Abstract
We have designed a doxycycline-regulated form of the H1 promoter of RNA polymerase III that allows the inducible knockdown of gene expression by small interfering RNAs (siRNAs). As a proof-of-principle, we have targeted β-catenin in colorectal cancer (CRC) cells. T-cell factor (TCF) target-gene expression is induced by accumulated β-catenin, and is the main transforming event in these cells. We have shown previously that the disruption of β-catenin/TCF4 activity in CRC cells by the overexpression of dominant-negative TCF induces rapid G1 arrest and differentiation. Stable integration of our inducible siRNA vector allowed the rapid production of siRNAs on doxycycline induction, followed by specific downregulation of β-catenin. In these CRC cells, TCF reporter-gene activity was inhibited, and G1 arrest and differentiation occurred. The inhibition of two other genes using this vector system shows that it should be useful for the inducible knockdown of gene expression.
Introduction
The transactivation of T-cell factor (TCF) target genes induced by wingless-related (WNT) pathway mutations is the main transforming event in colorectal cancer (CRC; Kinzler & Vogelstein, 1996; Bienz & Clevers, 2000). We have recently studied the TCF target-gene programme by the inducible overexpression of dominant-negative versions of TCF1 and TCF4 in CRC cell lines (van de Wetering et al., 2002; Batlle et al., 2002). This overexpression disrupted endogenous β-catenin/TCF4 activity in CRC cells. Importantly, it induced rapid G1 arrest and blocked a genetic programme that is physiologically active in the proliferative compartment of colon crypts. Consequently, an intestinal differentiation programme was induced. We concluded that the β-catenin/TCF4 complex is the master switch that controls the decision between proliferation versus differentiation in healthy and malignant intestinal epithelial cells.
As the overexpression of dominant-negative proteins might induce artefactual effects, we used a loss-of-function strategy to confirm our results. It is possible to carry out classical gene knockouts by homologous recombination in CRC cells (Shirasawa et al., 1993; Chan et al., 1999, 2002; Kim et al., 2002; Sekine et al., 2002), but this technology is relatively time-consuming. Moreover, as we expected a growth-arrest phenotype, clones would either fail to develop or would be selected for other growth-promoting events. More recently, RNA interference (RNAi), a well-established method for gene knockdown in model organisms (Sharp, 2001), can also be used for gene knockdown in mammalian cells (Elbashir et al. 2001). So-called small interfering RNAs (siRNAs) have been introduced into mammalian cells by the transient transfection of synthetic doublestranded RNA. Alternatively, promoters of genes transcribed by RNA polymerase III have been used to drive the expression of hairpin RNAs, which are very similar to siRNAs (Brummelkamp et al., 2002; McManus et al., 2002; Miyagishi & Taira, 2002; Paddison et al., 2002; Paul et al., 2002; Sui et al., 2002; Yu et al., 2002). These siRNA expression vectors have two advantages: they can be stably introduced into cells, either as selectable plasmids or as retroviruses, and they are relatively cheap to generate (for an overview, see McManus & Sharp, 2002; Paddison & Hannon, 2002; Shi, 2003). However, as with conventional knockout strategies, stably introduced siRNA vectors cannot be used when the target is essential for cellular survival.
To inducibly downregulate β-catenin/TCF4 activity in CRC cells, we have developed a doxycycline-inducible form of the RNA polymerase III H1 promoter to drive siRNA expression. The transfection of this plasmid into CRC cells carrying the tetracycline (Tet) repressor allowed the selection of clones that had stably integrated the vector in their genomes. The addition of doxycycline to the growth medium induced rapid downregulation of β-catenin messenger RNA and protein. This resulted in inhibition of TCF reporter-gene expression, G1 arrest and differentiation of the CRC cells, which confirmed our previous results.
Results and Discussion
Tet-inducible systems are widely used for mRNA expression driven by polymerase II promoters. One system uses a chimeric protein that consists of the transactivation domain of virion protein 16 (VP16) fused to the Tet repressor (Gossen & Bujard, 1992). This TetR–VP16 chimaera strongly enhances transcription from minimal promoters on binding to its cognate motif, the Tet operator. The addition of Tet inhibits binding of the repressor to its binding site. Another system makes use of the physiological function of the Tet repressor. Here, the Tet operator is situated between the promoter and the coding region of the gene of interest. Binding of the Tet repressor to the operator results in a transcriptional block of the promoter (Yao et al., 1998). Ohkawa and Taira have used the latter approach to inducibly express antisense RNA from the polymerase III promoter of the U6 gene (Ohkawa & Taira, 2000). For efficient expression of siRNAs, however, the Tet operator would have to be placed within the promoter region, as it is essential that transcription starts at the first nucleotide of the target sequence.
The RNA polymerase III promoter of the H1 gene has been studied in detail (Myslinski et al., 2001). The H1 gene has a well-defined transcriptional start site and terminates at a stretch of five thymidines, and is thus ideal for the expression of short, non-coding RNA molecules. It has been shown that sequences directly upstream of the transcription start site are generally dispensable for promoter activity in vivo (Myslinski et al., 2001). We reasoned that placing a Tet operator in this region might not affect promoter activity, whereas binding of the Tet repressor protein to this site would function as a 'road block', abrogating transcription.
As shown in Fig. 1, we created a binding site for the Tet repressor by replacing 19 bp from positions −23 to −5, relative to the transcription start site. BglII and HindIII sites were included to allow the insertion of doublestranded oligonucleotides. The pTER vector also contains a zeocin selectable marker. We inserted a 59-nucleotide human-β-catenin-derived sequence that, when transcribed, forms a hairpin consisting of a 19-bp stem and a 9-base loop. As described by Brummelkamp et al. (2002), the termination sequence that consists of five thymidines was included in the oligonucleotide. Two U residues are predicted to constitute the 3′ end of the corresponding RNA molecule.
Figure 1.
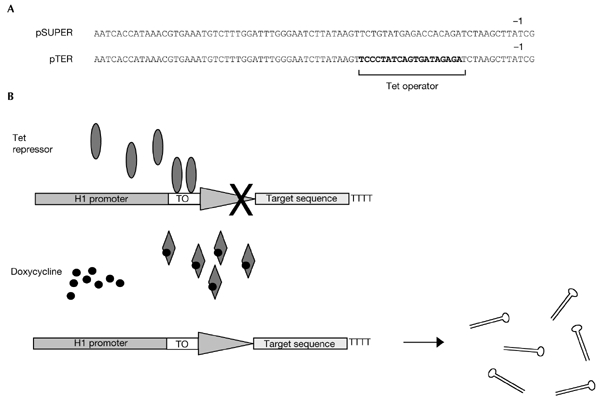
Overview of the system. (A) Alignment of the H1 promoter sequences in pSUPER and pTER. The tetracycline (Tet) operator (TO) sequence is shown in bold. (B) Schematic representation of the proposed pTER 'roadblock'. Transcription of the H1 promoter is blocked in cells expressing the Tet repressor (upper panel). Addition of doxycycline to the medium inhibits the binding of the Tet repressor and transcription is derepressed (lower panel).
We stably transfected the CRC cell line, LS174T, with a Tet-repressor expression construct using blasticidin selection (van de Wetering et al., 2002). Clones that stably expressed the Tet repressor were subsequently transfected with pTER–β-catenin and selected using zeocin. Selected clones were analysed for β-catenin expression by northern and western blotting before and after doxycline induction. Three clones showed various levels of reduction of β-catenin expression after 24–48 h, as shown in Fig. 2. To determine the efficiency of our inducible siRNA system, we performed northern blotting to visualize the production of the hairpin RNAs. As shown in Fig. 3, within 4 h of the addition of doxycycline, the hairpins were produced at significant levels. Moreover, the complete absence of short hairpin RNAs (shRNAs) in the uninduced state shows the robust transcriptional block mediated by the Tet repressor.
Figure 2.
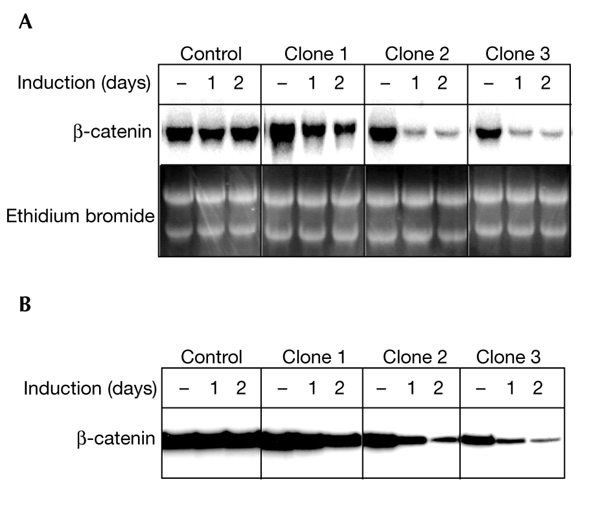
Downregulation of β-catenin in stable pTER transfectants. (A) A northern blot, showing β-catenin expression at the time points indicated. Equal amounts of RNA were loaded and analysed for β-catenin expression (data not shown). Clones two and three, in particular, strongly downregulate β-catenin messenger RNA on doxycycline treatment. (B) Western blot analysis of β-catenin protein levels at the indicated timepoints after induction. Equal loading was confirmed by β-actin staining (data not shown).
Figure 3.
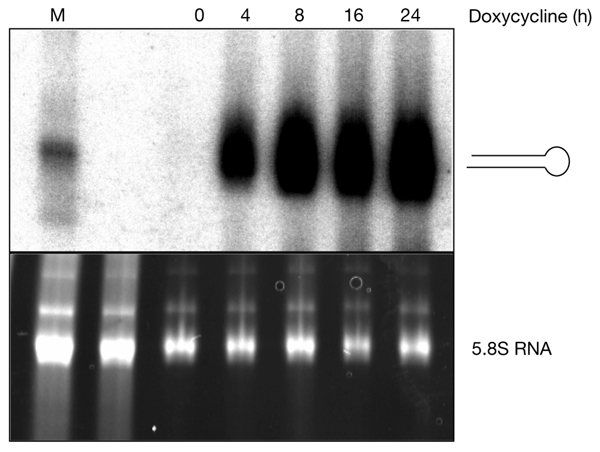
Production of β-catenin hairpin RNAs in stable pTER transfectants. Northern blot showing the expression of hairpin RNAs directed against β-catenin at the timepoints indicated. Without induction, no hairpin RNAs were detected, demonstrating the tight regulation by doxycyline (top panel). Equal amounts of RNA were loaded, as shown by the ethidium bromide staining of the 5.8S RNA (lower panel).
We then investigated the effects of β-catenin knockdown on TCF reporter activity. Most CRC cell lines spontaneously activate TCF reporters because of the constitutive activity of the WNT cascade by mutations in APC (Korinek et al., 1997) or β-catenin (Morin et al., 1997). LS174T cells carry a mutant, oncogenic β-catenin allele and, as a consequence, actively transcribe TCF reporter constructs. The induced expression of dominant-negative versions of TCF4 or TCF1 reduces this spontaneous activity to the background transcription levels of the control reporter (van de Wetering et al., 2002). Consistent with this result, the spontaneous activity of the TCF reporter, pTopglow, was reduced to background (pFopglow) levels on reduction of β-catenin levels by the induced expression of pTER–β-catenin by doxycycline in two of the three clones (Fig. 4).
Figure 4.
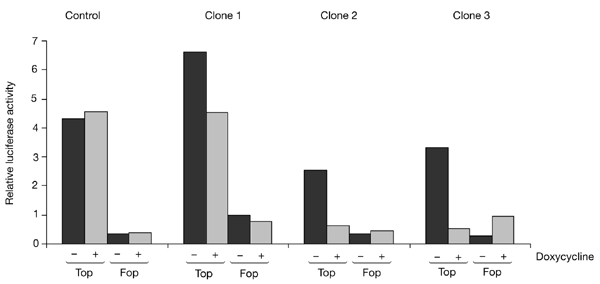
T-cell factor/β-catenin-driven transcription is abrogated on knockdown of β-catenin. The activity of the T-cell factor (TCF) reporter, pTopglow (Top), and the control, pFopglow (Fop), after 48 h with or without doxycyline treatment is shown. Parental cells that expressed the tetracycline (Tet) repressor were used as controls. Renilla luciferase levels were used as transfection controls. Clones 2 and 3 show complete inhibition of TCF reporter activity.
The removal of β-catenin function at 48 h occurred after the disappearance of β-catenin mRNA at 24 h. This delay allowed us to determine the specificity of the β-catenin siRNAs. At 20 h, mRNA expression was only affected by the production of siRNAs, and not by secondary effects resulting from loss of β-catenin. We therefore performed DNA microarray analysis to visualize the changes in gene expression 20 h after induction. In the 2 duplicate sets of measurements made in the experiment, using 17-K oligo arrays, 7 and 13 genes, respectively, were found to be downregulated more than two-fold (data not shown). More importantly, the only gene in common between the duplicate sets was β-catenin, which confirmed the specificity of the siRNAs that were used.
Inhibition of β-catenin/TCF activity in LS174T cells by the induced expression of dominant-negative TCF proteins results in a rapid G1 cell-cycle arrest and differentiation. Cell-cycle analysis of the three clones showed that the reduction in β-catenin to levels that are not sufficient to drive TCF/β-catenin transcription produced similar results. G1 arrest was already seen after 48 h of induction, but became more prominent after three days (Fig. 5A,B). The G1 arrest coincided with the differentiation of the cells. Fig. 6A shows the results of northern blot analysis of the expression of one of the differentiation markers, galectin 4, that we previously defined in the system (van de Wetering et al., 2002). This differentiation marker was rapidly induced in cells that had switched off TCF/β-catenin transcription, but was unaffected in the control cells. As a more global demonstration of the differentiation of CRC cells, we also used a histochemical stain for complex carbohydrates (mucins), periodic-acid/Schiff (PAS) stain. Fig. 6B shows the increase in PAS reactivity in induced LS174T cells on knockdown of β-catenin, which indicates the production of mucins in these differentiating cells.
Figure 5.
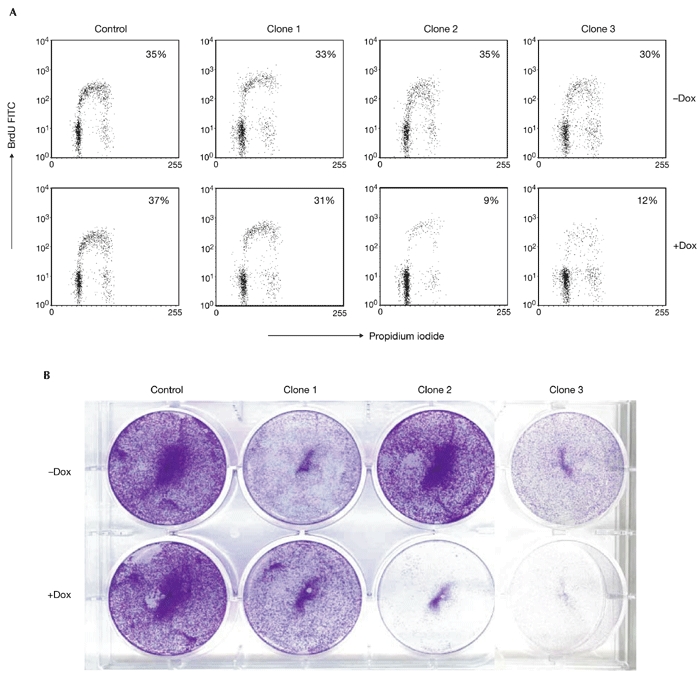
Knockdown of β-catenin results in cell-cycle arrest and growth arrest. (A) Ls174T pTER–β-catenin clones show a marked reduction in S-phase cells on β-catenin downregulation. The fluorescence activated cell sorting profiles of cells after 48 h with or without doxycycline (Dox) are shown. The percentages of cells in S phase for each clone analysed are indicated. Only clones in which TCF transcription is completely abrogated (clones 2 and 3 in Fig. 3) show a cell-cycle block in G1. The results are representative of several independent experiments. (B) Proliferation was arrested in Ls174T pTER–β-catenin transfectants. This was visualized by crystal-violet staining of cell cultures after β-catenin knockdown for five days. BrdU, 5-bromo-2-deoxyuridine.
Figure 6.
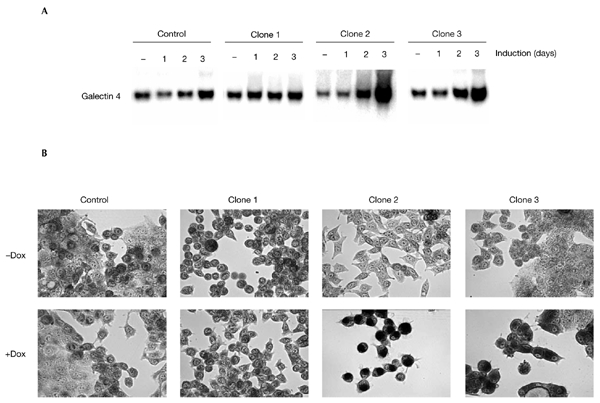
Knockdown of β-catenin induces differentiation of LS174T cells. (A) Northern blots showing expression of galectin 4, a marker of differentiated intestinal cells. Expression is strongly induced on β-catenin knockdown. (B) Periodic-acid/Schiff (PAS) staining of pTER–β-catenin transfectants and control cells shows a marked increase in mucin production on inhibition of β-catenin/T-cell factor signalling. Dox, doxycycline.
To support the results of our experiments using inducible siRNA, we attempted the knockdown of other genes. We transfected LS174Tr cells (LS174T cells expressing the Tet repressor) with c-MYC and transfected human embryonic kidney (HEK) cells that expressed Tet repressor (HEK-Tr cells) with pygopus-directed pTER vectors. Zeocin-resistant clones were tested by northern blotting. Between one in two clones (for β-catenin and pygopus) and one in ten clones (for c-MYC) efficiently downregulated the indicated genes. For each gene, two clones are shown in Fig. 7. For the pTER–β-catenin clones (Fig. 2A), strong genespecific reduction of RNA was seen after induction with doxycyline, whereas no effects on expression of other genes were seen (for example, β-catenin expression in the pTER–MYC and pTER–pygopus clones; Fig. 7).
Figure 7.
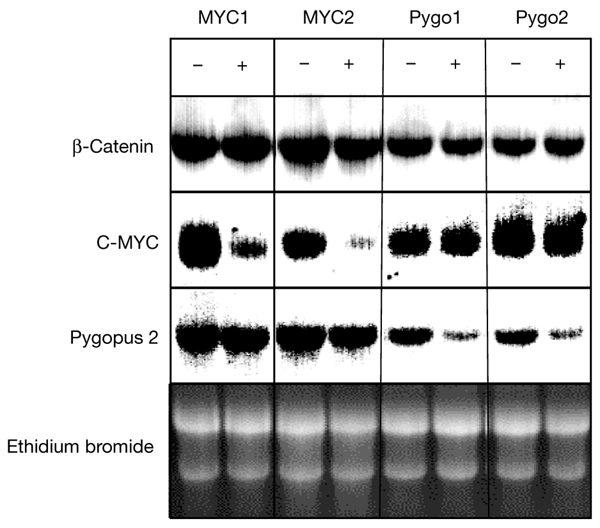
Specific downregulation of c-MYC and pygopus in stable pTER transfectants. Northern blot showing the analysis of two stable pTER–MYC and pTER–pygopus clones. 48 h after induction, the levels of the corresponding messenger RNA are markedly decreased. No effects on unrelated mRNAs (for example, β-catenin) were seen.
We conclude that knockdown of β-catenin/TCF activity by a loss-of-function approach reproduces the phenotypic changes that occur in CRC cells in which dominant-negative versions of TCF proteins are overexpressed. Moreover, the successful knockdown of three independent genes shows that our vector system is widely applicable for the inducible knockdown of gene expression. This should be particularly useful for the analysis of genes of which the functions are difficult to evaluate due to phenotypic effects on the growth or differentiation characteristics of cells (cell-cycle regulators, oncogenes and tumour suppressor genes), and for genes that control apoptosis.
Methods
Construction of pTER.
The oligonucleotides supertet (5′-CGATAAGCTTAGAT CTCTATCACTGATAGGGAACTTATAAGATTCCCAAATCC-3′) and T7 were used to amplify the H1 promoter from pSUPER (Brummelkamp et al., 2002) by PCR, thereby introducing a Tet operator sequence into the promoter. PCR fragments were cloned into pGEM-T and were confirmed by sequencing. The promoter fragment was cut out of pGEM-T using BamHI and HindIII and was ligated into pCDNA3.1-ZEO that had been digested with BglII and HindIII, thereby generating pTER (for Tet-inducible RNAi).
To make pTER–β-catenin, pTER–MYC and pTER–pygopus, gene specific oligonucleotides (100 pmol of each) were phosphorylated using T4 polynucleotide kinase in a total volume of 50 μl for 30 min. To anneal the oligonucleotides, the mixture was incubated at 95 °C for 5 min, and was cooled slowly.
1 μl of this mixture was ligated into pTER vector that had been digested with BglII and HindIII and treated with calf intestinal phosphatase. The oligonucleotides used were as follows: for β-catenin, 5′-GATCCCGTGGGTGGTATAGAGGCTCTTC AAGAGAGAGCCTCTATACCACCCACTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAGTGGGTGGTATAGAGGC TCTCTCTTGAAGAGCCTCTATACCACCCACGG-3′; for MYC, 5′-GATCCCGATGAGGAAGAAATCGATGTT CAAGAGACATCGATTTCTTCCTCATCTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAGATGAGGAAGAAATC GATGTCTCTTGAACATCGATTTCTTCCTCATCGG-3′; for pygopus, 5′-GATCCCTCCACCTGCTTCTACTGCTTTCA AGAGAAGCAGTAGAAGCAGGTGGATTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAATCCACCTGCTT CTACTGCTTCTCTTGAAAGCAGTAGAAGCAGGTGGAGG-3′.
Generation of stable pTER cell lines.
Ls174T cells were cultured in RPMI with 10% FCS. PCDNA6TR (Invitrogen) was used in accordance with the manufacturer's instructions to generate clones expressing the Tet repressor (van de Wetering et al., 2002). Tet repressor clone 1 (Tr1) was then transfected with pTER–β-catenin. Zeocin-resistant clones were tested for their ability to downreguate β-catenin by northern blotting. The parental LS174 Tet-repressor cell line, Tr1, was used as a negative control in all experiments. HEK-Tr cells were purchased from Invitrogen.
Cell-cycle analysis and cell staining.
5 × 106 Ls174T cells were seeded in 9-cm dishes and doxycycline was added to a concentration of 1 μg ml−1. After 24, 48 or 72 h, 5-bromo-2-deoxyuridine (BrdU; Roche) was added for 20 min and the cells were trypsinized and fixed in 70% ethanol. Nuclei were isolated and incubated with anti-BrdU–FITC (purchased from BD), and cell cycle profiles were determined by fluorescence-activated cell sorting analysis. For crystal-violet staining, 1 × 105 cells were seeded in wells of a six-well plate in the presence or absence of doxycycline. After five days, the cells were fixed in methanol, washed with water and stained with crystal violet for 5 min. Excess dye was washed away with water. For PAS staining, cells were washed with PBS, incubated in 1% periodic acid and stained with SCHIFFS Reagent (Merck).
Luciferase reporter assay.
To measure TCF/β-catenin-driven transactivation, a luciferase reporter assay was performed using the TCF reporter constructs pTopglow and pFopglow (van de Wetering et al., 2001). pTopglow contains TCF binding sites upstream of a minimal E1b TATA box that drives the expression of luciferase. The control plasmid, pFopglow, contains mutated TCF binding sites. For transient transfections, 2.5 × 105 cells per well were seeded in six-well plates and transfected with 500 ng of TCF reporter and 50 ng TKRenilla (Promega) using Fugene-6 (Roche). Luciferase activity was determined 48 h after transfection, using the Dual-luciferase reporter assay system (Promega).
Northern blotting.
10 μg of total RNA was separated on 1.25% agarose gels and transferred to Zeta-Probe membranes (Biorad). Loading was checked by ethidium bromide staining. Hybridization was performed using ExpressHyb Hybridization Solution (Clontech). Probes were labelled using the RadPrime DNA labelling system (Invitrogen).
For shRNA visualization, 30 μg of total RNA was separated on 9% polyacrylamide gels containing 8 M urea. The RNAs were then transferred to Zeta-Probe membranes by electroblotting. After backing at 80 °C, hybridization was performed at 52 °C in 0.25 M phosphate buffer, pH 7.2, and 7% SDS. Oligonucleotides of 19 nucleotides were labelled with [32P]γATP and used as probes.
DNA microarray analysis.
Total RNA treated with DNAse was used to generate complementary DNA labelled with CyDye (Amersham Bioscience). Equal amounts of probe were hybridized against 17-K oligonucleotide arrays. Dye swaps were performed for each experiment. The whole procedure is described in detail in van de Peppel et al. (2003).
Western blotting.
3 × 106 Ls174T cells were seeded in 9-cm dishes in the presence or absence of doxycycline (1 μg ml−1). Cells were harvested after 24 and 48 h, and 1 × 106 cells per sample were lysed in 1% nonidet P40 (NP40) lysis buffer (1% NP40, 30 mM Tris-HCl, pH 8.0, 150 mM NaCl, proteinase inhibitor (Boehringer)) on ice for 30 min. Lysates were cleared by centrifugation at 14,000g for 30 min and supernatants were collected. A Bradford test was used to determine the protein concentration. Equal amounts of protein were boiled, electrophoresed on 10% SDS–polyacrylamide gels and transferred to Immobilon-P membranes (Millipore). Blots were incubated with anti-β-catenin antibody (Transduction Laboratories), using RAMPO (Pierce) as the secondary antibody. Immune complexes were visualized using an enhanced chemiluminescence kit (Amersham Life Science).
Acknowledgments
The authors thank R. Bernards for sharing information before publication, F. Vervoordeeldonk and J. Heinen for photographic assistance, and the members of the Clevers lab for helpful discussions. F.C.P.H. and D.v.L. were supported by grants from The Netherlands Organization for Scientific Research (NOW; 05050205, 90101238, 90101226; 016026009 and 90104219) and by the EU Fifth Framework grant, TEMBLOR.
References
- Batlle E. et al. (2002) β-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell, 111, 251–263. [DOI] [PubMed] [Google Scholar]
- Bienz M. & Clevers H. (2000) Linking colorectal cancer to Wnt signaling. Cell, 103, 311–320. [DOI] [PubMed] [Google Scholar]
- Brummelkamp T.R., Bernards R. & Agami R. (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science, 296, 550–553. [DOI] [PubMed] [Google Scholar]
- Chan T.A., Hermeking H., Lengauer C., Kinzler K.W. & Vogelstein B. (1999) 14-3-3σ is required to prevent mitotic catastrophe after DNA damage. Nature, 401, 616–620. [DOI] [PubMed] [Google Scholar]
- Chan T.A., Wang Z., Dang L.H., Vogelstein B. & Kinzler K.W. (2002) Targeted inactivation of CTNNB1 reveals unexpected effects of β-catenin mutation. Proc. Natl Acad. Sci. USA, 99, 8265–8270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., and Tuschl T. (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature, 411, 494–498. [DOI] [PubMed] [Google Scholar]
- Gossen M. & Bujard H. (1992) Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl Acad. Sci. USA, 89, 5547–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.S., Crooks H., Foxworth A. & Waldman T. (2002) Proof-of-principle: oncogenic β-catenin is a valid molecular target for the development of pharmacological inhibitors. Mol. Cancer Ther., 1, 1355–1359. [PubMed] [Google Scholar]
- Kinzler K.W. & Vogelstein B. (1996) Lessons from hereditary colorectal cancer. Cell, 87, 159–170. [DOI] [PubMed] [Google Scholar]
- Korinek V., Barker N., Morin P.J., van Wichen D., de Weger R., Kinzler K.W., Vogelstein B. & Clevers H. (1997) Constitutive transcriptional activation by a β-catenin-Tcf complex in APC−/− colon carcinoma. Science, 275, 1784–1787. [DOI] [PubMed] [Google Scholar]
- McManus M.T. & Sharp P.A. (2002) Gene silencing in mammals by small interfering RNAs. Nature Rev. Genet. 3, 734–747. [DOI] [PubMed] [Google Scholar]
- McManus M.T., Petersen C.P., Haines B.B., Chen J. & Sharp P.A. (2002) Gene silencing using micro-RNA designed hairpins. RNA, 8, 842–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishi M. & Taira K. (2002) U6 promoter-driven siRNAs with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nature Biotechnol., 20, 497–500. [DOI] [PubMed] [Google Scholar]
- Morin P.J., Sparks A.B., Korinek V., Barker N., Clevers H., Vogelstein B. & Kinzler K.W. (1997) Activation of β-catenin-Tcf signaling in colon cancer by mutations in β-catenin or APC. Science, 275, 1787–1790. [DOI] [PubMed] [Google Scholar]
- Myslinski E., Ame J.C., Krol A. & Carbon P. (2001) An unusually compact external promoter for RNA polymerase III transcription of the human H1RNA gene. Nucleic Acids Res., 29, 2502–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkawa J. & Taira K. (2000) Control of the functional activity of an antisense RNA by a tetracycline-responsive derivative of the human U6 snRNA promoter. Hum. Gene Ther., 11, 577–585. [DOI] [PubMed] [Google Scholar]
- Paddison P.J. & Hannon G.J. (2002) RNA interference: the new somatic cell genetics? Cancer Cell, 2, 17–23. [DOI] [PubMed] [Google Scholar]
- Paddison P.J., Caudy A.A., Bernstein E., Hannon G.J. & Conklin D.S. (2002) Short hairpin RNAs (shRNAs) induce sequence specificsilencing in mammalian cells. Genes Dev., 16, 948–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul C.P., Good P.D., Winer I. & Engelke D.R. (2002) Effective expression of small interfering RNA in human cells. Nature Biotechnol., 20, 505–508. [DOI] [PubMed] [Google Scholar]
- Sekine S., Shibata T., Sakamoto M. & Hirohashi S. (2002) Target disruption of the mutant β-catenin gene in colon cancer cell line HCT116: preservation of its malignant phenotype. Oncogene, 38, 5906–5911. [DOI] [PubMed] [Google Scholar]
- Sharp P.A. (2001) RNA interference—2001. Genes Dev., 15, 485–490. [DOI] [PubMed] [Google Scholar]
- Shi Y. (2003) Mammalian RNAi for the masses. Trends Genet., 19, 9–12. [DOI] [PubMed] [Google Scholar]
- Shirasawa S., Furuse M., Yokoyama N. & Sasazuki T. (1993) Altered growth of human colon cancer cell lines disrupted at activated Ki-ras. Science, 260, 85–88. [DOI] [PubMed] [Google Scholar]
- Sui G., Soohoo C., el Affar B., Gay F., Shi Y., Forrester W.C. & Shi Y. (2002) A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl Acad. Sci. USA, 99, 5515–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Peppel J., Kemmeren P., van Bakel H., Radonjic M., van Leenen D. & Holstege F.C.P. (2003) Monitoring global messenger RNA changes in externally controlled microarray experiments. EMBO Rep., 4, 387–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering M., Barker N., Harkes I.C., van der Heyden M., Dijk N.J., Hollestelle A., Klijn J.G., Clevers H. & Schutte M. (2001) Mutant E-cadherin breast cancer cells do not display constitutive Wnt signaling. Cancer Res., 61, 278–284. [PubMed] [Google Scholar]
- van de Wetering M. et al. (2002) The β-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell, 111, 241–250. [DOI] [PubMed] [Google Scholar]
- Yao F., Svensjo T., Lu M., Eriksson C. & Eriksson E. (1998) Tetracycline repressor, tetR, rather than the tetR-mammalian cell transcription factor fusion derivatives, regulates inducible gene expression in mammalian cells. Hum. Gene Ther., 9, 1939–1950. [DOI] [PubMed] [Google Scholar]
- Yu J.Y., DeRuiter S.L. & Turner D.L. (2002) RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc. Natl Acad. Sci. USA, 99, 6047–6052. [DOI] [PMC free article] [PubMed] [Google Scholar]


