Abstract
The c-MYC oncoprotein regulates various aspects of cell behaviour by modulating gene expression. Here, we report the identification of the cAMP-response-element-binding protein (CBP) as a novel c-MYC binding partner. The two proteins interact both in vitro and in cells, and CBP binds to the carboxy-terminal region of c-MYC. Importantly, CBP, as well as p300, is associated with E-box-containing promoter regions of genes that are regulated by c-MYC. Furthermore, c-MYC and CBP/p300 function synergistically in the activation of reporter-gene constructs. Thus, CBP and p300 function as positive cofactors for c-MYC. In addition, c-MYC is acetylated in cells. This modification does not require MYC box II, suggesting that it is independent of TRRAP complexes. Instead, CBP acetylates c-MYC in vitro, and co-expression of CBP with c-MYC stimulates in vivo acetylation. Functionally, this results in a decrease in ubiquitination and stabilization of c-MYC proteins. Thus, CBP and p300 are novel functional binding partners of c-MYC.
Introduction
The expression of genes is controlled by DNA-sequence-specific transcriptional regulators that recruit cofactors to modulate the activity of the polymerase complex (Narlikar et al., 2002). Cofactors function at least in part by affecting chromatin structure through their associated enzymatic activities, including ATPases/helicases, histone acetyl-transferases (HATs) and histone deacetylases (HDACs) (Jenuwein & Allis, 2001; Sudarsanam & Winston, 2000). Targets of the latter include histone tails that, depending on their modification status, function at least partly as interaction surfaces for proteins involved in the regulation of chromatin structure and gene transcription (Jenuwein & Allis, 2001).
MYC proteins, including c-MYC, N-MYC and L-MYC, are transcriptional regulators that were first identified as transforming factors. Subsequently, it was shown that MYC stimulates cell proliferation, inhibits differentiation and induces apoptosis in many cell types. Structure–function analyses have revealed several regions that are important for the ability of MYC to control cell behaviour. These include the amino-terminal transactivation domain (TAD) and the carboxy-terminal bHLHZip (basic helix–loop–helix zipper) domain, which is responsible for dimerization with its essential partner, MAX, and for sequence-specific DNA binding, respectively (Lüscher, 2001; Oster et al., 2002).
Little detailed information is available as to how c-MYC affects the activity of the polymerase II (Pol II) complex (Amati et al., 2001; Lüscher, 2001). Among the many proteins that have been shown to interact with c-MYC (Oster et al., 2002), TRRAP, a component of HAT-containing complexes, attracted particular interest (McMahon et al., 1998, 2000; Park et al., 2001). TRRAP binds to the MYC box II (MBII) motif in the N-terminal TAD of c-MYC, and is recruited to c-MYC-regulated promoters (Bouchard et al., 2001; Frank et al., 2001). The activation of some genes, including cyclin D2, telomerase reverse-transcriptase and nucleolin, by c-MYC requires MBII (Bouchard et al., 2001; Frank et al., 2001; Nikiforov et al., 2002). However, several other genes can be activated by c-MYCΔMBII, that is, in the absence of TRRAP recruitment (Nikiforov et al., 2002). These findings suggest that TRRAP is important for c-MYC-dependent regulation of some MYC target genes, whereas others may depend on additional MYC-associated activities.
c-MYC-dependent activation of target genes is associated with an increase in histone H3 and histone H4 acetylation (Bouchard et al., 2001; Frank et al., 2001). At least three distinct HATs, GCN5, PCAF and TIP60, can associate with TRRAP complexes. For two of these, GCN5 and TIP60, an interaction with c-MYC has been demonstrated (McMahon et al., 2000; B. Amati, personal communication). Thus, recruitment of these HATs to TRRAP-containing complexes is probably significant for gene regulation by c-MYC.
In this study, we identified the co-activator CREB (cAMP response element)-binding protein (CBP) as a novel c-MYC interaction partner. CBP and its close relative p300 are important transcriptional mediators for many transcription factors. CBP functions partly through its intrinsic HAT activity, as well as through associated HAT enzymes. In addition, CBP and p300 have been shown to interact with components of the Pol II complex, which may also contribute to co-activator function. CBP/p300 has several functions in the control of cell behaviour, including cell proliferation (Chan & La Thangue, 2001; Goodman & Smolik, 2000). Our findings show that CBP interacts directly with c-MYC and stimulates its function. Furthermore, CBP and p300 are recruited to MYC-regulated genes. Last, CBP acetylates c-MYC, thereby affecting its ubiquitination and half-life. Together, these findings identify CBP and p300 as novel functional binding partners of c-MYC.
Results and Discussion
In vivo and in vitro interaction of c-MYC with CBP
In our attempt to identify novel c-MYC interacting proteins, we metabolically labelled human embryonic kidney (HEK) 293 cells with [35S]methionine, and immunoprecipitated c-MYC and associated proteins under low stringency conditions. We saw several labelled protein bands that were not immunoprecipitated with control antibodies. One of these bands co-migrated with CBP/p300, which have been identified previously as binding partners of the adenoviral protein E1A (Fig. 1A). We tested the possibility that c-MYC interacts with CBP/p300. CBP was detected in the eluate of c-MYCspecific but not MAD1-specific immunoprecipitates (Fig. 1B). Furthermore, CBP binding to c-MYC was seen in co-immunoprecipitation and western blot analysis in human Jurkat T cells and in HL60 and U937 myeloid cells. Importantly, CBP was also detected in MAXspecific immunoprecipitates, showing that the c-MYC/MAX heterodimeric complex interacts with CBP (Fig. 1C,D; and data not shown).
Figure 1.
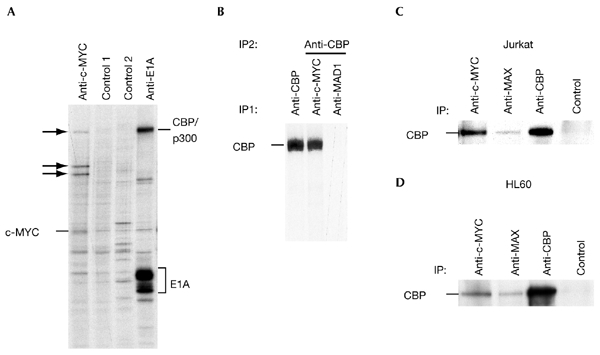
Association of endogenous c-MYC and CBP in cells.(A) Human embryonic kidney 293 (HEK293) cells were labelled with [35S]methionine for 2 h before lysis in F buffer. Immunoprecipitations (IPs) were performed with the rat monoclonal antibody 6A10 (specific for c-MYC), protein-G–sepharose beads only (control 1), monoclonal antibody 5C9 (specific for MAD1; control 2) and the mouse monoclonal antibody M73 specific for E1A. The immunoprecipitates were analysed by SDS–polyacrylamide gel electrophoresis (SDS–PAGE). Arrows indicate co-immunoprecipitated proteins. (B) HEK293 cells were treated as in (A). IPs were first performed with C20 anti-cAMP-response-element-binding protein (CBP), monoclonal antibody 6A10 or monoclonal antibody 5C9. Co-immunoprecipitated proteins were eluted in AB buffer and CBP was immunoprecipitated (using the C20 antibody) and analysed by SDS–PAGE. (C) Exponentially growing Jurkat T cells were lysed in F buffer and IPs were performed as indicated using N262 to immunoprecipitate c-MYC, C17 for MAX, AC238 for CBP and SC577 for Gal4, as a control. CBP was then detected by immunoblotting using C20. (D) Co-IPs were performed as in (C), using lysates from promyelocytic HL60 cells.
The interaction of c-MYC with CBP was further confirmed using a mammalian two-hybrid system. Transactivation by Gal4–CBP was strongly activated by co-expressing a c-MYC–VP16-TAD fusion protein (Fig. 2A). In addition, glutathione-S-transferase (GST) pull-down experiments showed a direct interaction between c-MYC and CBP (Fig. 2B,C). In CBP, the regions encompassing amino acids 451–721, which includes the KIX domain, and to a significantly lesser degree, amino acids 1–451, interacted with c-MYC. These CBP domains have been implicated in binding to several different transcription factors (Chan & La Thangue, 2001). Conversely, CBP binds to the C-terminal domain of c-MYC, but not to the TAD (Fig. 2B,C).
Figure 2.
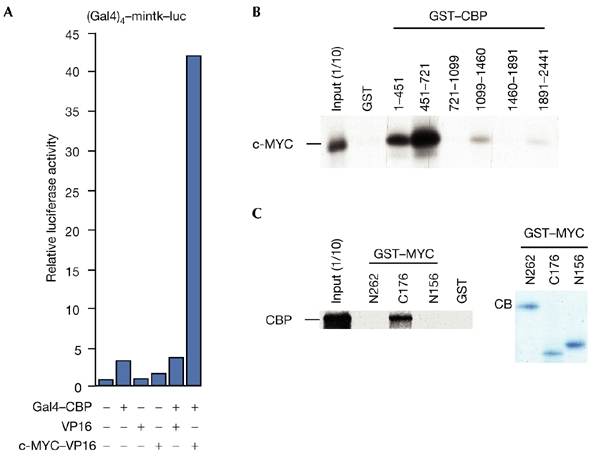
Interaction of MYC and CBP in overexpression experiments and in vitro. (A) Interaction of c-MYC and CBP (cAMP-response-element-binding protein) detected using a mammalian two-hybrid system. Constructs expressing Gal4–CBP (50 ng), c-MYC–VP16 (2 μg) and VP16 (2 μg) were co-expressed, as indicated, with the Gal4–mintk–luc (a fusion of Gal4, the minimal thymidine-kinase promoter and luciferase) reporter gene construct (2 μg) in U2OS cells. (B) Glutathione-S-transferase (GST)–CBP fusion proteins (1–2 μg) were used to detect interactions with in vitro transcribed and translated c-MYC. One-tenth of the in vitro transcribed/translated material was loaded as a control (input (1/10)). (C) Binding of in vitro transcribed and translated CBP to GST–MYC fusion proteins was analysed as indicated (left panel). A Coomassie-blue (CB)-stained gel containing the three GST–MYC fusion proteins is shown (right panel).
CBP stimulates c-MYC-dependent transactivation
Next, we tested whether CBP can modulate c-MYC-specific transactivation. c-MYC weakly activated a reporter gene construct containing c-MYC/MAX binding sites (Fig. 3A). However, this reporter was activated co-operatively by c-MYC and CBP, whereas a reporter with no binding sites was not regulated (Fig. 3A). In addition, p300 also co-operated efficiently with c-MYC (Fig. 3A), although the interaction of c-MYC with p300 appeared weaker than with CBP, as inferred from co-immunoprecipitation experiments (data not shown). The HAT domain of CBP was partly responsible for the co-operativity with c-MYC (Fig. 3B). As CBP bound to the C-terminal half of c-MYC, we tested whether CBP could stimulate MYC proteins lacking the N-terminal TAD. MYCΔN147 and MYCΔN177 were unable to activate a reporter construct, but co-operated efficiently with CBP (Fig. 3C). Thus, the C-terminal domain of c-MYC is sufficient to mediate a functional interaction with the co-activator CBP.
Figure 3.
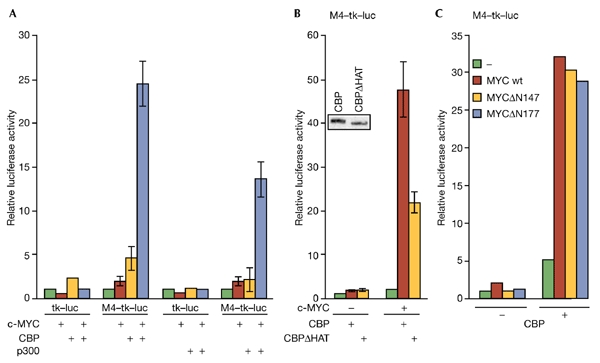
CBP stimulates c-MYC-dependent transactivation and is recruited to MYC-regulated promoters. (A) Reporter gene constructs with (M4–tk–luc; 2 μg) or without (tk–luc; 2 μg) four MYC/MAX binding sites (M4) were co-transfected with expression plasmids for c-MYC (1 μg), Gal4–CBP (5 μg) and Gal4–p300 (5 μg) into U2OS cells as indicated. (B) Transient transfections were performed as in (A). The inset shows the expression of Gal4–CBP and Gal4–CBPΔHAT, as determined by immunoblotting using the SC577 antibody. (C) Transient transfections were performed as in (A). In MYCΔN147 and MYCΔN177, the transactivation domain is deleted. CB,. cAMP-response-element-binding protein; Luc, luciferase; tk, thymidine kinase promoter; wt, wild type.
To test further the interaction of c-MYC with CBP and p300, we performed chromatin immunoprecipitations (ChIPs) and analysed the cyclin D2 promoter. We have previously demonstrated binding of c-MYC/MAX and MAD1/MAX in a differentiation-specific pattern, correlating with high and low levels of histone acetylation and promoter activity, respectively (Bouchard et al., 2001). This is consistent with the interpretation that c-MYC/MAX and MAD1/MAX complexes recruit HAT and HDAC activities, respectively, to the promoter. CBP bound to the cyclin D2 promoter in exponentially growing HL60 cells, but not in 12-O-tetra-decanoyl-phorbol 13-acetate (TPA)-treated cells (Fig. 4A). This correlated with a reduction in c-MYC binding, whereas MAD1 binding was induced. Low and constant signals were obtained with control antibodies against cytochrome c (Fig. 4A). In addition, CBP was localized to the promoter of the c-MYC-regulated ODC gene in HL60 cells (data not shown). Furthermore, p300 was crosslinked to the cyclin D2 promoter, but not to its 3′ untranslated region (UTR) in U937 cells (Fig. 4B). Proliferation of P493-6 B cells is dependent on exogenous c-MYC that is expressed from a tetracycline-regulated transgene (Schuhmacher et al., 1999). Induction of c-MYC results in an increase in its recruitment to the eIF2-α and ODC promoters (Fig. 4C). This is accompanied by an increase in both CBP and p300 binding. Together, our data show that there is recruitment of CBP/p300 to target genes by c-MYC/MAX complexes.
Figure 4.
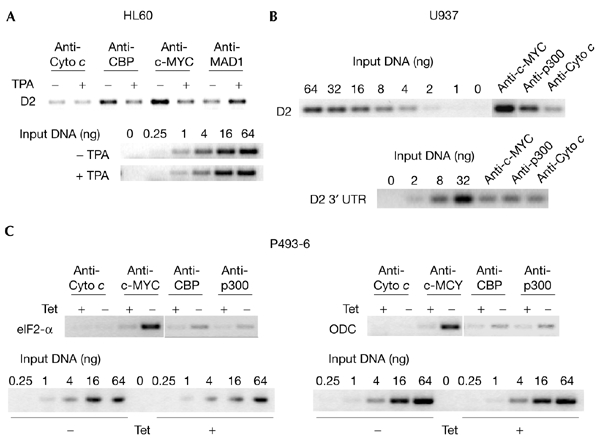
CBP and p300 are recruited to MYC target genes. (A) HL60 cells were grown exponentially (−) or induced to differentiate with 12-O-tetra-decanoyl-phorbol 13-acetate (TPA) (+) for 12 h. Chromatin immunoprecipitations were performed with antibodies recognizing the indicated proteins (c-MYC, N262; MAD1, C20; CBP, A22; Cyto c, SC7159). For PCR reactions, primers that amplify the region of the cyclin D2 promoter that contains the MYC/MAX binding site were used. For comparison, serial dilutions of input chromatin DNA were analysed. (B) Chromatin immunoprecipitations were performed on lysates of exponentially growing U937 cells. Specific antibodies were used as indicated (c-MYC, N262; p300, C22; Cyto c, SC7159). PCR reactions were carried out as in (A). (C) P493-6 cells were released from a tetracycline (Tet) block (−) or were mock treated (+). Cells were then crosslinked and processed. Immunoprecipitations and PCR reactions were carried out as in (A) and (B). CBP, cAMP-response-element-binding protein; D2, cyclin D2; ODC, ornithine decarboxylase; UTR, untranslated region.
Acetylation of c-MYC by CBP
CBP/p300 acetylate not only all four core histones, but also several transcriptional regulators, cofactors and general transcription factors (Chan & La Thangue, 2001; Sterner & Berger, 2000). Therefore, we tested whether c-MYC or MAX are substrates for CBP/p300. Recombinant CBP acetylated a maltose-binding protein (MBP)– c-MYC fusion protein, but did not acetylate either MBP or GST–MAX (Fig. 5A,B; and data not shown). Acetylation was detected by the incorporation of radioactive acetate or by immunoblotting using antibodies specific for acetylated lysine residues (anti-Ac–Lys). To determine whether acetylation also occurs in cells, FLAG-tagged MYC proteins were immunoprecipitated from transiently transfected COS7 cells, and the proteins were detected with anti-Ac–Lys (Fig. 5C). Acetylation of c-MYC, MYCΔMBI and MYCΔMBII was detected. This suggests that HAT activities associated with TRRAP are not responsible for MYC acetylation. Additional evidence for a role of CBP/p300 in c-MYC acetylation was obtained from co-expression experiments. Acetylation of c-MYC was increased when c-MYC was co-expressed with CBP, as compared with the control or with co-expressed CBPΔHAT (Fig. 5D). These results show that CBP is responsible for c-MYC acetylation in cells.
Figure 5.
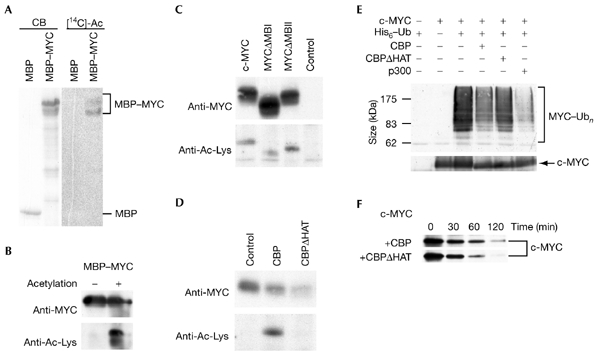
c-MYC is acetylated by CBP. (A) Maltose-binding protein (MBP)–MYC and MBP were incubated with His–CBP in the presence of [14C]acetyl-coenzyme A (acetyl-CoA). A Coomassie-blue-stained gel (CB) and the corresponding autoradiograph ([14C]Ac) are shown. (B) MBP–MYC was acetylated in the presence of His–CBP and acetyl-CoA or was mock-treated. Proteins were detected by western blotting using a MYC-specific antibody (9E10) or acetyl-lysine (Ac-Lys)-specific antibodies (upper and lower panels, respectively). (C) FLAG-tagged c-MYC and MYC mutants were expressed transiently in COS7 cells (10 μg of each construct), immunoprecipitated from F-buffer lysates using FLAG-specific antibodies and analysed by western blotting using monoclonal antibody 9E10 (upper panel) or Ac-Lys-specific antibodies (lower panel). (D) FLAG-tagged c-MYC was co-expressed in COS7 cells with Gal4–CBP, Gal4–CBPΔHAT or vector only. Immunoprecipitations and western blot analyses were performed as in (C). (E) The proteins indicated were expressed in human embryonic kidney (HEK) 293 cells and purified over Talon beads, and MYC proteins were identified using monoclonal antibody 9E10 (upper panel). Unmodified c-MYC proteins are shown in the lower panel. (F) The half-life of the c-MYC protein was analysed in transiently transfected HEK293 cells (transfected with 0.5 μg of pcDNA3–FLAG–MYC) after pulse–chase labelling using [35S]methionine. The chase times are indicated. CBP, cAMP-response-element-binding protein; HAT, histone acetyl-transferase; MBI and MBII, MYC boxes I and II; Ub, ubiquitin.
To address the functional relevance of c-MYC acetylation, we tested whether DNA binding is affected, as acetylation of several transcription factors, including c-MYB, GATA1 and E2F, has been shown to stimulate DNA binding (Chan & La Thangue, 2001). However, no effect was detectable in electrophoretic gel mobilityshift assays (data not shown). Another possibility is that c-MYC acetylation affects protein half-life. c-MYC is degraded in the ubiquitin–proteasome pathway (Lüscher, 2001). As ubiquitination and acetylation both occur at Lys residues, competition between the two types of modification is possible. Indeed, coexpression of c-MYC with CBP or p300, but not with CBPΔHAT, reduced the ubiquitination and turnover of c-MYC (Fig. 5E,F). Similarly, other transcription factors, including E2F1 and SMAD7, were shown to be stabilized on acetylation by CBP/p300 (Gronroos et al., 2002; Martinez-Balbas et al., 2000).
Functional relevance of CBP binding to c-MYC
The finding that CBP binds to the C-terminal domain of c-MYC was unanticipated. Previous mapping using Gal4 fusion proteins identified a TAD within the N-terminal one-third of c-MYC, whereas the C-terminal two-thirds of the protein showed no transactivating activity (Kato et al., 1990). However, several findings have indicated that the C-terminal region, in addition to its crucial function in interacting with MAX and binding to DNA, has other functions relevant to the regulation of gene transcription. The recruitment of the SWI/SNF complex has been suggested to stimulate c-MYC transcriptional activity (Cheng et al., 1999). The binding of other factors, including YY1, may interfere negatively with c-MYC transcriptional activity (Austen et al., 1998; Shrivastava et al., 1993). MIZ1 and SMAD2/SMAD3 are other factors that bind to the C terminus of MYC. Although the way in which these proteins affect MYC function has not been determined, both are repressed by c-MYC, suggesting that when MYC is bound to MIZ1 or SMAD2/SMAD3, the MYC TAD is not active (Feng et al., 2002; Staller et al., 2001). These findings suggest that both positive and negative functions are associated with the C-terminal domain of c-MYC. This may explain why, at first, no transactivating activity was found to be associated with the C-terminal half of c-MYC (Kato et al., 1990). Thus, the binding of CBP/p300 to the c-MYC C terminus provides an additional link from this region of c-MYC to the control of gene transcription.
In addition to GCN5 and TIP60, which preferentially acetylate core histones, we provide evidence for a third HAT activity that is recruited by c-MYC to promoters. Although CBP and p300 can modify core histones in vitro, it is not known whether this occurs in cells (Chan & La Thangue, 2001; Goodman & Smolik, 2000). Indeed, it has been argued that the CBP/p300 HAT activity is not involved in regulating chromatin by modifying histones (Agalioti et al., 2002; Li et al., 1999). Importantly, CBP/p300 acetylate other substrates, including transcription factors and components of the Pol II complex (Chan & La Thangue, 2001; Goodman & Smolik, 2000). Thus, whereas the primary targets of GCN5 and TIP60 are probably histones associated with c-MYC-regulated genes, our findings demonstrate that CBP/p300 affect c-MYC itself. Acetylation seems to be linked to ubiquitination, and may, in addition to altering c-MYC stability, influence its interaction with other factors, and thus modulate transcription. Furthermore, CBP/p300 may also function by contacting basal transcription factors, and thus perform a bridging function (Chan & La Thangue, 2001). In summary, the functions of CBP/p300 as c-MYC cofactors seem to be complex, and involve direct modification of c-MYC and, potentially, communication with the Pol II complex and/or chromatin remodelling.
Methods
Cell culture and transient transfection assays.
Culture and differentiation of cells and transient transfections have been described previously (Bouchard et al., 2001; Lüscher-Firzlaff et al., 1999). P493-6 cells (provided by D. Eick) were grown for 3 days in the presence of tetracycline to arrest the cells in G0/G1 phase. To induce c-MYC expression, tetracycline was washed out and the cells were cultured for 5 h before harvesting.
Plasmids and recombinant proteins.
pVR1012–Gal4, pVR1012– Gal4–CBP and pVR1012–Gal4–p300 were provided by N. Perkins; pSV–FLAG–VP16, pSV–FLAG–c-MYC–VP16, pMBP–Pre and pMBP– Pre-MYC were provided by H. Ariga; GST–CBP–HAT and pcDNA3–Gal4–CBPΔHAT were provided by T. Kouzarides; a recombinant baculovirus expressing His–CBP was provided by D. Thanos; pCMV–His6–ubiquitin was provided by D. Bohmann.
GST–CBP fragments (Janknecht & Nordheim, 1996), p(Gal4)4–mintk–luc (a fusion of Gal4, the minimal thymidine-kinase promoter and luciferase; Lüscher-Firzlaff et al., 1999), pEQ176P2– c-MYC, tk–luc, M4–tk–luc (a fusion of four MYC/MAX binding sites (M4) to tk and luc) and the β-galactosidase expression vector pEQ176 (Austen et al., 1998) have been described previously.
Interaction assays.
For in vivo interaction assays, whole-cell lysates were prepared in F buffer (10 mM Tris-HCl, pH 7.05, 50 mM NaCl, 30 mM Na4P2O7, 50 mM NaF, 5 μM ZnCl2, 100 μM Na3VO4, 1% Triton X-100, 1 mM phenylmethylsulphonyl fluoride, 5 units ml−1 α2-macroglobulin, 2.5 units ml−1 pepstatin A, 2.5 units ml−1 leupeptin, 0.15 mM benzamidin). Co-immunoprecipitated proteins were detected either by western blot analysis or by eluting the proteins in AB buffer (20 mM Tris–HCl, pH 7.5, 50 mM NaCl, 0.5% NP-40, 0.5% deoxycholate, 0.5% SDS, 0.5% aprotinin, 1 mM EDTA) and performing a second immunoprecipitation (Krippner-Heidenreich et al., 2001; Lüscher-Firzlaff et al., 1999).
The following antibodies were used for immunoprecipitations and/or western blot analyses (as indicated in the figure legends): monoclonal antibodies 6A10 and 9E10 (human c-MYC); monoclonal antibody 5C9 (human MAD1; Sommer et al., 1997); monoclonal antibody M73 (against E1A, provided by P. Whyte); anti-c-MYC N262; anti-MAX C17; anti-MAD1 C20; anti-CBP A22; anti-p300 C20; anti-cytochrome c SC7159; anti-Gal4 SC577 (Santa Cruz); anti-CBP AC238 (NeoMarkers); anti-acetyl-lysine (New England Biolabs); and anti-FLAG M2–agaroseslurry (Sigma).
Fusion proteins were prepared as described previously (Kitaura et al., 2000; Lüscher-Firzlaff et al., 1999). For binding reactions, 3 μg of GST fusion protein was bound to glutathione–agarose beads and incubated with [35S]methionine-labelled in vitro transcribed and translated proteins in binding buffer (Oelgeschlager et al., 1996).
Purification of His–CBP and acetyltransferase assays.
Purification of His–CBP was performed as described previously (Chen et al., 2001). Purified MBP or MBP–MYC (3 μg) was incubated in 30 ml of HAT buffer (50 mM Tris-HCl, pH 8.0, 2 mM EDTA, 1 mM dithiothreitol, 1 mM PMSF, 10 mM sodium butyrate) with 50 ng of purified His–CBP and 1 μl [14C]acetyl-coenzyme A (55 mCi mol−1; NEN) for 1 h at 30 °C. Alternatively, acetylation was detected by immunoblotting with an acetyl-lysinespecific antibody.
Chromatin crosslinking and immunoprecipitation (ChIP).
ChIP assays were performed as described previously (Bouchard et al., 2001; Strutt & Paro, 1999). The following PCR primers were used: hCycD23-UTRf, 5′-ATCAGACCCTATTCTCGGCTCA GG-3′ hCycD2-3-UTRr, 5′-CAGTCAGTAAGGCACTTTATTTCCCC-3′ hCycD2prom1, 5′-CCCCTTCCTCCTGGAGTGAAATAC-3′ hCycD2prom2, 5′-CGTGCTCTAACGCATCCTTGAGTC-3′ eIF2a-h1, 5′-TTCTCGGAGGACCCAGACTCTATG-3′ eIF2a-h2, 5′-TCA CAGAGACCAGACTTGCTTCCC; hODCprom-h1, 5′-GAGCAGAGC GCACCGGGATCA; and hODCprom-h2, 5′-CAGTACCTCGTGCC CGAGAGC.
Ubiquitination assays.
Ubiquitination assays were performed as described in Campanero & Flemington (1997). Transiently transfected HEK293 cells were treated with 25 mM MG132 for 2 h before lysis in 8 M urea, 0.1 M sodium phosphate, pH 8.0, 10 mM imidazole. His-tagged proteins were bound to Talon beads and eluted in SDS sample buffer containing 200 mM imidazole. Modified c-MYC proteins were detected using monoclonal antibody 9E10.
Acknowledgments
We thank H. Ariga, D. Bohmann, D. Eick, T. Kouzarides, N. Perkins and P. Whyte for reagents, B. Amati for communicating unpublished results, and H. Burkhardt for excellent technical assistance. Discussions with L.-G. Larsson are particularly appreciated. This work was supported by grants from the Deutsche Forschungsgemeinschaft and from the Fonds der Chemischen Industrie to B.L.
References
- Agalioti T., Chen G. & Thanos D. (2002) Deciphering the transcriptional histone acetylation code for a human gene. Cell, 111, 381–392. [DOI] [PubMed] [Google Scholar]
- Amati B., Frank S.R., Donjerkovic D. & Taubert S. (2001) Function of the c-Myc oncoprotein in chromatin remodeling and transcription. Biochim. Biophys. Acta, 1471, M135–M145. [DOI] [PubMed] [Google Scholar]
- Austen M., Cerni C., Lüscher-Firzlaff J.M. & Lüscher B. (1998) YY1 can inhibit c-Myc function through a mechanism requiring DNA binding of YY1 but neither its transactivation domain nor direct interaction with c-Myc. Oncogene, 17, 511–520. [DOI] [PubMed] [Google Scholar]
- Bouchard C., Dittrich O., Kiermaier A., Dohmann K., Menkel A., Eilers M. & Lüscher B. (2001) Regulation of cyclin D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP recruitment and histone acetylation at the cyclin D2 promoter. Genes Dev., 15, 2042–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanero M.R. & Flemington E.K. (1997) Regulation of E2F through ubiquitin-proteasome-dependent degradation: stabilization by the pRB tumor suppressor protein. Proc. Natl Acad. Sci. USA, 94, 2221–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan H.M. & La Thangue N.B. (2001) p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci., 114, 2363–2373. [DOI] [PubMed] [Google Scholar]
- Chen C.J., Deng Z., Kim A.Y., Blobel G.A. & Lieberman P.M. (2001) Stimulation of CREB binding protein nucleosomal histone acetyltransferase activity by a class of transcriptional activators. Mol. Cell. Biol., 21, 476–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S.W., Davies K.P., Yung E., Beltran R.J., Yu J. & Kalpana G.V. (1999) c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nature Genet., 22, 102–105. [DOI] [PubMed] [Google Scholar]
- Feng X.H., Liang Y.Y., Liang M., Zhai W. & Lin X. (2002) Direct interaction of c-Myc with Smad2 and Smad3 to inhibit TGF-β-mediated induction of the CDK inhibitor p15(Ink4B). Mol. Cell, 9, 133–143. [DOI] [PubMed] [Google Scholar]
- Frank S.R., Schroeder M., Fernandez P., Taubert S. & Amati B. (2001) Binding of c-Myc to its target sites in chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev., 15, 2069–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman R.H. & Smolik S. (2000) CBP/p300 in cell growth, transformation and development. Genes Dev., 14, 1553–1577. [PubMed] [Google Scholar]
- Gronroos E., Hellman U., Heldin C.H. & Ericsson J. (2002) Control of Smad7 stability by competition between acetylation and ubiquitination. Mol. Cell, 10, 483–493. [DOI] [PubMed] [Google Scholar]
- Janknecht R. & Nordheim A. (1996) Regulation of the c-fos promoter by the ternary complex factor Sap-1a and its coactivator CBP. Oncogene, 12, 1961–1969. [PubMed] [Google Scholar]
- Jenuwein T. & Allis C.D. (2001) Translating the histone code. Science, 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- Kato G.J., Barrett J., Villa-Garcia M. & Dang C.V. (1990) An amino-terminal c-Myc domain required for neoplastic transformation activates transcription. Mol. Cell. Biol., 10, 5914–5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitaura H., Shinshi M., Uchikoshi Y., Ono T., Iguchi-Ariga S.M. & Ariga H. (2000) Reciprocal regulation via protein–protein interaction between c-Myc and p21cip1/waf1/sdi1 in DNA replication and transcription. J. Biol. Chem., 275, 10477–10483. [DOI] [PubMed] [Google Scholar]
- Krippner-Heidenreich A., Talanian R.V., Sekul R., Kraft R., Thole H., Ottleben H. & Lüscher B. (2001) Targeting of the transcription factor Max during apoptosis: phosphorylation-regulated cleavage by caspase-5 at an unusual glutamic acid in position P1. Biochem. J., 358, 705–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Imhof A., Collingwood T.N., Urnov F.D. & Wolffe A.P. (1999) p300 stimulates transcription instigated by ligand-bound thyroid hormone receptor at a step subsequent to chromatin disruption. EMBO J., 18, 5634–5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüscher B. (2001) Function and regulation of the transcription factors of the Myc/Max/Mad network. Gene, 277, 1–14. [DOI] [PubMed] [Google Scholar]
- Lüscher-Firzlaff J.M., Westendorf J.M., Zwicker J., Burkhardt H., Henriksson M., Muller R., Pirollet F. & Lüscher B. (1999) Interaction of the fork head domain transcription factor MPP2 with the human papilloma virus 16 E7 protein: enhancement of transformation and transactivation. Oncogene, 18, 5620–5630. [DOI] [PubMed] [Google Scholar]
- Martinez-Balbas M.A., Bauer U.M., Nielsen S.J., Brehm A. & Kouzarides T. (2000) Regulation of E2F1 activity by acetylation. EMBO J., 19, 662–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon S.B., Van Buskirk H.A., Dugan K.A., Copeland T.D. & Cole M.D. (1998) The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell, 94, 363–374. [DOI] [PubMed] [Google Scholar]
- McMahon S.B., Wood M.A. & Cole M.D. (2000) The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol. Cell. Biol., 20, 556–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narlikar G.J., Fan H.Y. & Kingston R.E. (2002) Cooperation between complexes that regulate chromatin structure and transcription. Cell, 108, 475–487. [DOI] [PubMed] [Google Scholar]
- Nikiforov M.A., Chandriani S., Park J., Kotenko I., Matheos D., Johnsson A., McMahon S.B. & Cole M.D. (2002) TRRAP-dependent and TRRAP-independent transcriptional activation by Myc family oncoproteins. Mol. Cell. Biol., 22, 5054–5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelgeschlager M., Janknecht R., Krieg J., Schreek S. & Lüscher B. (1996) Interaction of the co-activator CBP with Myb proteins: effects on Myb- specific transactivation and on the cooperativity with NF-M. EMBO J., 15, 2771–2780. [PMC free article] [PubMed] [Google Scholar]
- Oster S.K., Ho C.S., Soucie E.L. & Penn L.Z. (2002) The myc oncogene: MarvelouslY Complex. Adv. Cancer Res., 84, 81–154. [DOI] [PubMed] [Google Scholar]
- Park J., Kunjibettu S., McMahon S.B. & Cole M.D. (2001) The ATM-related domain of TRRAP is required for histone acetyltransferase recruitment and Myc-dependent oncogenesis. Genes Dev., 15, 1619–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuhmacher M., Staege M.S., Pajic A., Polack A., Weidle U.H., Bornkamm G.W., Eick D. & Kohlhuber F. (1999) Control of cell growth by c-Myc in the absence of cell division. Curr. Biol., 9, 1255–1258. [DOI] [PubMed] [Google Scholar]
- Shrivastava A., Saleque S., Kalpana G.V., Artandi S., Goff S.P. & Calame K. (1993) Inhibition of transcriptional regulator Yin-Yang-1 by association with c-Myc. Science, 262, 1889–1892. [DOI] [PubMed] [Google Scholar]
- Sommer A., Hilfenhaus S., Menkel A., Kremmer E., Seiser C., Loidl P. & Lüscher B. (1997) Cell growth inhibition by the Mad/Max complex through recruitment of histone deacetylase activity. Curr. Biol., 7, 357–365. [DOI] [PubMed] [Google Scholar]
- Staller P. et al. (2001) Repression of p15INK4b expression by Myc through association with Miz-1. Nature Cell Biol., 3, 392–399. [DOI] [PubMed] [Google Scholar]
- Sterner D.E. & Berger S.L. (2000) Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev., 64, 435–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutt H. & Paro R. (1999) Mapping DNA target sites of chromatin proteins in vivo by formaldehyde crosslinking. Methods Mol. Biol., 119, 455–467. [DOI] [PubMed] [Google Scholar]
- Sudarsanam P. & Winston F. (2000) The Swi/Snf family nucleosome-remodeling complexes and transcriptional control. Trends Genet., 16, 345–351. [DOI] [PubMed] [Google Scholar]


