ line structures, could regenerate functional gonads from the remaining somatic tissue. During de-growth (the planarian's response to starvation) (16-18), the reproductive organs are resorbed (19, 20); they can be regenerated after feeding has resumed and the animal reaches an appropriate size. After amputation of the head and ovaries of a sexually mature planarian, the testes are resorbed and are only reformed after regeneration of the head is complete (10). Thus, the plasticity of the planarian reproductive organs provides a unique opportunity to examine the specification and maintenance of germ cells, and the signals co-ordinating the removal of the reproductive structures during de-growth.
Understanding how the planarian stem cells are specified to make germ cells will require identifying the genes that are differentially expressed during sexual development and analyzing their functions. Although some genes expressed in the reproductive organs have been identified from several different planarian species (21-26), mechanistic studies are lacking. The planarian Schmidtea mediterranea provides several advantages as a model for studying epigenetic germ cell specification. There are two strains of this species: hermaphroditic, sexually reproducing worms and asexual worms that reproduce strictly by transverse fission, without developing gonads or a copulatory apparatus (13). The sexual and asexual strains can be distinguished genetically by a chromosomal translocation present in the asexuals (27). A collection of ≈3,200 unique ESTs has already been generated from the asexual strain (28, 29).
Here, we report the sequencing and analysis of 27,161 ESTs from normalized/subtracted cDNA libraries from a clonal line of the sexual strain of S. mediterranea; these ESTs represent ≈10,000 unique transcripts. The predicted products of the assembled ESTs were analyzed for similarity to sequences in the public databases, annotated by using Gene Ontology (GO) terms, and assigned conserved protein domains. Finally, we tested the validity of the GO annotation by performing whole-mount in situ hybridization on reproductively mature hermaphrodites to determine the expression patterns of ESTs annotated to the category “reproduction.” We found that 87% of these ESTs were expressed in the reproductive organs, validating the utility of the GO annotation. The planarian EST collection reported here, combined with microarray analysis and the ability to perform large-scale functional analyses using RNA interference (13, 29), will help elucidate the mechanisms by which inductive interactions can specify germ cell fate and the extent to which these mechanisms have been conserved evolutionarily.
Methods
RNA Isolation. Total RNA was isolated from sexually mature worms (n = 25) or juveniles (n = 141) from a clonal line of the hermaphroditic strain of S. mediterranea by using RNAlater and ToTALLY RNA (Ambion, Austin, TX) and then precipitated with LiCl. Poly(A)+-RNA was isolated from total RNA by using the Oligotex Direct mRNA kit (Qiagen).
cDNA Synthesis, Size Selection, and Cloning. The poly(A)+-RNA from mature planarians was converted to double-stranded cDNA by using the SuperScript Choice system (Invitrogen). First-strand cDNA synthesis was primed by using a modified oligo(dT) primer, 5′-AACTGGAAGAATTCGCGGCCGCTCGCA(T)18V-3′. cDNAs ≥500 bp were selected by agarose gel electrophoresis. EcoRI adaptors (5′-AATTCCATTGTGTTGGG-3′, Invitrogen) were ligated to the cDNAs, which were digested with NotI and directionally cloned into the EcoRI and NotI sites of pBS II SK(+) (Stratagene). Cloned cDNAs were electroporated into DH10B cells (Invitrogen) and amplified overnight in LB medium plus 75 μg/ml carbenicillin at 37°C. The primary library consisted of 4 × 106 clones. The background of empty clones was <1%.
Normalization and Subtraction of the Primary Library. The primary cDNA library was normalized as described in ref. 30. A single-stranded DNA version of the library was created by digestion with Gene II and Exonuclease III enzymes (Invitrogen). Purified single-stranded DNAs were used as template for PCR amplification using the T7 and T3 priming sites flanking the cDNA inserts. The purified PCR products were used as a driver for subtractive hybridization. Unhybridized single-stranded DNA circles were separated from hybridized DNA duplexes by hydroxyapatite. Purified single-stranded circles were rendered partially double-stranded by M13 reverse primer extension and electroporated into DH10B cells. This normalized library was plated, and 192 clones were picked and sequenced to determine redundancy. The titer of the normalized library was 7 × 106 clones. To allow further isolation of less abundant transcripts, the normalized cDNA library was subtracted by using as driver PCR products from a pool of 7,974 previously sequenced cDNAs. The titer of the subtracted library was 1 × 106 clones.
Juvenile cDNA Library. The library from sexually immature planarians was prepared as described above by using different EcoRI adaptors (5′-AATTCCGTTGCTGTCG-3′, Promega). Library normalization was performed as described above, except that PCR was performed on a pool of purified cDNAs from 6,505 unique clones sequenced from the first library. The titers were 5 × 106 clones in the primary library and 1 × 106 clones in the normalized/subtracted library.
EST Sequencing. Individual transformed bacterial colonies were robotically picked and racked as glycerol stocks in 384-well plates. After overnight growth of the glycerol stocks, bacteria were inoculated into 96-well deep cultures and grown overnight. Plasmid DNA was purified with Qiagen 8000 and 9600 BioRobots. Sequencing was performed by using standard T7 (5′ reads) or M13 reverse (3′ reads) primers and ABI BigDye terminator chemistry on ABI 3700 and 3730xl capillary systems (Applied Biosystems).
Sequence Analysis. The sequences were assigned quality values by calling bases with phred (31). Quality trimming (phred ≥ 20) and vector trimming were performed in sequencher 4.2 (Gene Codes, Ann Arbor, MI). After trimming, sequences <100 bp were omitted from further analysis, then checked for contaminants by blastn against the National Center for Biotechnology Information's (NCBI) Nucleotide (www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Nucleotide) and UniVec (www.ncbi.nlm.nih.gov/VecScreen/UniVec.html) databases. The resulting ESTs were assembled with cap3 (32) or sequencher (minimum 40-bp overlap and 95% identity). Redundancy was estimated by using the formula, 1 - ((no. contigs + no. singlets)/total no. sequences) × 100. The assembled ESTs were compared with the nonredundant sequence protein database (NCBI) by using stand-alone blast (33). ORF analysis was performed with flip 2.0.2 software (http://megasun.bch.umontreal.ca/ogmp).
Annotation of the EST Assembly. Based on the closest GO-annotated blastx homologue, sequences were assigned a biological process, molecular function, or cellular component from the GO database (34, 35). Domain searches were performed with rps-blast (E value ≤ 1 × 10-4) against the Conserved Domain Database (www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=cdd) (36).
Whole-Mount in Situ Hybridization to Sexual Planarians. Planarians starved for at least 1 week were fixed and bleached as described in ref. 37. Samples were loaded into a BioLane HTI automated in situ hybridization instrument (Hölle & Hüttner, Tübingen, Germany) and processed as described in ref. 38 except that hybridization was carried out for 48 h. Planarians were imaged with a Leica MZ125 stereomicroscope and a MicroFire digital camera (Optronics International, Chelmsford, MA).
Results and Discussion
Generation and Assembly of ESTs From Hermaphroditic S. mediterranea. Normalized and subtracted, directionally cloned cDNA libraries were generated from two separate developmental stages of the sexual strain of S. mediterranea: reproductively mature animals and “juveniles” that had not yet reached reproductive maturity, as judged by their smaller size and lack of a gonopore. First, we performed 5′-end sequencing on clones from the sexually mature planarian cDNA library until the redundancy reached 50% (see Methods) and thereby obtained 7,974 clones from the normalized library. These clones were then subtracted from the mature planarian cDNA library, allowing us to sequence an additional 8,448 clones before reaching 50% redundancy. To maximize the likelihood of finding new clones, the juvenile cDNA library was normalized and the clones obtained from the sexually mature library were used as subtraction drivers; a total of 7,872 clones were obtained from this adult-subtracted juvenile library. In addition, we estimated the total number of putative transcripts (see below) resulting from sequencing of the mature worm library, re-arrayed 6,505 unique clones from this library, and obtained additional sequences from their 3′ ends. The resulting final set of 27,161 ESTs (88% of the total after trimming and removal of contaminating sequences) with an average read length of 630 bp was considered high-quality and suitable for contiguous DNA sequence (contig) assembly (Table 1). Using PCR amplification of clones selected randomly from the mature and juvenile cDNA libraries, we estimated that the insert length averaged 1 kbp and ranged from 0.5 to 2.5 kbp.
Table 1. Hermaphroditic S. mediterranea EST sequencing project summary.
| Total sequences | 30,799 |
| Mature library | 22,927 |
| Juvenile library | 7,872 |
| 5′ end reads | 24,102 |
| 3′ end reads* | 6,697 |
| Total high-quality sequences† | 27,161 |
6,505 are 3′ reads of clones from the mature library previously sequenced from the 5′ end; 192 are from the normalized library check and sequenced from 3′ end
Total number after quality/vector trimming and eliminating contaminating sequences
The high-quality ESTs were assembled by using either cap3 or sequencher; these different assemblies produced comparable results, and we selected the cap3 assembly for further analyses (Table 2). Of the total of 27,161 ESTs, 23,164 assembled into 6,488 contigs; 3,997 remained as single sequences (singlets). The total number of contigs and singlets combined was 10,485. We identified 631 singlets that were unassembled 3′ reads of clones previously sequenced from the 5′ end; it is likely that they did not assemble because of short read length and/or large insert size. Excluding these 3′ reads, we estimate that the EST assembly represents 9,854 different transcripts. Most of the contigs in the assembly (3,263; ≈30%) consisted of two ESTs (Table 2). There was a single large contig comprised of 390 ESTs (1.4% of the total number of ESTs) corresponding to mitochondrial rRNA. This transcript accounted for >15% of clones sequenced from the primary cDNA library before normalization. Therefore, normalization was effective, reducing the frequency of this clone to <2% of the total clones sequenced.
Table 2. Hermaphroditic S. mediterranea EST assembly.
| cap3 | sequencher | |
|---|---|---|
| No. of sequences analyzed | 27,161 | 27,161 |
| No. post-assembly | 10,485 | 10,942 |
| Total no. of putative transcripts* | 9,854 | 10,520 |
| No. of contigs | 6,488 | 6,655 |
| No. of singlets | 3,997 | 4,287 |
| Distribution of contigs containing | ||
| 2 ESTs | 3,263 | 3,619 |
| 3 ESTs | 1,325 | 1,337 |
| 4–5 ESTs | 1,103 | 951 |
| 6–10 ESTs | 613 | 571 |
| 11–15 ESTs | 108 | 115 |
| >16 ESTs | 76 | 62 |
The assembly parameters used for both cap3 and sequencher were 40-bp minimum overlap and 95% identity. Contig, contiguous sequences composed of two or more overlapping EST sequences.
Unassembled 3′-end reads of clones previously sequenced from the 5′ end were excluded from the predicted number of unique transcripts
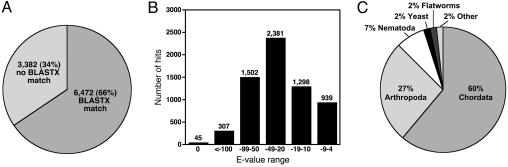
blast Analysis of the EST Assembly. Of the 9,854 assembled sequences, 6,472 (66%) were similar to protein sequences in the nonredundant protein database (Fig. 1A). We binned by significance the number of blastx hits and found that 85% had E values smaller than 1 × 10-10 (Fig. 1B). Furthermore, of the 3,382 putative transcripts with no blastx matches, 1,705 (50%) were predicted to have ORFs of 450 bp (150 aa) or greater. By combining the number of assembled sequences with matches in the nonredundant protein database and those with predicted ORFs, we estimate that at least 8,177 (83%) of the assembled sequences likely encode proteins.
Fig. 1.
blastx analysis of the sexual S. mediterranea ESTs. (A) Results of blastx analysis (E value ≤ 1 × 10-4) comparing the unique set of 9,854 assembled ESTs to the nonredundant protein database. (B) Distribution of blastx matches by E value. The number of ESTs for the E value ranges is indicated above each bar. (C) Organization of the phyla representing the top blastx hits. Of the 6,470 ESTs that were assigned a taxon ID, ≈60% had their top matches with Chordates.
We also surveyed the species represented in the best hit found by blastx; the majority of the assembled ESTs had matches to sequences from Chordates (60%) and Arthropods (27%) (Fig. 1C). Top matches to Chordata were all represented by vertebrate sequences, and 26% of these matches were to human sequences. Because planarians are known to share genes with vertebrates that have been lost from both C. elegans and Drosophila (28), we looked for more such sequences in this EST collection. The assembled ESTs were compared with the proteomes of Saccharomyces cerevisiae, Schizosaccharomyces pombe, C. elegans, Drosophila melanogaster, Anopheles gambiae, Danio rerio, Gallus gallus, Sus scrofa, Bos taurus, Mus musculus, Rattus norvegicus, Pan troglodytes, and Homo sapiens. A secondary screen to remove sequences that hit Caenorhabditis briggsae, Drosophila yakuba, and/or Drosophila pseudoobscura identified 316 planarian sequences that matched sequences from vertebrates but were not found in yeast and other invertebrate proteomes (Data Set 1, which is published as supporting information on the PNAS web site). The functions of most of these conserved genes are not known: 44% (139/316) of them encode hypothetical proteins. We speculate that these conserved sequences may play roles in processes such as long-term tissue maintenance and cell turnover that are likely less important for short-lived organisms like nematodes and insects.
In addition, we compared the EST assembly to a collection of 287 genes associated with human diseases (39) by tblastn. We found that 142 planarian transcripts encoded predicted proteins with significant similarity to these human sequences. Given that our EST collection does not represent the entire planarian genome, it seems likely that the vast majority of human disease genes will have homologues in planarians. Because planarians are susceptible to RNA interference (38), they will provide a complementary model invertebrate for studying the functions of conserved genes implicated in human biology and disease (28, 29).
At the time of analysis, there were 3,202 S. mediterranea ESTs available in the public databases (28, 29). We downloaded these sequences and asked what percentage is represented in our EST assembly. For these searches, we used the entire set of unique sequences resulting from the cap3 assembly (10,485) to maximize the likelihood of finding matches and to produce a conservative estimate of the number of newly identified genes. blastn analysis (E value ≤ 1 × 10-20) showed that of the 3,202 sequences, 1,738 are represented in our assembly. Therefore, our EST data augment the currently available S. mediterranea EST data with ≈8,116 new sequences (of 9,854 predicted to be unique).
We also compared this collection of ESTs with sequences from the trematode Schistosoma mansoni, a parasitic flatworm that is the primary causative agent of schistosomiasis (40). An EST project for S. mansoni produced a set of 30,988 assembled sequences (41). Using tblastx (E value ≤ 1 × 10-4), we found that 4,957 (47%) planarian transcripts share similarity with S. mansoni sequences, including 11/28 genes suggested as candidate vaccine targets for schistosomiasis (41) (Data Set 1). Verjovski-Almeida et al. (41) speculated that some of these candidate genes could encode receptors that bind host factors (e.g., VLDL, stomatin, and activin IIB). Identification of homologues of these receptors in a free-living flatworm suggests that such factors are likely to play roles in endogenous signaling processes. Investigating the function of the planarian homologues should help to identify genes that are required for flatworm viability. Similarly, it should be possible to examine genes that are shared between planarians and parasitic flatworms, yet absent from the human genome, and thus identify potential targets for the treatment and prevention of parasitic flatworm infections (28).
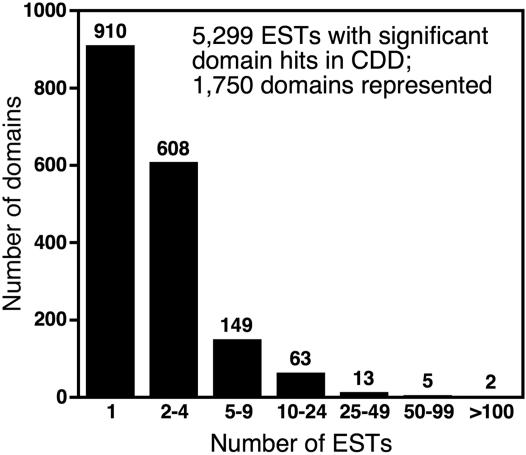
Conserved Protein Domains Commonly Encountered in the EST Assembly. To identify predicted protein domains in the EST assembly, we performed rps-blast searches against the Conserved Domain Database (36) and found 5,299 (54%) sequences with significant matches. The domains most highly represented in our EST collection were Serine/Threonine protein kinase catalytic domain and RNA recognition motif (Table 3). However, these domains only account for 2.4% and 1.9% of ESTs with Conserved Domain Database matches, respectively. When we analyzed the distribution of the domains with the highest rps-blast significance assigned to ESTs in our collection, we found that there were ≈1,750 different domains represented; 910 (≈9%) ESTs were assigned a unique domain (Fig. 2). The diversity of domains represented is likely due to the normalization and subtraction techniques used to generate the cDNA libraries, resulting in a wide representation of gene classes or families. For example, this collection contains a large number of domains associated with transcription factors that are likely to be expressed at fairly low levels and have not been found in previous planarian EST collections (28, 42).
Table 3. Conserved domains most frequently encountered in hermaphroditic S. mediterranea ESTs.
| Conserved domain | No. of hits | % |
|---|---|---|
| Serine/Threonine protein kinases | 135 | 2.4 |
| RRM (RNA recognition motif) | 103 | 1.9 |
| Ankyrin repeat | 68 | 1.2 |
| EF-hand calcium binding motif | 64 | 1.2 |
| WD40 | 64 | 1.2 |
| TUBULIN | 60 | 1.1 |
| Smc (Chromosome segregation ATPases) | 59 | 1.1 |
| RAB (Rab subfamily of small GTPases) | 44 | 0.8 |
| RING-finger | 38 | 0.7 |
| LRR (leucine-rich repeat) | 35 | 0.6 |
| Transmembrane 4 | 35 | 0.6 |
| 7tm_1 (rhodopsin family) | 31 | 0.6 |
| Ubiquitin | 30 | 0.5 |
Fig. 2.
Frequency of protein domains found in the hermaphroditic S. mediterranea EST assembly. The assembled sequences were analyzed by performing rps-blast (E value ≤ 1 × 10-4) in all six reading frames against the Conserved Domain Database. The total number of domains per EST number range is indicated above each bar.
Assignment of GO Terms to the EST Assembly. The predicted transcripts in the hermaphroditic S. mediterranea EST assembly were assigned a biological process, molecular function, and cellular component from the GO database (34, 35). We analyzed the results for the 9,854 unique transcripts and have assigned a biological process to 3,076 (31% of the total and 48% of those sharing homology in the nonredundant protein database), a molecular function to 3,013 (31% and 47%, respectively), and a cellular component to 1,066 (11% and 16%, respectively) sequences. We assigned parent terms in the biological process ontology and found that the most abundant categories were “metabolism” (19%), “protein metabolism” (14%), “transport” (13%), and “signal transduction” (10%) (Table 4). The most commonly assigned term in the molecular function category was “ATP binding,” and the most common cellular components were “integral to membrane” and “nucleus” (Tables 5 and 6, which are published as supporting information on the PNAS web site).
Table 4. GO terms in the Biological Process category associated with S. mediterranea ESTs.
| Biological process | No. of hits | % |
|---|---|---|
| Metabolism* | 599 | 19 |
| Protein metabolism | 441 | 14 |
| Transport | 388 | 13 |
| Signal transduction | 301 | 10 |
| Development | 239 | 8 |
| RNA metabolism | 134 | 4 |
| Response to stimulus | 132 | 4 |
| Cytoskeleton organization and biogenesis | 120 | 4 |
| Cell proliferation | 112 | 4 |
| Translation | 78 | 3 |
| Cell motility | 72 | 2 |
| Transcription | 57 | 2 |
| Reproduction | 53 | 2 |
| Cell growth and/or maintenance | 53 | 2 |
| Cell death | 47 | 2 |
| Cell adhesion | 42 | 1 |
| Behavior | 41 | 1 |
| Nuclear organization and biogenesis | 36 | 1 |
| Synaptic transmission | 31 | 1 |
| DNA metabolism | 25 | 1 |
| Other (terms represented <0.5% of total) | 75 | 2 |
Includes amino acid (43), carbohydrate (121), lipid (69), and nucleotide (39) metabolism, biosynthesis (85), and electron transport (73)
Generation of New Molecular Markers for the Planarian Reproductive Organs. One of the aims for creating this collection of ESTs is to investigate epigenetic germ cell specification in planarians. To facilitate these studies, specific markers of the reproductive structures are needed. One approach to develop such markers would be to analyze the expression patterns of all of the ESTs by in situ hybridization. Although such screens are feasible using the asexual planarian strain (28), they are less practical with the hermaphroditic strain because of their larger size and slow generation time. Therefore, other criteria are necessary to identify candidate markers. The annotation of gene products using GO terms (34, 35) provides a useful resource for identifying candidate genes by putative function. Thus, ESTs annotated under the biological process of “reproduction” (Table 7, which is published as supporting information on the PNAS web site) were selected for in situ hybridization analysis. Only one of these genes has been studied in sexual planarians: PL06004A2E04 shares similarity with DjvlgA (blastn, E value = 1 × 10-18) from Dugesia japonica (22) and is related to the vasa-like genes that are involved in germ cell development (43). In addition, a planarian homologue of pumilio, a member of the PUF protein family involved in germline stem cell maintenance (44), has been shown to be important for neoblast maintenance in asexual D. japonica (45); its role in planarian germ cell specification has yet to be investigated.
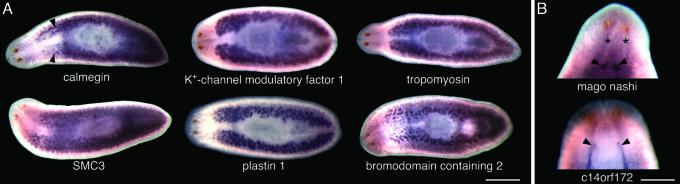
We tested the validity of the GO annotation by analyzing the expression patterns for all 53 of these ESTs in sexually mature planarians and found that 46/53 transcripts (87%) were expressed in the reproductive organs (Fig. 3). All 46 ESTs were expressed in the testes, dorsolateral clusters that run from behind the head to the tail (Fig. 3A). For example, clone PL06015A2C10 is homologous to calmegin, a testis-specific endoplasmic reticulum resident chaper-one required for the binding of sperm to egg plasma membrane and, thus, sperm fertility in mice (46). PL06019A2G01 shares homology with SMC3, a core component of the cohesin complex responsible for sister chromatid cohesion in mitosis and meiosis (47, 48). PL06020B1C11, a bromodomain containing 2 homologue, is expressed in mitotic somatic cells and meiotic germ cells in the mouse testes (49) as well as in ovaries, where it might play a role in mitotic and meiotic cell cycle regulation (50).
Fig. 3.
Whole-mount in situ hybridization to mature S. mediterranea hermaphrodites using ESTs annotated to the GO term of reproduction. (A) Representative results of annotated ESTs expressed in testes (arrowheads). From the top left: PL06015A2C10, calmegin; PL03020B1B02, K+-channel modulatory factor 1; PL04019B2G03, cytoskeletal tropomyosin; PL06019A2G01, SMC3; PL06020B1C06, plastin 1; PL06020B1C11, bromodomain containing 2. (B) ESTs that are expressed in the ovaries (arrowheads), which can be viewed ventrally and are located posterior to the cephalic ganglia (asterisk). PL04006A1H09, mago nashi; PL06003X1E11, c14orf172. Detailed information for each homologue can be found in Table 7. Anterior is to the left in A and at the top in B. (Scale bars: A, 1 mm; B, 0.5 mm.)
Expression in the ovaries was observed less frequently, but 15 (28%) ESTs were clearly detected in this organ (Fig. 3B). For example, PL04006A1H09, is homologous to mago nashi, a gene required for germ-plasm assembly and axis determination in Drosophila (51-53). The ovaries in planarians are small, and because of their location in the animal and thickness of the specimen, the ability to detect these structures unambiguously by in situ hybridization typically requires high levels of expression. In addition, the cDNA libraries reported in this study were prepared from whole animals; it seems likely that the abundance of testes tissue relative to the ovaries resulted in an under-representation of ovary-specific transcripts. Future studies will benefit from preparation of cDNA libraries from ovary-enriched tissues. Furthermore, 35 of the ESTs expressed in the reproductive tissues were also detected in other cell types, including: central nervous system, gastrovascular system, mesenchyme, or neoblasts (Table 7).
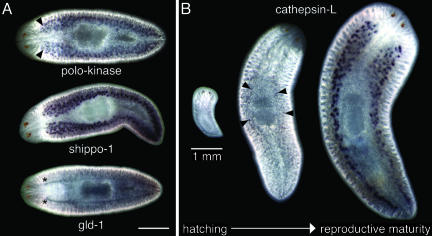
For in situ analysis, we also chose additional candidate genes implicated in reproductive processes based on blastx similarity; these genes were not annotated to the GO term “reproduction.” For example, we studied planarian homologues of cdc-5/polo-like kinases, important regulators of cell cycle checkpoints (54) also implicated in the coordination of chromosome segregation during meiosis (55); the mammalian sperm tail protein, Shippo-1 (56); and Cathepsin-L, a protein implicated in the maturation of sperm during mammalian spermatogenesis (57). All of these genes were expressed in the planarian testes. A planarian homologue of gld-1, a gene required for oocyte development in C. elegans (58), was detected in the ovaries and oviducts (Fig. 4). The expression of cathepsin-L mRNA was used to monitor the development of the testes (Fig. 4B). Cathepsin-L mRNA was not detected in 2- to 3-day-old hatchlings; the transcript was first detectable in the testes primordia of juvenile planarians and strongly expressed in the testes of mature worms (Fig. 4B). Cathepsin-L mRNA was not detected in asexual worms; consistent with this observation, Northern blot analysis showed that this gene is expressed at high levels in sexual worms but is undetectable in asexual animals (data not shown). These results provide additional evidence that the planarian reproductive organs are formed postembryonically (5, 8-11, 14).
Fig. 4.
Whole-mount in situ hybridization to developing and mature S. mediterranea hermaphrodites. (A) Selected ESTs that are expressed in the reproductive structures. From top to bottom: PL06001X1H06, cdc5-like (R. norvegicus, NP_445979, blastx = 7 × 10-10) expression is detected in the testes (arrowheads); PL030013A20C06 (Contig894), sperm tail protein SHIPPO1 (H. sapiens, NP_444510, blastx = 3 × 10-22) also expressed in the testes; PL06016A2E04, gld-1 (C. elegans, NP_492143, blastx = 2 × 10-29) expression can be detected in the ovaries (asterisks) and oviducts. (B) Developmental expression of clone PL010001001G06 (Contig184), Cathepsin-L (D. rerio, NP_997749, blastx = 1 × 10-99). This transcript is undetectable in hatchlings (left), first becomes apparent in the developing testes of juveniles (arrowheads), and is strongly expressed in the testes of mature worms (right). Anterior is to the left in A and at the top in B. (Scale bars: 1 mm.)
The Hermaphroditic S. mediterranea EST Database. We have designed a relational database similar to that created for the Axolotl EST project (59) for easy access and browsing of our EST collection. The database can be searched by contig or clone name, gene description, GO terms, conserved domains, or gene expression patterns, which have direct web links to their respective databases, simplifying browsing of the information pertaining to each sequence. In addition, the user can download the EST sequences and/or chromatograms. The database is available at www.life.uiuc.edu/planaria.
Conclusions
The annotated ESTs discussed in this paper will provide a useful resource for studies on germ cell determination, regeneration, and other areas of research. Our in situ hybridization results validate the GO annotation and provide >50 new markers of the planarian reproductive system; such markers will be useful for analyzing the development, regression, and regeneration of these structures. Moreover, the hermaphroditic strain of S. mediterranea is the focus of an on-going genome sequencing project; the collection of ESTs described here will be particularly important for annotating the planarian genome. In combination with high-throughput in situ hybridization and RNA interference screens (28, 29), these genomic-level analyses should generate new insights into many aspects of planarian biology. Given the critical role of stem cells in tissue maintenance and regeneration in planarians, these studies should also help us identify evolutionarily conserved mechanisms that regulate stem cell proliferation and differentiation (28).
Supplementary Material
Acknowledgments
We thank Francesc Cebrià, Tingxia Guo, and Gene Robinson for helpful comments on the manuscript; Naomi Thompson (Gene Codes) for help with Sequencher; Jeffrey Haas, Phil Anders, and Daniel Davidson for computer support; Ryan Kim, Peter Schweitzer, and the high-throughput sequencing staff of the UIUC Keck Center; Claire Miller for help with the human disease homologue analysis; Maria Pala for providing the sexual strain of S. mediterranea; and Alejandro Sánchez Alvarado, in whose laboratory P.A.N. generated the clonal line used here. R.M.Z. is a Fellow of the Jane Coffin Childs Memorial Fund for Medical Research. This work was supported by National Science Foundation CAREER Award IBN-0237825 and National Institutes of Health Grant R01 HD043403 (to P.A.N.). P.A.N. is a Damon Runyon Scholar supported by the Damon Runyon Cancer Research Foundation (DRS 33-03).
Author contributions: P.A.N. and R.M.Z. designed research; R.M.Z., A.H., Y.W., J.M.S., and P.A.N. performed research; B.H. contributed new reagents/analytic tools; R.M.Z., B.H., Y.W., J.M.S., and P.A.N. analyzed data; R.M.Z. and P.A.N. wrote the paper; and B.H. contributed to creating the EST relational database.
Conflict of interest statement: No conflicts declared.
Abbreviation: GO, Gene Ontology.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. DN289353-DN316518).
References
- 1.Nieuwkoop, P. D. & Sutasurya, L. A. (1979) Primordial Germ Cells in the Chordates (Cambridge Univ. Press, London).
- 2.Nieuwkoop, P. D. & Sutasurya, L. A. (1981) Primordial Germ Cells in the Invertebrates: From Epigenesis to Preformation (Cambridge Univ. Press, London).
- 3.Extavour, C. G. & Akam, M. (2003) Development (Cambridge, U.K.) 130, 5869-5884. [DOI] [PubMed] [Google Scholar]
- 4.Johnson, A. D., Drum, M., Bachvarova, R. F., Masi, T., White, M. E. & Crother, B. I. (2003) Evol. Dev. 5, 414-431. [DOI] [PubMed] [Google Scholar]
- 5.Baguñá, J., Saló, E. & Auladell, C. (1989) Development (Cambridge, U.K.) 107, 77-86. [Google Scholar]
- 6.Newmark, P. & Sánchez Alvarado, A. (2000) Dev. Biol. 220, 142-153. [DOI] [PubMed] [Google Scholar]
- 7.Reddien, P. W. & Sánchez Alvarado, A. (2004) Annu. Rev. Cell Dev. Biol. 20, 725-757. [DOI] [PubMed] [Google Scholar]
- 8.Curtis, W. C. (1902) Proc. Boston Soc. Nat. Hist. 30, 515-559. [Google Scholar]
- 9.Fedecka-Bruner, B. (1965) in Regeneration in Animals and Related Problems, eds. Kiortsis, V. & Trampusch, H. A. L. (North-Holland, Amsterdam), pp. 185-192.
- 10.Ghirardelli, E. (1965) in Regeneration in Animals and Related Problems, eds. Kiortsis, V. & Trampusch, H. A. L. (North-Holland, Amsterdam), pp. 177-184.
- 11.Kobayashi, K. & Hoshi, M. (2002) Zool. Sci. 19, 661-666. [DOI] [PubMed] [Google Scholar]
- 12.Hyman, L. H. (1951) The Invertebrates: Platyhelminthes and Rhynchocoela, The Acoelomate Bilateria (McGraw-Hill, New York).
- 13.Newmark, P. A. & Sánchez Alvarado, A. (2002) Nat. Rev. Genet. 3, 210-219. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi, K., Arioka, S. & Hoshi, M. (2002) Zool. Sci. 19, 1267-1278. [DOI] [PubMed] [Google Scholar]
- 15.Morgan, T. H. (1902) Arch. Entwicklungsmech. Org. 13, 179-212. [Google Scholar]
- 16.Abeloos, M. (1930) Bull. Biol. 1, 1-140. [Google Scholar]
- 17.Baguñá, J. & Romero, R. (1981) Hydrobiologia 84, 181-194. [Google Scholar]
- 18.Oviedo, N. J., Newmark, P. A. & Sanchez Alvarado, A. (2003) Dev. Dyn. 226, 326-333. [DOI] [PubMed] [Google Scholar]
- 19.Berninger, J. (1911) Zool. Jahrb. 30, 181-216. [Google Scholar]
- 20.Schultz, E. (1904) Arch. Entwicklungsmech. Org. 18, 555-577. [Google Scholar]
- 21.Ogawa, K., Wakayama, A., Kunisada, T., Orii, H., Watanabe, K. & Agata, K. (1998) Biochem. Biophys. Res. Commun. 248, 204-209. [DOI] [PubMed] [Google Scholar]
- 22.Shibata, N., Umesono, Y., Orii, H., Sakurai, T., Watanabe, K. & Agata, K. (1999) Dev. Biol. 206, 73-87. [DOI] [PubMed] [Google Scholar]
- 23.Salvetti, A., Lena, A., Rossi, L., Deri, P., Cecchettini, A., Batistoni, R. & Gremigni, V. (2002) Gene Expression Patterns 2, 195-200. [DOI] [PubMed] [Google Scholar]
- 24.Hase, S., Kobayashi, K., Koyanagi, R., Hoshi, M. & Matsumoto, M. (2003) Dev. Genes Evol. 212, 585-592. [DOI] [PubMed] [Google Scholar]
- 25.Simoncelli, F., Sorbolini, S., Fagotti, A., Di Rosa, I., Porceddu, A. & Pascolini, R. (2003) Biochim. Biophys. Acta 1629, 26-33. [DOI] [PubMed] [Google Scholar]
- 26.Orii, H., Sakurai, T. & Watanabe, K. (2005) Dev. Genes Evol. 215, 143-157. [DOI] [PubMed] [Google Scholar]
- 27.Baguñá, J., Carranza, S., Pala, M., Ribera, C., Giribet, G., Arnedo, M. A., Ribas, M. & Riutort, M. (1999) Ital. J. Zool. 66, 207-214. [Google Scholar]
- 28.Sánchez Alvarado, A., Newmark, P. A., Robb, S. M. & Juste, R. (2002) Development (Cambridge, U.K.) 129, 5659-5665. [DOI] [PubMed] [Google Scholar]
- 29.Reddien, P. W., Bermange, A. L., Murfitt, K. J., Jennings, J. R. & Sánchez Alvarado, A. (2005) Dev. Cell 8, 635-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonaldo, M. F., Lennon, G. & Soares, M. B. (1996) Genome Res. 6, 791-806. [DOI] [PubMed] [Google Scholar]
- 31.Ewing, B., Hillier, L., Wendl, M. C. & Green, P. (1998) Genome Res. 8, 175-185. [DOI] [PubMed] [Google Scholar]
- 32.Huang, X. & Madan, A. (1999) Genome Res. 9, 868-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., Davis, A. P., Dolinski, K., Dwight, S. S., Eppig, J. T., et al. (2000) Nat. Genet. 25, 25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.The Gene Ontology Consortium (2001) Genome Res. 11, 1425-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marchler-Bauer, A., Anderson, J. B., Cherukuri, P. F., DeWeese-Scott, C., Geer, L. Y., Gwadz, M., He, S., Hurwitz, D. I., Jackson, J. D., Ke, Z., et al. (2005) Nucleic Acids Res. 33, Database Issue, D192-D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Umesono, Y., Watanabe, K. & Agata, K. (1997) Dev. Growth Differ. 39, 723-727. [DOI] [PubMed] [Google Scholar]
- 38.Sánchez Alvarado, A. & Newmark, P. A. (1999) Proc. Natl. Acad. Sci. USA 96, 5049-5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fortini, M. E., Skupski, M. P., Boguski, M. S. & Hariharan, I. K. (2000) J. Cell Biol. 150, F23-F30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grevelding, C. G. (2004) Curr. Biol. 14, R545. [DOI] [PubMed] [Google Scholar]
- 41.Verjovski-Almeida, S., DeMarco, R., Martins, E. A., Guimaraes, P. E., Ojopi, E. P., Paquola, A. C., Piazza, J. P., Nishiyama, M. Y., Jr., Kitajima, J. P., Adamson, R. E., et al. (2003) Nat. Genet. 35, 148-157. [DOI] [PubMed] [Google Scholar]
- 42.Mineta, K., Nakazawa, M., Cebriá, F., Ikeo, K., Agata, K. & Gojobori, T. (2003) Proc. Natl. Acad. Sci. USA 100, 7666-7671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raz, E. (2000) Genome Biol. 1, REVIEWS1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wickens, M., Bernstein, D. S., Kimble, J. & Parker, R. (2002) Trends Genet. 18, 150-157. [DOI] [PubMed] [Google Scholar]
- 45.Salvetti, A., Rossi, L., Lena, A., Batistoni, R., Deri, P., Rainaldi, G., Locci, M. T., Evangelista, M. & Gremigni, V. (2005) Development (Cambridge, U.K.) 132, 1863-1874. [DOI] [PubMed] [Google Scholar]
- 46.Ikawa, M., Wada, I., Kominami, K., Watanabe, D., Toshimori, K., Nishimune, Y. & Okabe, M. (1997) Nature 387, 607-611. [DOI] [PubMed] [Google Scholar]
- 47.Jessberger, R. (2002) Nat. Rev. Mol. Cell Biol. 3, 767-778. [DOI] [PubMed] [Google Scholar]
- 48.Nasmyth, K. & Haering, C. H. (2005) Annu. Rev. Biochem. 74, 595-648. [DOI] [PubMed] [Google Scholar]
- 49.Rhee, K., Brunori, M., Besset, V., Trousdale, R. & Wolgemuth, D. J. (1998) J. Cell Sci. 111, 3541-3550. [DOI] [PubMed] [Google Scholar]
- 50.Trousdale, R. K. & Wolgemuth, D. J. (2004) Mol. Reprod. Dev. 68, 261-268. [DOI] [PubMed] [Google Scholar]
- 51.Newmark, P. A. & Boswell, R. E. (1994) Development (Cambridge, U.K.) 120, 1303-1313. [DOI] [PubMed] [Google Scholar]
- 52.Micklem, D. R., Dasgupta, R., Elliott, H., Gergely, F., Davidson, C., Brand, A., Gonzalez-Reyes, A. & St. Johnston, D. (1997) Curr. Biol. 7, 468-478. [DOI] [PubMed] [Google Scholar]
- 53.Newmark, P. A., Mohr, S. E., Gong, L. & Boswell, R. E. (1997) Development (Cambridge, U.K.) 124, 3197-3207. [DOI] [PubMed] [Google Scholar]
- 54.Xie, S., Xie, B., Lee, M. Y. & Dai, W. (2005) Oncogene 24, 277-286. [DOI] [PubMed] [Google Scholar]
- 55.Lee, B. H. & Amon, A. (2003) Science 300, 482-486. [DOI] [PubMed] [Google Scholar]
- 56.Egydio de Carvalho, C., Tanaka, H., Iguchi, N., Ventela, S., Nojima, H. & Nishimune, Y. (2002) Biol. Reprod. 66, 785-795. [DOI] [PubMed] [Google Scholar]
- 57.Wright, W. W., Smith, L., Kerr, C. & Charron, M. (2003) Biol. Reprod. 68, 680-687. [DOI] [PubMed] [Google Scholar]
- 58.Francis, R., Barton, M. K., Kimble, J. & Schedl, T. (1995) Genetics 139, 579-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Habermann, B., Bebin, A. G., Herklotz, S., Volkmer, M., Eckelt, K., Pehlke, K., Epperlein, H. H., Schackert, H. K., Wiebe, G. & Tanaka, E. M. (2004) Genome Biol. 5, R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.