Abstract
The modification of cellular proteins by ubiquitin (Ub) is an important event that underlies protein stability and function in eukaryotes. Protein ubiquitylation is a dynamic and reversible process; attached Ub can be removed by deubiquitylating enzymes (DUBs), a heterogeneous group of cysteine proteases that cleave proteins precisely at the Ub–protein bond. Two families of DUBs have been identified previously. Here, we describe new, highly specific Ub iso-peptidases, that have no sequence homology to known DUBs, but which belong to the OTU (ovarian tumour) superfamily of proteins. Two novel proteins were isolated from HeLa cells by affinity purification using the DUB-specific inhibitor, Ub aldehyde (Ubal). We have named these proteins otubain 1 and otubain 2, for OTU-domain Ubal-binding protein. Functional analysis of otubains shows that the OTU domain contains an active cysteine protease site.
Results and Discussion
Protein ubiquitylation is regulated by a complex interplay between the E2 ubiquitin (Ub)-conjugating enzymes, the E3 Ub–protein ligases and the deubiquitylating enzymes (DUBs). Substrate specificity is conferred by the E3s, and the reversibility of ubiquitylation is provided by the DUBs. The two families of deubiquitylating enzymes (DUBs) include Ub carboxy-terminal hydrolases (UCHs) and Ub-specific proteases (USPs/UBPs; D'Andrea & Pellman, 1998; Wilkinson, 1997). Since their discovery more than ten years ago, DUBs have emerged as key players in a variety of cellular processes, ranging from the control of protein degradation to tissue development. The most recent examples include the regulation of Drosophila eye development through the deubiquitylation of Liquid facets by Fat facets, and the regulation of p53 degradation by herpesvirus-associated ubiquitinspecific protease (USP7/HAUSP; Chen et al., 2002; M. Li et al., 2002). Both families of DUBs are cysteine proteases, which can be inhibited by Ub aldehyde (Ubal; Wilkinson, 1997). Recently, a new Ubal-insensitive deubiquitylating activity of the proteasome has been investigated, and has been ascribed to POH1 (also known as Rpn11, in yeast) subunit of the 19S proteasome complex (Yao & Cohen, 2002; Verma et al., 2002; Maytal-Kivity et al., 2002).
We have used Ubal to isolate DUBs from HeLa cells (Balakirev et al., 2002). We synthesized Ubal that was labelled with biotin (biot) on the ε-amino group of lysine residues (biot–Ubal). The cell extract (from 1 × 109cells) was incubated with biot–Ubal (or with biot–Ub as a control), and was loaded onto a streptavidin–agarose column. The column was rinsed extensively with 1.5 M NaCl and a Ub-containing buffer to remove non-covalently bound proteins. The proteins that were covalently bound to Ubal by thio-hemiacetal bonds were than eluted with dithiothreitol (DTT)-containing urea buffer and 1 × Laemmli buffer and were analysed by SDS–polyacrylamide gel electrophoresis (SDS–PAGE; Fig. 1A). The main protein bands were sequenced by mass spectrometry, and were found to contain the human Ub hydrolase UCH-L3 and two novel proteins that corresponded to previously uncharacterized complementary DNA clones (Fig. 1A; see also p36 and p34 in figure 2A, in Balakirev et al., 2002). The predicted proteins, with relative molecular masses of 31 kDa and 27 kDa, show no obvious similarity to any known DUB, but are similar to each other, and both have OTU domains. The OTU superfamily comprises a group of putative cysteine proteases that are homologous to the ovarian tumor gene product of Drosophila (Makarova et al., 2000). The ∼100 identified OTU family members include proteins from eukaryotes, viruses and pathogenic bacteria, and none of them have a known biochemical function. We have named our proteins otubain 1 (31 kDa) and otubain 2 (27 kDa), for OTU-domain Ubal-binding protein. By searching databases, we identified several sequences that are closely related to otubains. These proteins contain the conserved cysteine, histidine and aspartate residues that define the putative catalytic triad of cysteine proteases (Fig. 1B; see Makarova et al., 2000). Sequence analysis revealed signatures that resembled Ub-interaction motifs (UIMs; Hofmann & Falquet, 2001) and Ub-associated domains (UBAs; Hofmann & Bucher, 1996), which are commonly found in proteins of the Ub pathway (Fig. 1B). In addition, the proteins have putative nuclear localization signals (NLSs) and a consensus LxxLL motif (where x is any amino acid), which is known to mediate the binding of transcriptional co-activators to liganded nuclear receptors (Heery et al., 1997).
Figure 1.
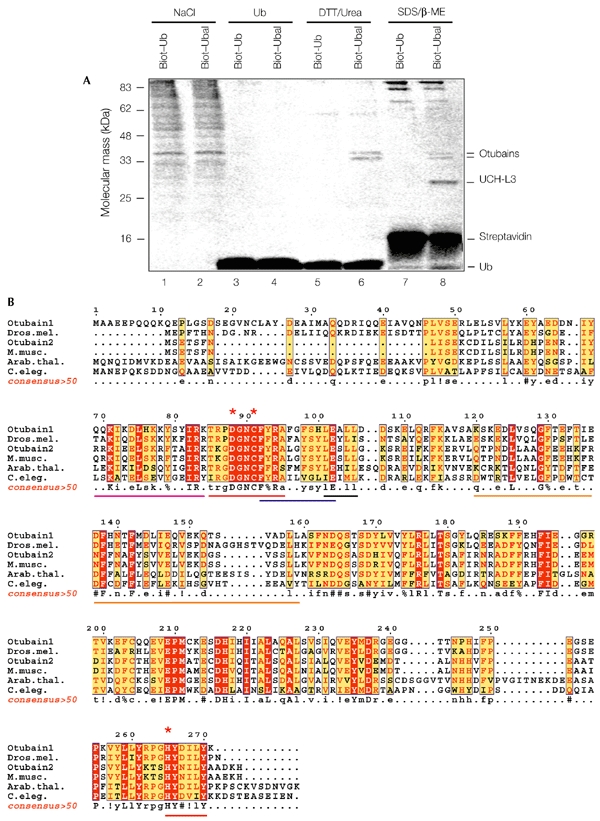
Identification of novel ubiquitin-aldehyde-binding proteins. (A) Ubiqutin aldehyde (Ubal)-affinity purification of otubains. Cell lysates were incubated with biotin–ubiquitin (biot–Ub) or biot–Ubal (20 ng mg−1 of protein; for 1 h at 4 °C), and loaded onto streptavidin–agarose columns. After extensive washing, the bound proteins were eluted with 1.5 M NaCl in suspension buffer (SB; lanes 1 and 2), 1 mg ml−1 Ub in SB (lanes 3 and 4), 100 mM dithiothreitol (DTT) and 4 M urea in SB (lanes 5 and 6), or 1 × Laemmli buffer (lanes 7 and 8). The eluted proteins were resolved by SDS–polyacrylamide gel electrophoresis and stained with Coomassie blue. The proteins identified by mass spectrometry are indicated. (B) Clustal W sequence alignment of the otubain-family proteins, edited with ESPript software (Gouet et al., 1999). The predicted proteins are from humans (GenBank accession numbers: AK000120 for otubain 1; AK025569 for otubain 2), Drosophila melanogaster (Dros. mel.; AY061382), Mus musculus (M. musc.; AK019830), Arabidopsis thaliana (Arab. thal.; AY084389) and Caenorhabditis elegans (C. eleg.; Z81039). The red asterisks indicate the putative catalytic triad of the cysteine protease, and the lines below the sequences mark the OTU (ovarian tumour) domain (red), putative nuclear localization signal (magenta), Ub interaction motif (UIM)-like motif Φ-xx-A-xxxs-xx-Ac (where Φ indicates an aromatic amino acid, x indicates any amino acid and Ac indicates an acidic amino acid; blue), Ub-associated (UBA)-like domain (orange) and the LxxLL motif (black). β-ME, β-mercaptoethanol. A consensus >50 sequence (a sequence showing residues that are more than 50% conserved) was generated using ESPript software. Uppercase letters in the consensus>50 sequence and red shading in the other sequences indicate identity; lowercase letters in the consensus >50 sequence and yellow shading in the other sequences indicate a consensus level of >50%; ! indicates I or V; % indicates F or Y; # indicates N, D, Q or E.
Figure 2.
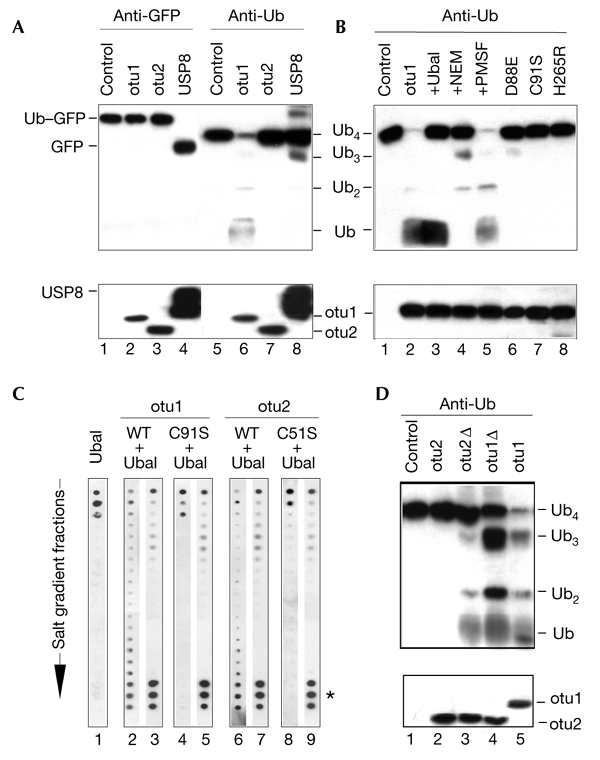
Otubains are deubiquitylating enzymes. (A) Proteolysis of ubiquitin–green fluorescent protein (Ub–GFP; lanes 1–4) and tetra-Ub (lanes 5–8) by otubains (otu1 and otu2) and ubiquitin-specific protease 8 (USP8). Assays were carried out at 37 °C for 30 min, in 20 μl of assay buffer containing the protein substrate (0.1–1.0 μM) and a protease (10–100 nM). Substrate cleavage was analysed by western blotting with anti-GFP (lanes 1–4) and anti-Ub (lanes 5–8) antibodies. The lower blot shows the proteases in each reaction probed with anti-His6 (for the otubains) and anti-glutathione-S-transferase (anti-GST; for GST–USP8) antibodies. (B) Protease inhibition profile of otubain 1. Effect of different inhibitors (lanes 1–5) and OTU (ovarian tumour)-domain mutations (lanes 6–8) on the processing of tetra-Ub by otubain 1. Reaction conditions were the same as in Fig. 2A, except that 0.5 μM Ub aldehyde (Ubal; lane 3), 0.5 μM NEM (N-ethylmaleimide; lane 4), or 1 mM PMSF (phenylmethylsulphonyl fluoride; lane 5) were added, as indicated. (C) Otubain–Ubal interaction. Wild-type otubains (otu1 and otu2: WT) and their activesite mutants (otu1 and otu2: C91/51S) were incubated with Ubal, and the reaction mixtures were separated by fast performance liquid chromatography, using an anion-exchange column. The fractions were eluted over a salt gradient and dot-blotted and probed in parallel, using the anti-Ub antibody (lanes 1, 2, 4, 6 and 8) and affinity-purified anti-otubain antibodies (lanes 3, 5, 7 and 9). Ubal alone was eluted at the beginning of the gradient (lane 1). The formation of otubain–Ubal complexes was seen by co-purification of Ubal with otubains (asterisk, lanes 2, 3, 6 and 7). Complex formation did not change the elution profile of the otubains (not shown). The active-site mutants of otubains did not interact with Ubal (lanes 4, 5, 8 and 9). (D) Effect of otubain truncations on tetra-Ub proteolysis. Reaction conditions were the same as in Fig. 2A. The lower panel shows the otubains blotted with the anti-His6 antibody. Otubain 1Δ (otu1Δ) shows slightly higher SDS–polyacrylamide gel electrophoreis mobility than otu 2 and otubain 2Δ1–229 (otu2Δ).
As otubains have an affinity for DUB-specific inhibitors, we tested whether they are deubiquitylating enzymes. The proteins were cloned by RT–PCR (polymerase chain reaction with reverse transcription), expressed in Escherichia coli and purified by nickel-affinity chromatography, and their primary structures were confirmed by MALDI–TOF (matrix-assisted laser desorption/ionization time-of-flight) mass spectrometry. Consistent with our observations about cellular otubains, the recombinant proteins showed retarded migration in SDS–PAGE experiments (36 kDa for otubain 1; 29 kDa for otubain 2), presumably due to their acidic nature (pIs 4.59 and 5.67, respectively). Both proteins have a tendency to oligomerize, as was shown by native gel-electrophoresis and analytical centrifugation (data not shown). The enzymatic properties of otubains were examined by using common DUB substrates. Unlike USP8 (a Ub-specific protease, which was used as a control), otubains did not cleave the peptide substrate LRGG-7-amido-4-methylcoumarin or the Ub fusion substrate Ub–GFP (green fluorescent protein). However, otubain 1 efficiently processed tetra-Ub (Fig. 2A). Mass spectrometry analysis of the products of proteolysis showed that cleavage occurred at the iso-peptide bond. Because otubain 1 did not cleave a Ub–peptide bond, we suggest that this is a highly specific Ub iso-peptidase. The protease inhibition profile suggests that otubains are cysteine proteases: the proteolysis of tetra-Ub by otubain 1 was inhibited by Ubal and by the thiol-blocking agent NEM (N-ethylmaleimide), whereas an inhibitor of serine proteases, PMSF (phenylmethylsulphonyl fluoride), had no effect (Fig. 2B). In addition, the metalloprotease inhibitors EDTA and 1,10-phenantroline did not inhibit otubain 1 (data not shown). The mutation of any of the putative catalytic residues in the OTU domain abolished the activity of otubain 1 (Fig. 2B), whereas the mutation of any of the other three cysteines in the sequence did not significantly affect proteolysis (data not shown). This is the first direct evidence that the OTU domain contains an active cysteine protease site.
No substrate cleavage was observed with otubain 2, despite its high level of sequence identity to otubain 1 (Fig. 2A). This was unexpected because, like otubain 1, otubain 2 formed a complex with Ubal (Fig. 2C). Moreover, Ubal binding to otubain 2 was inhibited by mutation of the activesite cysteine (C51S), suggesting that otubain 2 is a functional cysteine protease (Fig. 2C). The main differences between the two otubains include the long amino-terminal extension of otubain 1 and the short carboxy-terminal extension of otubain 2. The removal of five C-terminal amino acids downstream of the conserved OTU domain of otubain 2 produces the enzymatically active Ub iso-peptidase otubain 2Δ1–229 (Fig. 2D). This suggests that the C-terminal extension of otubain 2 inhibits its proteolytic activity, and may be involved in the regulation of otubain 2 in the cell. Truncation of otubain 1 at leucine 47 (otubain 1Δ) did not significantly inhibit its activity, suggesting that the N-terminal extension does not determine the specificity of otubain 1 for tetra-Ub (Fig. 2D).
Recently, Ploegh and co-workers have also identified an OTU-domain protein that corresponds to otubain 1 using a proteomics approach, with a panel of reactive Ub derivatives (Borodovsky et al., 2002). Although these authors did not perform functional characterizations, they showed that, of all the DUB inhibitors tested, only haemagglutinin (HA)–Ub1–75–bromoethylamide interacted with otubain 1. Similar to Ubal, which was used to isolate the otubains (Balakirev et al., 2002; and this work), this derivative has no extension in the putative P′ position of the DUB cleavage site. Because, in our assays, otubains cleave only an iso-peptide bond, these data suggest that otubains have a more rigid selectivity in the P′ position compared with other DUBs, and, probably, a different mechanism of substrate recognition (Johnston et al., 1999; Hu et al., 2002).
The otubains are encoded by two genes that are located at chromosomal positions 11q13.1 (otubain 1) and 14q32.12 (otubain 2; Ensembl genome database, Hubbard et al., 2002). RT–PCR analysis and western blotting, followed by detection with affinity-purified antibodies, showed a widespread expression of otubains in human tissues (Fig. 3A,B). We identified two forms of the otubain 1 transcript in leukocytes, and the highest levels of otubain 2 expression were seen in brain tissue. The overexpression of wild-type otubains and their activesite mutants in HeLa cells did not change the levels of intracellular Ub–protein conjugates; only a small decrease in the levels of conjugates was seen in otubain 1-transfected cells (Fig. 3C). Moreover, the knockout of otubains using siRNAs (small interfering RNAs; not shown) or antisense cDNAs had no effect on the pattern of ubiquitylated proteins in the cells (Fig. 3C). It seems unlikely, therefore, that otubains are involved in the general protein degradation machinery; but, instead, they may function in specific Ub-dependent pathways.
Figure 3.
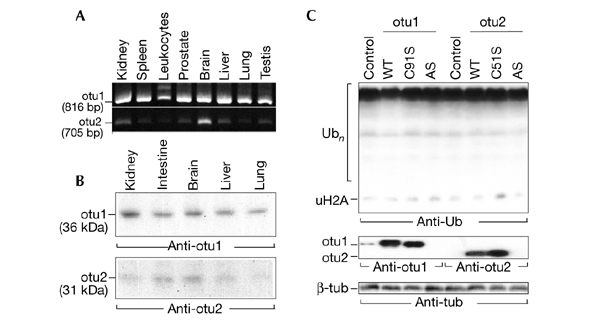
Expression of otubains in tissues and cultured cells. (A) RT–PCR (polymerase chain reaction with reverse transcription) analysis of otubains, performed on a panel of complementary DNAs from various human tissues (OriGene). Note the two forms of the otubain 1 (otu1) transcript from leukocytes, and that the highest level of otubain 2 (otu2) expression was in brain tissue. (B) Western blot analysis of otubains in human tissues (Protein Medley; Clontech) using affinity-purified anti-otubain antibodies. (C) Transfection of HeLa cells with wild-type otubains (otu1 and otu2: WT), their active-site mutants (otu1 and otu2: C91/51S), and antisense cDNAs (otu1 and otu2: AS) on intracellular ubiquitin (Ub)–protein conjugates. Note the slight decrease in Ub-conjugates in otubain 1-transfected cells (otu1, WT lane) and the disappearance of endogenous otubain 1 induced by antisense cDNA (otu1; AS; middle blot). Tub, tubulin; uH2A, monoubiquitylated histone 2A.
Some insights into these pathways might come from the observation that another human member of the OTU superfamily, the anti-inflammatory protein A20, inhibits tumour necrosis factor (TNF) signalling by an unknown mechanism that involves an interaction with the TNF-receptor-associated factors, TRAF2 and TRAF6 (Beyaert et al., 2000). Remarkably, signalling through TRAF2 is modulated by ubiquitylation and proteasome-mediated degradation of TRAF2, whereas signalling through TRAF6 requires its labelling with the non-canonical K63-linked polyubiquitin chain (X. Li et al., 2002; Deng et al., 2000; Wang et al., 2001). Because of the similarity of the activesite organization of A20 protein and otubains, which are Ub iso-peptidases (Fig. 4A), it is possible that A20 might inhibit TNF signalling by disassembling the polyubiquitin chain on TRAFs. It is relevant in this context that, first, the TRAF-binding domain (TRAFB) of A20 homologues exactly matches the OTU domain of otubains (Fig. 4A); and, second, in one of the A20 homologues, the activesite Asp is replaced by Ala (Fig. 4A); although this TRAFB protein (TRABID) is still able to interact with TRAF6, it functions as a dominant-negative mutant by stimulating TNF signalling (Evans et al., 2001).
Figure 4.
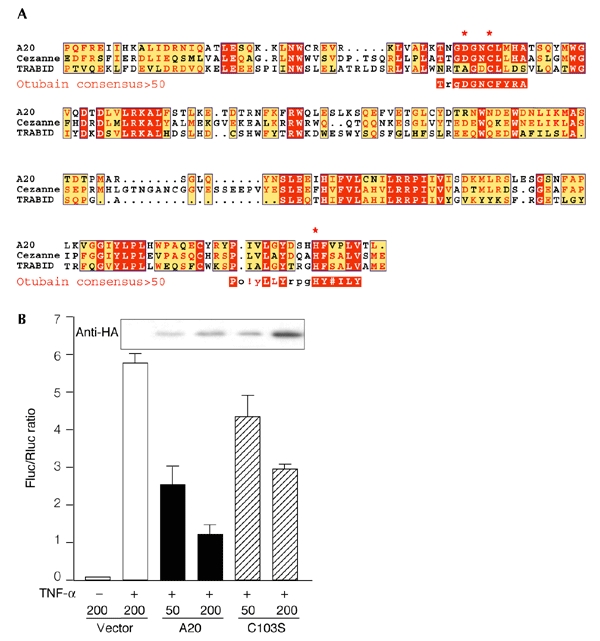
Role of the otubain-like domain of the A20 protein in the regulation of tumour necrosis factor signalling. (A) The TRAF (TNF-receptor-associated factor)-binding domain of A20 homologues (cellular zinc-finger anti-nuclear factor-κB (NF-κB) protein (Cezanne) and TRAF-binding-domain protein (TRABID); Evans et al., 2001) is shown, aligned with the otubain activesite (represented as a consensus>50 sequence (a sequence showing residues that are more than 50% conserved) as shown in Fig. 1B). The activesite Asp in the TRABID protein is replaced by an Ala residue. Uppercase letters in the consensus>50 sequences and red shading in the other sequences indicate identity; lowercase letters in the consensus >50 sequences and yellow shading indicate a consensus level of >50%; # indicates N, D, Q or E; red asterisks indicate the putative catalytic triad of the cysteine protease. (B) Effect of mutations in active-site cysteine residues on the NF-κB-inhibitory function of the A20 protein. A reporter-gene assay was performed using the firefly luciferase (Fluc) construct κB3–luc and the Renilla luciferase (Rluc) construct pRL–TK. HeLa cells were co-transfected with 100 ng of κB3–luc, 100 ng of pRL-TK, and the indicated amounts (in nanograms) of the haemagglutinin (HA)–A20 and HA–A20(C103S) constructs. The total amount of transfected DNA was kept constant at 400 ng by adding pHM6 empty vector. NF-κB was induced 18 h after transfection by stimulation with tumour necrosis factor (TNF)-α for 6 h, and its activity is represented as normalized Fluc/Rluc activity. The result shown is an average of two experiments. The blot above the graph shows the expression of A20 proteins in transfected cells as detected by the anti-HA antibody.
To test the role of the OTU domain in A20 protein function, we made the HA–A20(C103S) mutant, which has a point mutation of the active-site Cys. Reporter-gene experiments using the firefly luciferase gene, under the control of three tandem repeats of nuclear factor-κB (NF-κB) binding sites (κB3–luc), were then performed to measure the inhibitory effect of A20 and the A20(C103S) mutant on TNF-α-induced NF-κB activation. Dose-dependent inhibition of NF-κB activity in TNF-α-stimulated cells was seen with both constructs (Fig. 4B). However, a comparison of the efficiency of A20 versus A20(C103S) in regulating NF-κB showed that the mutant was about half as potent. Western blot analysis, performed in parallel, showed that similar amounts of the proteins were expressed in transfected cells, suggesting that this difference could not be attributed to different expression levels, but, instead, reflects a genuine difference in protein activity. It is likely that a putative deubiquitylating activity of A20 contributes to the inhibition of TNF signalling, but is not required. These data are in agreement with the observation that the C-terminal part of the A20 protein, which contains only zinc-finger motifs, also has an inhibitory effect on NF-κB activation (Song et al., 1996). Our continuing studies aim to characterize the proteolytic activity of A20 to define its role in this signalling mechanism. So far, we have found that, like otubains, the recombinant TRAFB domain of the A20 protein interacts with Ubal, although our attempts to obtain substrate cleavage with this protein have, so far, been unsuccessful.
In conclusion, we have described novel deubiquitylating enzymes that belong to the OTU superfamily of proteins. This is the first example of functional cysteine proteases from this family. Our preliminary results suggest that these proteases may be involved in specific signalling pathways.
Methods
Otubain identification.
Synthesis of biotin-labelled Ubal, Ubal affinity-isolation of otubains from extracts of Hela cells, and mass-spectrometry sequencing were performed as described in Balakirev et al. (2002). The suspension buffer for affinity chromatography contained 20 mM HEPES–K+, pH 7.5, 10 mM KCl, 5 mM MgCl2, 0.5 mM DTT, 0.5 mM EDTA, 0.1 mM phenylmethylsulphonyl fluoride, 0.5% NP-40.
Plasmid construction.
Otubain sequences were amplified from total messenger RNAs isolated from HeLa cells, and were cloned into the pET20b and pcDNA3.1–TOPO vectors. The primers used were 5′-ATACATATGCACCATCACCATCATCACATGGCGGCGAGGAACCTCAG-3′ and 5′-ATAAAGCTTTAATTTGTAGAGGATATCGTAGTGTCCAGG-3′ for otubain 1 and 5′-ATACATATGCACCATCACCATCATCACATGAGTGAAACATCTTTCAACC-3′ and 5′-ATAAAGCTTTTAATGTTTATCGGCTGCATAAAGG-3′ for otubain 2 (restriction sites are underlined). The pcDNA3.1 clones containing the otubain genes in the reverse orientation were used for the production of antisense cDNAs in transfected cells. The Ub–GFP cDNA was a gift from N.P. Dantuma. The pHA-A20 expression vector was provided by P.C. Evans. The mutants were produced by oligonucleotide-directed mutagenesis.
Protein expression and characterization.
The proteins were expressed in E. coli Bl21lysS, purified by Ni-affinity chromatography, and their primary structures were confirmed by MALDI–TOF mass spectrometry. The experiments carried out to analyse the proteolysis of different substrates by otubains, and the interaction with Ubal, were performed in assay buffer (150 mM NaCl, 0.5 mM DTT, 20 mM Tris, pH 8), as described in Balakirev et al., (2002). The antibodies used for western blotting were an anti-GFP antibody (ClonTech), anti-Ub monoclonal antibody P4D1 (Santa Cruz) and anti-glutathione-S-transferase (GST) IgG1 monoclonal antibody B-14 (Santa Cruz), which was used to detect the GST–USP8 construct (a gift from C.F. Draetta). Rabbit anti-otubain polyclonal antisera were raised against recombinant otubain 1 and otubain 2, and were affinity purified on Affigel-15 gels coupled to the corresponding otubain. For the analysis of otubain–Ubal complex formation, 20 μg of otubain were incubated with 10 μg of Ubal in 300 μl of 50 mM Hepes, pH 7.5. This was run through an anion-exchange column (NaCl gradient, 10 mM–1 M; 2 ml of Source Q (Pharmacia)). The fractions (1 ml) were collected, and were analysed by dot blotting with anti-Ub and anti-otubain antibodies.
Cell transfection and analysis.
Transfections of HeLa cells and western blot analyses were performed as described in Balakirev et al. (2002). The antibodies used for western blotting were anti-otubain polyclonal antisera, anti-Ub P4D1, anti-HA monoclonal antibody 12CA5 (Roche) and anti-β-tubulin monoclonal antibody TUB 2.1 (Sigma). For reporter gene experiments, cells were co-transfected with A20 constructs (or empty vector), the firefly luciferase gene construct under the control of three tandem repeats of NF-κB binding sites (κB3–luc plasmid; a gift from Z.J. Chen) and pRL-TK (encoding Renilla luciferase; Promega), to normalize transfection efficiency. The total amount of transfected DNA was kept constant at 400 ng by adding the pHM6 empty vector. Cells were grown overnight, stimulated with TNF-α (20 ng ml−1) for 6 h, and luciferase activities were measured using appropriate substrates (24–30 h after transfection).
Acknowledgments
This work was partly supported by a grant from L'Association pour la Recherche sur le Cancer to M.Y.B.
References
- Balakirev M., Jaquinod M., Haas A. & Chroboczek J. (2002) Deubiquitinating function of adenovirus proteinase. J. Virol., 76, 6323–6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyaert R., Heyninck K. & Van Huffel S. (2000) A20 and A20-binding proteins as cellular inhibitors of nuclear factor-κB-dependent gene expression and apoptosis. Biochem. Pharmacol., 60, 1143–1151. [DOI] [PubMed] [Google Scholar]
- Borodovsky A., Ovaa H., Kolli N., Gan-Erdene T., Wilkinson K.D., Ploegh H.L. & Kessler B.M. (2002) Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem. Biol., 9, 1149–1159. [DOI] [PubMed] [Google Scholar]
- Chen X., Zhang B. & Fischer J.A. (2002) A specific protein substrate for a deubiquitinating enzyme: Liquid facets is the substrate of Fat facets. Genes Dev., 16, 289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Andrea A. & Pellman D. (1998) Deubiquitinating enzymes: a new class of biological regulators. Crit. Rev. Biochem. Mol. Biol., 33, 337–352. [DOI] [PubMed] [Google Scholar]
- Deng L., Wang C., Spencer E., Yang L., Braun A., You J., Slaughter C., Pickart C. & Chen Z.J. (2000) Activation of the IκB kinase complex by TRAF6 requires a dimeric Ub-conjugating enzyme complex and a unique polyubiquitin chain. Cell, 103, 351–361. [DOI] [PubMed] [Google Scholar]
- Evans P.C., Taylor E.R., Coadwell J., Heyninck K., Beyaert R. & Kilshaw P.J. (2001) Isolation and characterization of two novel A20-like proteins. Biochem. J., 357, 617–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouet P., Courcelle B., Stuart D.I. & Metoz F. (1999) ESPript: multiple sequence alignments in PostScript. Bioinformatics, 15, 305–308. [DOI] [PubMed] [Google Scholar]
- Heery D.M., Kalkhoven E., Hoare S. & Parker M.G. (1997) A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature, 387, 733–736. [DOI] [PubMed] [Google Scholar]
- Hofmann K. & Bucher P. (1996) The UBA domain: a sequence motif present in multiple enzyme classes of the ubiquitination pathway. Trends Biochem. Sci., 21, 172–173. [PubMed] [Google Scholar]
- Hofmann K. & Falquet L. (2001) A Ub-interacting motif conserved in components of the proteasomal and lysosomal protein degradation systems. Trends Biochem. Sci., 26, 347–350. [DOI] [PubMed] [Google Scholar]
- Hu M., Li P., Li M., Li W., Yao T., Wu J.W., Gu W., Cohen R.E. & Shi Y. (2002) Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with Ub aldehyde. Cell, 111, 1041–1054. [DOI] [PubMed] [Google Scholar]
- Hubbard T. et al. (2002) The Ensembl genome database project. Nucl. Acids Res., 30, 38–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston S.C., Riddle S.M., Cohen R.E. & Hill C.P. (1999) Structural basis for the specificity of Ub C-terminal hydrolases. EMBO J., 18, 3877–3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Chen D., Shiloh A., Luo J., Nikolaev A.Y., Qin J. & Gu W. (2002) Deubiquitylation of p53 by HAUSP is an important pathway for p53 stabilization. Nature, 416, 648–653. [DOI] [PubMed] [Google Scholar]
- Li X., Yang Y. & Ashwell J.D. (2002) TNF-RII and c-IAP1 mediate ubiquitination and degradation of TRAF2. Nature, 416, 345–347. [DOI] [PubMed] [Google Scholar]
- Makarova K.S., Aravind L. & Koonin E.V. (2000) A novel superfamily of predicted cysteine proteases from eukaryotes, viruses and Chlamydia pneumoniae. Trends Biochem. Sci., 25, 50–52. [DOI] [PubMed] [Google Scholar]
- Maytal-Kivity V., Reis N., Hofmann K. & Glickman M.H. (2002) MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem., 3, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H.Y., Rothe M. & Goeddel D.V. (1996) The tumor necrosis factor-inducible zinc finger protein A20 interacts with TRAF1/TRAF2 and inhibits NF-κB activation. Proc. Natl Acad. Sci. USA, 93, 6721–6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R., Aravind L., Oania R., McDonald W.H., Yates J.R., Koonin E.V. & Deshaies R.J. (2002) Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science, 298, 611–615. [DOI] [PubMed] [Google Scholar]
- Wang C., Deng L., Hong M., Akkaraju G.R., Inoue J. & Chen Z.J. (2001) TAK1 is a Ub-dependent kinase of MKK and IKK. Nature, 412, 346–351. [DOI] [PubMed] [Google Scholar]
- Wilkinson K.D. (1997) Regulation of Ub-dependent processes by deubiquitinating enzymes. FASEB J., 11, 1245–1256. [DOI] [PubMed] [Google Scholar]
- Yao T. & Cohen R.E. (2002) A cryptic protease couples deubiquitination and degradation by the proteasome. Nature, 419, 403–407. [DOI] [PubMed] [Google Scholar]


