Abstract
The transcription factor MYC binds specific DNA sites in cellular chromatin and induces the acetylation of histones H3 and H4. However, the histone acetyltransferases (HATs) that are responsible for these modifications have not yet been identified. MYC associates with TRRAP, a subunit of distinct macromolecular complexes that contain the HATs GCN5/PCAF or TIP60. Although the association of MYC with GCN5 has been shown, its interaction with TIP60 has never been analysed. Here, we show that MYC associates with TIP60 and recruits it to chromatin in vivo with four other components of the TIP60 complex: TRRAP, p400, TIP48 and TIP49. Overexpression of enzymatically inactive TIP60 delays the MYC-induced acetylation of histone H4, and also reduces the level of MYC binding to chromatin. Thus, the TIP60 HAT complex is recruited to MYC-target genes and, probably with other other HATs, contributes to histone acetylation in response to mitogenic signals.
Introduction
The transcription factor MYC is expressed in proliferating and mitogen-stimulated cells, binds specific sites in genomic DNA and induces histone acetylation (Bouchard et al., 2001; Fernandez et al., 2003; Frank et al., 2001). This modification is catalysed by histone acetyl-transferases (HATs), which are recruited to chromatin through their interaction with sequencespecific transcription factors (Sterner & Berger, 2000). One possible mediator of MYC-induced acetylation is TRRAP, a protein that binds MYC (McMahon et al., 1998) and is recruited to MYC-binding sites in chromatin (Bouchard et al., 2001; Frank et al., 2001). TRRAP itself is not a HAT, but is found in distinct macromolecular complexes that contain HAT subunits, including GCN5, PCAF and TIP60 (Sterner & Berger 2000; Amati et al. 2001). GCN5 and PCAF are closely related proteins, and the complexes of which they are part contain essentially the same subunits (Brand et al., 1999; Martinez et al., 1998; Ogryzko et al., 1998; Vassilev et al., 1998). TIP60 belongs to a distinct class of HATs (the MYST family) and, except for TRRAP, the TIP60 and GCN5/PCAF complexes share no known common components (Ikura et al., 2000). Consistent with its interaction with TRRAP, MYC coprecipitates with GCN5 from cell lysates (McMahon et al., 2000; S.T., unpublished data). Here, we show that TIP60 is another cofactor for MYC in vivo.
Results and Discussion
To address whether MYC and TIP60 interact, 293 cells were transfected with vectors expressing MYC and FLAG–haemagglutinin (HA)-tagged TIP60. Immunoprecipitation with a TIP60-specific antibody followed by immunoblotting showed specific co-immunoprecipitation of MYC (Fig. 1A, upper panel). In addition, TIP60 was recovered in MYC immunoprecipitates (Fig. 1A, lower panel). This interaction was specific, as the MYST-family HAT, HBO1, did not coprecipitate with MYC (data not shown). We failed to detect coprecipitation of endogenous MYC and TIP60, possibly because of low expression levels of both proteins. However, this interaction is clearly observed on chromatin (see below).
Figure 1.
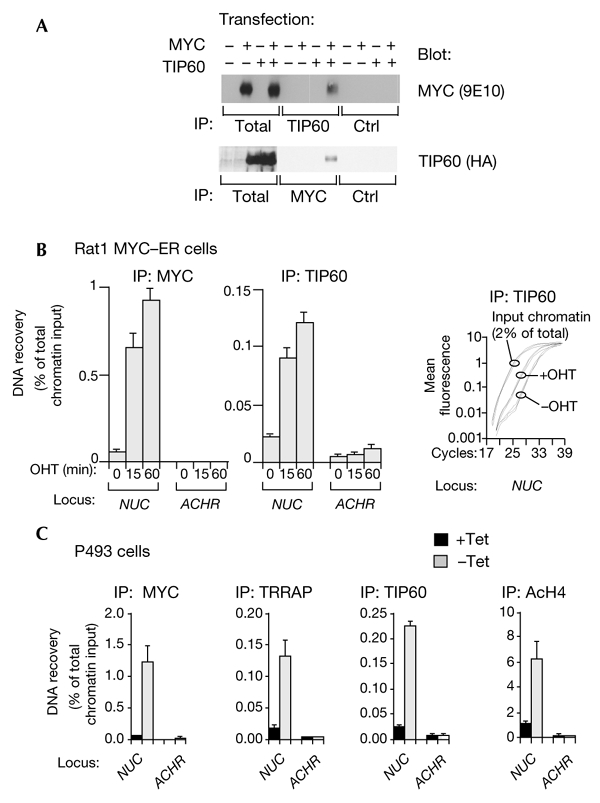
MYC binds TIP60 and recruits it to chromatin in vivo. (A) 293T cells were transfected with vectors expressing human MYC and FLAG–haemagglutinin (HA)-tagged TIP60. Whole-cell lysates (400 μg) were immunoprecipitated (IP) with antibodies against TIP60 (CLHF antiserum; upper panel) or MYC (C33 monoclonal; lower panel), followed by immunoblotting as indicated. 'Total' indicates that the whole-cell lysate (40 μg) was directly loaded for immunoblotting. 'Ctrl' indicates that control IPs were performed with pre-immune serum or with a monoclonal antibody of the same isotype as C33. (B) Quiescent Rat1 cells expressing a MYC–oestrogen-receptor (ER) chimaera were analysed by chromatin precipitation with MYC and TIP60 antibodies before or after 4-hydroxytamoxifem (OHT) treatment. PCR analysis was performed with primers that recognise MYC-bound E-boxes in NUC intron 1 or with primers that amplify the ACHR promoter, which was used as a control. The right panel shows representative PCR amplification curves from TIP60 immunoprecipitations, which were performed either before treatment with OHT (−OHT) or 1 h after treatment (+OHT). (C) P493-6 cells were growth-arrested by the addition of tetracycline (Tet) to serum-containing medium. To induce MYC expression, cells were washed and resuspended in fresh serum-containing medium without Tet (−Tet). Control cells were treated in an identical manner, but in the continuous presence of Tet (+Tet). Chromatin immunoprecipitation analysis was performed with antibodies against MYC, TRRAP, TIP60 or acetylated histone H4 (AcH4), as indicated.
To determine whether MYC recruits TIP60 to chromatin, quiescent Rat1 cells that expressed a conditionally active MYC–oestrogen-receptor (ER) chimaera were treated with 4-hydroxytamoxifen (OHT) and analysed by quantitative chromatin immunoprecipitation (ChIP). Both MYC–ER and TIP60 appeared rapidly on nucleolin (NUC) intron 1, but not on the ACHR promoter, which was used as a control (Fig. 1B). The right panel in Fig. 1B shows the NUC PCR-amplification curves for TIP60 immunoprecipitations performed in triplicate, which were carried out either before or 1 h after MYC–ER activation with OHT. DNA recovery was quantified as a percentage of the total input chromatin, as described in Frank et al. (2001). To confirm that a non-chimeric MYC protein also recruits TIP60, we performed ChIP on a human B-cell line (P493-6) that carries a tetracycline (Tet)-repressible c-MYC transgene. On removal of Tet, MYC associated with NUC intron 1, accompanied by the binding of TRRAP and TIP60, H4 acetylation (Fig. 1C), H3 acetylation, and the induction of NUC messenger RNA expression (Fernandez et al., 2003; Schuhmacher et al., 2001). None of these changes were observed for a control promoter (ACHR; Fig. 1C). Thus, MYC is sufficient for the recruitment of endogenous TIP60 to chromatin.
We then showed that TIP60 is also recruited by endogenous MYC. Following serum stimulation of Rat1 cells, MYC bound chromatin and induced H4 acetylation at several previously characterized target loci (Frank et al., 2001; upper panels in Fig. 2A). TIP60 associated with the same sites (lower panels). The NUC promoter was the most highly enriched in both MYC and TIP60 immunoprecipitations, whereas the control ACHR promoter was not enriched. The amplification of adjacent sequences in the NUC locus (upper panel in Fig. 2B) showed that MYC, TIP60 and TRRAP colocalized over the E-boxes in intron 1 (amplicon 3). Moreover, the kinetics of MYC and TIP60 binding and H4 acetylation at this site were essentially superimposable on each other (Fig. 2C), consistent with a function of MYC in the recruitment of TRRAP and TIP60. To prove this formally, we used Myc−/− Rat1 cells. In these cells, the induction of H4 acetylation and NUC expression by serum is defective, but is readily restored by infection of the cells with a MYC-expressing adenovirus (AdMYC; Frank et al., 2001). Similarly, serum did not induce binding of TRRAP or TIP60 to NUC. This was the case for MYC−/− cells that were uninfected (data not shown) or infected with the control adenovirus, AdGFP (expressing green fluorescent protein; Fig. 2D, black bars). By contrast, AdMYC restored the recruitment of TIP60 and TRRAP (Fig. 2D, shaded bars) to levels similar to those seen in parental Rat1 cells (Fig. 2A). Both cofactors were expressed at similar levels in Rat1 and Myc−/− cells, whether or not cells had been treated with serum (data not shown). In conclusion, MYC is required for recruitment of TRRAP and TIP60 to chromatin during a normal mitogenic response.
Figure 2.
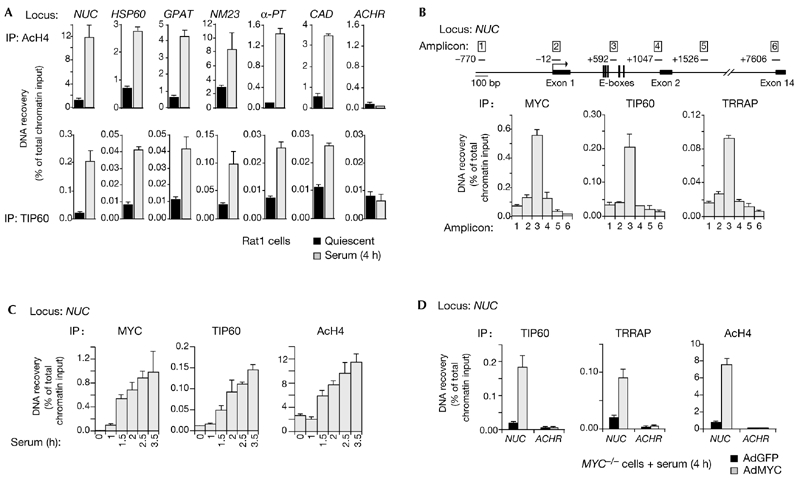
TIP60 associates with MYC target-loci. (A) Chromatin immunoprecipitation (ChIP) analysis of quiescent and serum-stimulated Rat1 cells using antibodies against acetylated histone H4 (AcH4; upper panels) and TIP60 (lower panels). The MYC binding sites (NUC, HSP60, GPAT, NM23-H2, α-PT and CAD) and the control site (ACHR) are the same as were used previously (Frank et al., 2001). (B) MYC, TIP60 and TRRAP colocalize on the NUC gene in Rat1 cells stimulated with serum for 4 h. DNA that was recovered by ChIP using the antibodies indicated was amplified with different primer pairs to give amplicons 1–5 in NUC, as indicated in the map. Numerical positions on the map are relative to the cap site, and correspond to the 5′ ends of the amplicons. (C) Kinetics of MYC binding, TIP60 recruitment and H4 acetylation at NUC intron 1 after serum stimulation of Rat1 cells. (D) Serumstimulated MYC−/− cells were infected with either a control adenovirus that expresses green fluorescent protein (AdGFP; black bars) or a MYC adenovirus (AdMYC, stippled bars). ChIP analysis showed that AdMYC restored binding of TIP60 and TRRAP and H4 acetylation at NUC intron 1.
TRRAP and TIP60 are part of a multi-subunit complex that also includes, among other proteins, p400, TIP48 (also known as TAP54-β) and TIP49 (also known as TAP54-α; Fuchs et al., 2001; Ikura et al., 2000). ChIP analysis showed that these proteins were recruited to NUC intron 1 in P493-6 cells on Tet removal (Fig. 3A). The same result was obtained after serum stimulation in Rat1 cells, in which TIP48, TIP49 and p400 colocalized with MYC on intron 1 (Fig. 3B). Thus, as tested by ChIP, MYC recruits at least five subunits of the TIP60 complex. MYC can also be coprecipitated from cell lysates with seven individual subunits, including TIP60 (Fig. 1A), TRRAP, p400, TIP48, TIP49, BAF53 and β-actin (Fuchs et al., 2001; McMahon et al., 1998; Park et al., 2002; Wood et al., 2000). Although it has been suggested that MYC interacts with TIP48/49 and TRRAP/BAF53 within different complexes (Park et al., 2002; Wood et al., 2000), it is able to bind the purified p400 complex (which contains all the subunits except TIP60; Fuchs et al., 2001). Our data are consistent with the latter result, suggesting that MYC recruits the whole p400/TIP60 complex to chromatin.
Figure 3.

MYC recruits the TIP60 complex subunits TIP48, TIP49 and p400 to chromatin. (A) MYC expression was induced in P493-6 cells by the removal of tetracycline (Tet), as described in Fig. 1C. Chromatin immunoprcipitation (ChIP) analysis was performed with antibodies against TIP48, TIP49 or p400, as indicated. (B) Quiescent and serumstimulated Rat1 cells were analysed by ChIP using the same antibodies as in (A). Amplicons 1–5 located along the rat NUC gene are the same as those shown in Fig. 2B.
To determine whether TIP60 has a role in MYC-induced acetylation, we constructed adenoviruses that express either wild-type TIP60 (AdTIP60wt) or an enzymatically inactive mutant (AdTIP60mut; Ikura et al., 2000). We tested whether expression of AdTIP60mut would interfere with the induction of H4 acetylation that was observed after serum stimulation and simultaneous infection of Myc−/− cells with AdMYC. Under these conditions, both TIP60 proteins accumulated at similar levels and did not influence MYC expression (data not shown). AdTIP60mut reproducibly resulted in a delay of ∼1 h in H4 acetylation at MYC target loci, whereas AdTIP60wt or AdGFP did not cause any significant changes in H4 acetylation (Fig. 4, top panels). Unexpectedly, TIP60mut also reduced the level of MYC binding to several sites (Fig. 4, lower panel), suggesting that TIP60 HAT activity somehow stabilizes MYC/MAX binding on chromatin. This may occur through the acetylation of MYC/MAX, histones or other proteins. Consistent with the second of these possibilities, histone acetylation correlates positively with MYC binding (Fernandez et al., 2003). In addition, the delay in H4 acetylation was longer than that for MYC binding at some target sites, consistent with the possibility that TIP60 acetylates histone H4.
Figure 4.
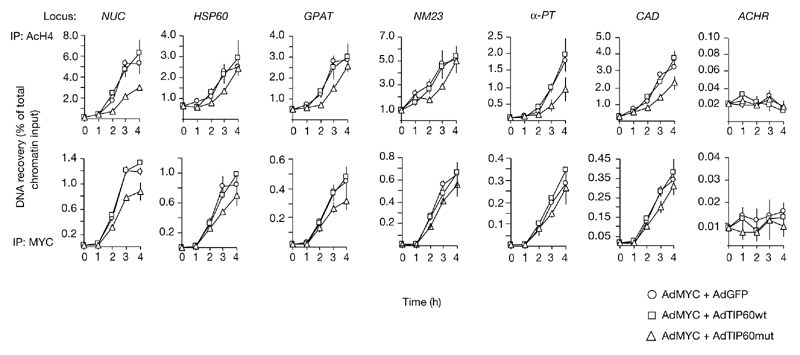
Expression of catalytically inactive TIP60 delays MYC-dependent H4 acetylation. MYC−/− cells were stimulated with serum and infected with a MYC-expressing adenovirus (AdMyc) in combination with viruses that express either green fluorescent protein (AdGFP), wild-type TIP60 (AdTIP60wt) or catalytically inactive TIP60 (AdTIP60mut). Chromatin immunopreciptiation analysis was performed at the indicated timepoints, using antibodies against acetylated H4 (AcH4; upper panel) or MYC (lower panel). PCR amplification was performed on the same sites as for Fig. 2A.
MYC can induce the acetylation of histones H3 and H4 in vivo (Bouchard et al., 2001; Fernandez et al., 2003; Nikiforov et al., 2002). In vitro, TIP60 and its yeast orthologue, Esa1, preferentially acetylate H4, whereas GCN5, PCAF and yeast Gcn5 preferentially target H3 (Allard et al., 1999; Brand et al., 1999; Ikura et al., 2000; Smith et al., 1998; Vignali et al., 2000; Xu et al., 1998). Furthermore, deletion of the yeast Esa1 and Gcn5 genes selectively decreases the in vivo acetylation levels of H4 and H3, respectively (Kuo et al., 2000; Reid et al., 2000; Vogelauer et al., 2000). Like TIP60, GCN5 can be co-immunoprecipitated with MYC from cell lysates (McMahon et al., 2000; S.T., unpublished data). Although evidence for the recruitment of GCN5 to MYC target genes has not yet been found, this is probably due to technical limitations. Based on their enzymatic specificities, we speculate that MYC recruits both of the TRRAP-associated HATs, GCN5/PCAF and TIP60, to mediate the acetylation of H3 and H4, respectively. We note, however, that our data do not rule out either distinct roles for TIP60 or the participation of GCN5 or other HATs in H4 acetylation. As we may not have achieved a quantitative blockade of TIP60 activity in our experiments, this question remains unanswered.
So far, we have been unable to determine conclusively whether TIP60 is required for the transcriptional activation of MYC target genes. In particular, TIP60mut did not interfere with mRNA accumulation (data not shown). However, given the slight delay in H4 acetylation (Fig. 4), an effect on mRNA levels might be too subtle to detect. Notably, the ability of TIP60mut to inhibit doublestrand DNA-break repair after γ-irradiation in HeLa cells was also transient (Ikura et al., 2000), suggesting that this mutant may be a weak dominant-negative allele. We also tried to suppress TIP60 expression by using small interfering RNAs, but we did not achieve full suppression (see supplementary Fig. 1B), and did not reveal any effects on MYC activity. The determination of the relative roles of TIP60, GCN5 and other HATs, such as C/EBP-binding protein (CBP)/p300 (Vervoorts et al., 2003), in MYC-induced acetylation and transcription will depend on the complete inhibition or depletion of these enzymes in vivo.
It has been proposed that MYC regulates some target genes (CAD in particular) by a mechanism that is distinct from histone acetylation and downstream from RNA polymerase (pol) II loading (Eberhardy & Farnham, 2001). However, all MYC target genes that were tested here, including CAD, were subject to Myc-induced H4 acetylation (Figs 2A, 4; Frank et al., 2001) and had RNA pol II loaded onto their cap sites before MYC induction (S.R.F., unpublished data). Although these observations do not preclude additional mechanisms of action for MYC (for example, see Cheng et al., 1999; Eberhardy & Farnham, 2001; Nikiforov et al., 2002), they argue against the classification of MYC-activated genes into distinct categories. Instead, for the genes characterized so far, acetylation (at least of H4) is a shared mechanism that seems to take place after RNA pol II loading. Understanding the contribution of acetylation to transcription awaits the mapping of the lysine residues that are targeted by MYC-recruited HATs in the H3 and H4 tails, and the identification of the molecular events that occur after these modifications.
Methods
Cells and adenoviruses.
Rat1 cells were cultured in DMEM with 10% FCS. Cells were made quiescent by contact inhibition followed by serum removal for two days. To induce cell-cycle entry, cells were trypsinized and reseeded onto plates containing DMEM and 10% FCS at a dilution of 1:3. P493-6 cells were cultured as described previously (Schuhmacher et al., 2001). The recombinant adenoviruses AdTIP60wt and AdTIP60mut were prepared as described previously for AdMYC. Myc−/− cells were infected with 1,000 particles per cell (∼50–150 particles per infectious unit) using a cationic-lipid-enhanced method (Frank et al., 2001). For co-expression studies, cells were infected with equal amounts of each virus, resulting in a total viral load of 2,000 particles per cell.
Antibodies.
TIP60-specific antisera were raised against keyhole-limpet-haemocyanin-conjugated peptides, which comprised the following residues of human TIP60: CRLPVLRRNQDNEDEWPLAE (RLPV antiserum), CLGTDEDSQDSSDGIPSAPRM (CLGT antiserum) and CLHFTPKDWSKRGKW (CLHF or CT antiserum). Antisera were purified by binding to a peptide-conjugated Sulfolink gel (Pierce), and pre-immune sera were purified on a protein A column. Each antibody specifically recognised a transfected TIP60 protein in immunoblotting or immunoprecipitation assays (see supplementary information online; and data not shown), as well as recognising endogenous TIP60 in immunoblots from various cell lines (data not shown; see supplementary information online for an example). In ChIP experiments, each TIP60 antibody specifically enriched the MYC target site in NUC intron 1 (data not shown). As the TIP60 antibody pool consistently yielded the best signal, it was used for all experiments.
Antisera against TIP48 and TIP49 were raised against peptides that were similar to those described previously in Wood et al. (2000), and that differed only in the presence of carboxy-terminal cysteines, which were added to enable crosslinking. The p400 antibody, F21, was raised against a peptide that comprised the C terminus of p400 (Fuchs et al., 2001). All antibodies were affinity purified as described above. Other antibodies used for ChIP were anti-MYC (2 μg; N262; Santa Cruz (catalogue number SC164)), anti-acetylated-histone-H4 (3 μl; Upstate Biotech (catalogue number 06-866)) and affinity-purified anti-TRRAP-CT (Frank et al., 2001).
Co-immunoprecipitation assays.
Human MYC and FLAG–HA– TIP60 proteins were expressed in 293T cells by transient transfection using the Fugene transfection reagent (Roche). Cells were lysed in IPH buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 5 mM EDTA, 0.5% v/v NP40, 1 mM dithiothreitol, 20 mM NaF, and protease inhibitors). For immunoprecipitation and immunoblotting (Fig. 1), we used the TIP60 antibody pool (see above), rabbit anti-HA (Babco), and the anti-MYC monoclonals 9E10 (Babco) and C33 (Santa Cruz).
Chromatin immunoprecipitation assays.
ChIP assays and quantitative PCR were performed as described in Frank et al. (2001). 2 μg of each antibody (see above) was used to precipitate chromatin from 1–4 × 107 cells. The PCR primers listed below were used to amplify genomic sequences. For rat NUC (Genbank AH002217) amplicon 1: 5′-CAGGCAGGCCGAGTACTTCT-3′ and 5′-TCCAGGTTCAGAGAGGTGACTTC-3′; for amplicon 2: 5′-CAAGCTCAGTCTTTTGCCTCAGA-3′ and 5′-GGATCGGCGGGTAATGAAG-3′; for amplicon 3: 5′-GGAGTGGGAGTACACGGGAA-3′ and 5′-GGAGTGGGAGTACACGGGAA-3′; for amplicon 4: 5′-GAGGTGGTGGCCGAGGA-3′ and 5′-GCCGGCCTACAAATGCAC-3′; for amplicon 5: 5′-CCACGACAAATGACAAGTTTAGTGA-3′ and 5′-AAGCTGGACCCTGATTCCATCT-3′; for amplicon 6: 5′-TCCTAACTGTACCAGCCTTTCATG-3′ and 5′-CGCCTCGGAAGCCTCC-3′; for human NUC intron 1 (Genbank M60858): 5′-TTTTGCGACGCGTACGAG-3′ and 5′-ACTAGGGCCGATACCGCC-3′; for the human ACHR promoter (Genbank Y00508): 5′-CCTTCATTGGGATCACCACG-3′ and 5′-AGGAGATGAGTACCAGCAGGTTG-3′. Other primers were described in Frank et al. (2001).
Supplementary information is available at EMBO reports online (http://www.nature.com/embor/journal/vaop/ncurrent/extref/4-embor861-s1.pdf).
Supplementary Material
Supplementary Figure 1
Acknowledgments
We thank G. Evan, J. Sedivy, Y. Nakatani, T. Ikura, D. Eick and C. Grandori for reagents, S. Zurawski for the purification of adenoviruses, Z. Herceg and Z.-Q. Wang for thoughtful comments on the manuscript, B. Luscher for communicating unpublished data, and E. Lees for her support. DNAX is supported by Schering-Plough Corp.
References
- Allard S., Utley R.T., Savard J., Clarke A., Grant P., Brandl C.J., Pillus L., Workman J.L. & Cote J. (1999) NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J., 18, 5108–5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amati B., Frank S.R., Donjerkovic D. & Taubert S. (2001) Function of the c-Myc oncoprotein in chromatin remodeling and transcription. Biochim. Biophys. Acta, 1471, M135–M145. [DOI] [PubMed] [Google Scholar]
- Bouchard C., Dittrich O., Kiermaier A., Dohmann K., Menkel A., Eilers M. & Lüscher B. (2001) Regulation of cyclin D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP recruitment and histone acetylation at the cyclin D2 promoter. Genes Dev., 15, 2042–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand M., Yamamoto K., Staub A. & Tora L. (1999) Identification of TATA-binding protein-free TAFII-containing complex subunits suggests a role in nucleosome acetylation and signal transduction. J. Biol. Chem., 274, 18285–18289. [DOI] [PubMed] [Google Scholar]
- Cheng S.W., Davies K.P., Yung E., Beltran R.J., Yu J. & Kalpana G.V. (1999) c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nature Genet., 22, 102–105. [DOI] [PubMed] [Google Scholar]
- Eberhardy S.R. & Farnham P.J. (2001) c-Myc mediates activation of the cad promoter via a post-RNA polymerase II recruitment mechanism. J. Biol. Chem., 276, 48562–48571. [DOI] [PubMed] [Google Scholar]
- Fernandez P., Frank S.R., Wang L., Schroeder M., Liu S., Greene J., Cocito A. & Amati B. (2003) Genomic targets of the human c-Myc protein. Genes Dev., 17, 1115–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S.R., Schroeder M., Fernandez P., Taubert S. & Amati B. (2001) Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev., 15, 2069–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs M., Gerber J., Drapkin R., Sif S., Ikura T., Ogryzko V., Lane W.S., Nakatani Y. & Livingston D.M. (2001) The p400 complex is an essential E1A transformation target. Cell, 106, 297–307. [DOI] [PubMed] [Google Scholar]
- Ikura T., Ogryzko V.V., Grigoriev M., Groisman R., Wang J., Horikoshi M., Scully R., Qin J. & Nakatani Y. (2000) Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell, 102, 463–473. [DOI] [PubMed] [Google Scholar]
- Kuo M.H., vom Baur, Struhl K. & Allis C.D. (2000) Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol. Cell, 6, 1309–1320. [DOI] [PubMed] [Google Scholar]
- Martinez E., Kundu T.K., Fu J. & Roeder R.G. (1998) A human SPT3–TAFII31–GCN5-L acetylase complex distinct from transcription factor IID. J. Biol. Chem., 273, 23781–23785. [DOI] [PubMed] [Google Scholar]
- McMahon S.B., Van Buskirk H.A., Dugan K.A., Copeland T.D. & Cole M.D. (1998) The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell, 94, 363–374. [DOI] [PubMed] [Google Scholar]
- McMahon S.B., Wood M.A. & Cole M.D. (2000) The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol. Cell. Biol., 20, 556–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforov M.A., Chandriani S., Park J., Kotenko I., Matheos D., Johnsson A., McMahon S.B. & Cole M.D. (2002) TRRAP-dependent and TRRAP-independent transcriptional activation by Myc family oncoproteins. Mol. Cell. Biol., 22, 5054–5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogryzko V.V., Kotani T., Zhang X., Schlitz R.L., Howard T., Yang X.J., Howard B.H., Qin J. & Nakatani Y. (1998) Histone-like TAFs within the PCAF histone acetylase complex. Cell, 94, 35–44. [DOI] [PubMed] [Google Scholar]
- Park J., Wood M.A. & Cole M.D. (2002) BAF53 forms distinct nuclear complexes and functions as a critical c-Myc-interacting nuclear cofactor for oncogenic transformation. Mol. Cell. Biol., 22, 1307–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid J.L., Iyer V.R., Brown P.O. & Struhl K. (2000) Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol. Cell, 6, 1297–1307. [DOI] [PubMed] [Google Scholar]
- Schuhmacher M. et al. (2001) The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Res., 29, 397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.R., Eisen A., Gu W., Sattah M., Pannuti A., Zhou J., Cook R.G., Lucchesi J.C. & Allis C.D. (1998) ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc. Natl Acad. Sci. USA, 95, 3561–3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner D.E. & Berger S.L. (2000) Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev., 64, 435–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev A., Yamauchi J., Kotani T., Prives C., Avantaggiati M.L., Qin J. & Nakatani Y. (1998) The 400 kDa subunit of the PCAF histone acetylase complex belongs to the ATM superfamily. Mol. Cell, 2, 869–875. [DOI] [PubMed] [Google Scholar]
- Vervoorts J., Luscher-Firzlaff J.M., Rottmann S., Lilischkis R., Walsemann G., Dohmann K., Austen M. & Luscher B. (2003) Stimulation of c-MYC transcriptional activity and acetylation by recruitment of the cofactor CBP. EMBO Rep., 4, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignali M., Steger D.J., Neely K.E. & Workman J.L. (2000) Distribution of acetylated histones resulting from Gal4-VP16 recruitment of SAGA and NuA4 complexes. EMBO J., 19, 2629–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelauer M., Wu J., Suka N. & Grunstein M. (2000) Global histone acetylation and deacetylation in yeast. Nature, 408, 495–498. [DOI] [PubMed] [Google Scholar]
- Wood M.A., McMahon S.B. & Cole M.D. (2000) An ATPase/helicase complex is an essential cofactor for oncogenic transformation by c-Myc. Mol. Cell, 5, 321–330. [DOI] [PubMed] [Google Scholar]
- Xu W., Edmondson D.G. & Roth S.Y. (1998) Mammalian GCN5 and P/CAF acetyltransferases have homologous amino-terminal domains important for recognition of nucleosomal substrates. Mol. Cell. Biol., 18, 5659–5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1


