Abstract
IL-13 dysregulation plays a critical role in the pathogenesis of a variety of inflammatory and remodeling diseases. In these settings, STAT6 is believed to be the canonical signaling molecule mediating the tissue effects of IL-13. Signaling cascades involving MAPKs have been linked to inflammation and remodeling. We hypothesized that MAPKs play critical roles in effector responses induced by IL-13 in the lung. We found that Tg IL-13 expression in the lung led to potent activation of ERK1/2 but not JNK1/2 or p38. ERK1/2 activation also occurred in mice with null mutations of STAT6. Systemic administration of the MAPK/ERK kinase 1 (MEK1) inhibitor PD98059 or use of Tg mice in which a dominant-negative MEK1 construct was expressed inhibited IL-13–induced inflammation and alveolar remodeling. There were associated decreases in IL-13–induced chemokines (MIP-1α/CCL-3, MIP-1β/CCL-4, MIP-2/CXCL-1, RANTES/CCL-5), MMP-2, -9, -12, and -14, and cathepsin B and increased levels of α1-antitrypsin. IL-13–induced tissue and molecular responses were noted that were equally and differentially dependent on ERK1/2 and STAT6 signaling. Thus, ERK1/2 is activated by IL-13 in the lung in a STAT6-independent manner where it contributes to IL-13–induced inflammation and remodeling and is required for optimal IL-13 stimulation of specific chemokines and proteases as well as the inhibition of specific antiproteases. ERK1/2 regulators may be useful in the treatment of IL-13–induced diseases and disorders.
Introduction
IL-13 is a pleiotropic 12-kDa cytokine product of a gene on chromosome 5 at q31 that is 1 of the major effector molecules at sites of Th2 inflammation and remodeling (1, 2). Studies from our laboratory and others have used overexpression Tg modeling and other approaches to define the effector properties of IL-13. These studies demonstrated that IL-13 is a potent stimulator of eosinophil-, lymphocyte-, and macrophage-rich inflammation, mucus metaplasia, tissue fibrosis, and parenchymal proteolysis (2–6). In the lung, IL-13 induces asthma-like airway hyperresponsiveness after methacholine challenge (5, 7). In accord with these findings, IL-13 dysregulation is thought to play an important role in the pathogenesis of a variety of diseases, including asthma, pulmonary and hepatic fibrosis, fungal pneumonitis, viral pneumonia, nodular sclerosing Hodgkin disease, and chronic obstructive pulmonary disease (4, 5, 8–11). In all of these cases, activation of the STAT6 signal transduction pathway is believed to mediate the biologic effects of IL-13 (7, 12–14). However, the possibility that other signal transduction pathways also contribute to IL-13–induced tissue responses has not been adequately addressed.
MAPKs are a family of serine-threonine kinases that mediate the nuclear response of cells to a wide variety of extracellular stresses such as inflammatory cytokines, growth factors, UV light, and osmotic stress (15, 16). MAPKs are divided into 3 major subfamilies: the ERKs, of which ERKs 1 and 2 are the most abundant in mammalian cells, the JNKs, and the p38 MAPKs (17). Each MAPK is activated through dual phosphorylation via a specific upstream phosphorylation cascade. MAPKs have been implicated in the pathogenesis of asthma-like Th2 inflammation with chemical inhibition of ERK1/2 MAPK–reducing lung eosinophilia in ovalbumin-sensitized and -challenged rats and mice (18, 19). MAPKs have also been implicated in IL-13–induced eotaxin production, IL-13 receptor regulation, mucus secretion, and 15 lipoxygenase stimulation in vitro (20–25). However, the role of MAPKs in mediating the biologic effects of IL-13 in vivo has not been investigated.
We hypothesized that activated MAPKs play an important role in the pathogenesis of IL-13–induced lung inflammation and remodeling. To test this hypothesis, we characterized lung MAPK activation in inducible and lung epithelial cell–specific, IL-13 overexpressing Tg mice (CC10–rtTA–IL-13) (CC10, Clara cell 10; rtTA, reverse tetracycline transactivator) and defined the effects of MAPK inactivation on the IL-13–induced tissue responses in these animals. These studies demonstrate that IL-13 is a potent and selective stimulator of ERK1/2 MAPK activation and that this activation occurs in the absence of STAT6. They also demonstrate that chemical and genetic interventions that selectively inactivate ERK1/2 significantly abrogate IL-13–induced inflammation and lung remodeling. In addition, these studies provide mechanistic insights by demonstrating that ERK1/2 MAPK activation is required for optimal IL-13 stimulation of chemokines (MIP-1α/CCL-3, MIP-1β/CCL-4, MIP-2/CXCL-1, RANTES/CCL-5), MMP-2, -9, -12, and -14, and cathepsin B and IL-13 inhibition of β1-antitrypsin. Lastly, by comparing the effects of IL-13 in mice in which ERK1/2- or STAT6-mediated signaling responses were absent, we identified tissue and molecular effects of IL-13 that were equally and differentially dependent on STAT6 and ERK1/2.
Results
Effect of IL-13 on ERK1/2 MAPK.
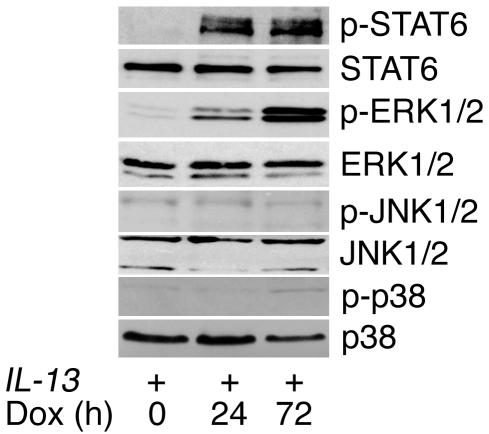
We first determined whether IL-13 activates MAPKs using CC10–rtTA–IL-13 mice in which the IL-13 Tg can be inducibly expressed in the adult murine lung by the administration of doxycycline-containing (dox-containing) water. As expected, increased levels of phospho-STAT6 were readily detected by Western blot analysis after Tg activation (Figure 1). Three major MAPK signaling pathways have been identified in mammalian systems: the ERK1/2, JNK1/2, and p38 pathways. After 24 hours of IL-13 induction, phosphorylation of ERK1 (p44) and ERK2 (p42) but not JNK1/2 or p38 MAPK was observed (Figure 1). These studies demonstrate that, in addition to STAT6, ERK1/2 is also activated by IL-13 in vivo.
Figure 1.
Effect of IL-13 on MAPK activation. Western blots for phosphorylated (p) STAT6, ERK1/2, JNK1/2, and p38 were performed on lung lysates from CC10–rtTA–IL-13 mice (IL-13+) fed dox water for 0 to 3 days. Antibodies against the relevant total protein were used to control for loading. Data are representative of n = 4 for each group.
Role of STAT6 in IL-13–induced ERK1/2 MAPK activation.
Studies were next undertaken to determine whether IL-13 activated ERK1/2 via STAT6-dependent or -independent pathways. To answer this question, we bred constitutive CC10–IL-13 Tg mice and inducible CC10–rtTA–IL-13 Tg with mice with homozygous null mutations of STAT6 and compared the effects of IL-13 in mice with wild-type and null STAT6 loci. As noted above, Tg IL-13 was a potent activator of ERK1/2 in mice with wild-type STAT6 loci in both Tg systems (Figure 2, A and B). STAT6 phosphorylation was present in IL-13 Tg+ mice with wild-type STAT6 loci but absent in IL-13 Tg+ mice with homozygous null mutations of STAT6, indicating appropriate STAT6 responses in these mice (Figure 2, A and B). Importantly, ERK1/2 phosphorylation was readily apparent in IL-13 Tg+ mice with homozygous null mutations of STAT6 (Figure 2, A and B). This activation was comparable in magnitude to that seen in the IL-13 Tg+ mice with wild-type STAT6 loci (Figure 2). Thus, IL-13 activation of ERK1/2 does not require functional STAT6.
Figure 2.
IL-13–induced ERK1/2 activation occurs in the absence of STAT6. Western blots for phosphorylated ERK1/2 and STAT6 were performed on lung lysates from (A) constitutive CC10–IL-13+ or (B) CC10–rtTA–IL-13+ Tg mice. In each, we compare Tg– (–) and Tg+ (+) IL-13 mice with wild-type (+/+) and homozygous null (–/–) STAT6 loci. β-tubulin was used as loading control. Data are representative of n = 4 for each group.
Chemical and genetic manipulations of ERK1/2 MAPK.
In order to define the role(s) of ERK1/2 in IL-13–induced tissue responses, chemical and genetic manipulations were undertaken. In the chemical intervention studies, we compared the effects of Tg IL-13 in mice treated with the MAPK/ERK kinase 1 (MEK1) inhibitor PD98059 (PD) or its vehicle control. We also bred CC10–rtTA–IL-13 Tg mice with CC10–dominant-negative MEK1 (CC10-dnMEK1) Tg mice. Because MEK1 is the dominant upstream kinase that activates ERK1/2, this allowed us to compare the effects of Tg IL-13 in mice in which the epithelial activation of ERK1/2 MAPK was specifically inhibited. In accord with our expectations, STAT6 and ERK1/2 phosphorylation were readily apparent in lungs from IL-13 Tg+ mice that were treated with the vehicle control of PD and IL-13 Tg+ mice that did not contain the dnMEK1 construct (Figure 3, A and B). In contrast, in PD-treated IL-13 Tg+ mice and IL-13 Tg+ mice that expressed dnMEK1, the ability of Tg IL-13 to phosphorylate ERK1/2 was abrogated while STAT6 phosphorylation was not altered (Figure 3, A and B). In order to demonstrate that PD or dnMEK1 did not alter IL-13 levels in IL-13 Tg mice, we also quantified the levels of bronchoalveolar lavage (BAL) fluid IL-13. These studies demonstrated that BAL-fluid IL-13 levels were not significantly changed in PD-treated IL-13+ or IL-13+/dnMEK1+ mice (Figure 3C).
Figure 3.
ERK1/2 inhibition in IL-13 Tg mice. Western blots for phosphorylated ERK1/2 and STAT6 were performed on lung lysates from IL-13– and CC10–rtTA–IL-13 Tg+ (+) mice. (A) Mice treated with PD or its vehicle control. (B) Mice with and without the CC10–dnMEK1 Tg construct. Data are representative of n = 4 for each group. (C) Levels of IL-13 protein in BAL fluids from IL-13– and IL-13+ mice with and without PD or dnMEK1. Data are expressed as mean ± SEM. n = 4–5 for each group. *P < 0.05 compared with IL-13+.
Role of ERK1/2 MAPK in IL-13–induced inflammation.
Studies were next undertaken to define the significance of ERK1/2 activation in IL-13–mediated inflammation. In these experiments, we first assessed the inflammation in BAL fluids and lung tissues from CC10–rtTA–IL-13 Tg+ mice treated with PD or its vehicle control. As expected, BAL fluids from IL-13+ mice on dox water for 10 days exhibited significant increases in total eosinophil, lymphocyte, and macrophage recovery in BAL fluids (Figure 4A). ERK1/2 appeared to play an important role(s) in this response because PD-treated IL-13 Tg+ mice manifested a significant decrease in BAL-fluid total cell recovery (Figure 4A). This was associated with a significant decrease in BAL-fluid eosinophils (Figure 4A). Significant decreases in neutrophil or lymphocyte recovery were not noted (Figure 4A). A marked decrease in BAL-fluid eosinophilia without significant changes in neutrophils or lymphocytes was also seen in IL-13 Tg+ mice that expressed epithelial targeted dnMEK1 (Figure 4B). With both ERK1/2-based interventions, a similar decrease in lung-tissue eosinophilia was seen with antimajor basic protein (anti-MBP) immunostaining and morphometric eosinophil quantification (Figure 4C). Thus, these chemical and genetic interventions demonstrate that ERK1/2 plays an important role in IL-13–induced pulmonary eosinophilic inflammation.
Figure 4.
Role of ERK1/2 MAPK in IL-13–induced inflammation. BAL fluids and lungs were obtained from IL-13– and CC10–rtTA–IL-13 Tg+ (IL-13+) mice. (A) BAL-fluid cellularity of mice treated with PD or vehicle control (veh). (B) BAL-fluid cellularity of IL-13+ mice with and without dnMEK1. (C) MBP immunostaining and morphometric eosinophil quantification were used in comparing lungs from IL-13– and IL-13+ mice after PD administration or with and without dnMEK1. Data are expressed as mean ± SEM and are representative of n = 4–16 for each group. *P < 0.05 compared with IL-13+.
Role of STAT6 in IL-13–induced inflammation.
To further define the mechanisms of IL-13–induced inflammation, we also defined the inflammatory response in CC10–rtTA–IL-13 mice with null mutations of STAT6. In the absence of STAT6, BAL-fluid inflammatory cell recovery was decreased by approximately 65% (Figure 5A). This was associated with a significant decrease in BAL fluids and tissue eosinophilia (Figure 5, A and B). These decreases were comparable to those achieved with PD or dnMEK1 (Figure 4). In contrast to the chemical and genetic ERK1/2-based interventions, however, decreases in BAL-fluid neutrophil and lymphocyte counts were seen in the absence of STAT6 (Figure 5A). These studies demonstrate that ERK1/2 and STAT6 have comparable effects on eosinophilic inflammation. They also highlight interesting differences in the roles of ERK1/2 and STAT6 in the pathogenesis of IL-13–induced neutrophil and lymphocyte accumulation.
Figure 5.
Role of STAT6 in IL-13–induced inflammation. BAL fluids and lung tissues were obtained from IL-13– and CC10–rtTA–IL-13 Tg+ (IL-13+) mice with wild-type or null STAT6 loci. (A) BAL fluids were obtained for total cell counts and differentials. (B) MBP immunostaining and morphometric lung tissue eosinophil quantification were undertaken. Data are expressed as mean ± SEM and are representative of n = 4–5 for each group. *P < 0.05 compared with IL-13+/STAT6+/+ mice.
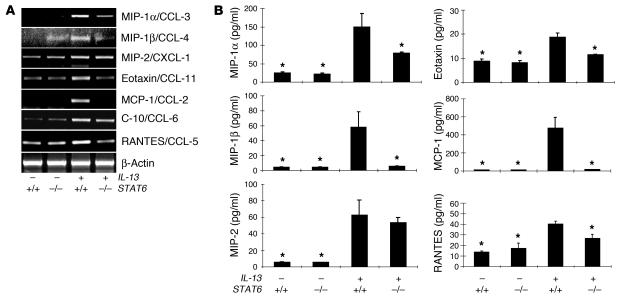
Role of ERK1/2 MAPK in IL-13–induced chemokine elaboration.
Previous studies from our laboratory demonstrated that IL-13 generates tissue inflammation by inducing a powerful chemokine response (6, 26). Thus, studies were undertaken to determine whether the ERK1/2 pathway played an important role(s) in this inductive event. In accord with our previous studies (6, 26), Tg IL-13 was a potent stimulator of a variety of chemokines, including MIP-1α/CCL-3, MIP-1β/CCL-4, MIP-2/CXCL-1, eotaxin/CCL-11, C10/CCL-6, MCP-1/CCL-2, and RANTES/CCL-5 (Figure 6). Interestingly, PD-treated IL-13 Tg+ mice on dox water had lower levels of MIP-1α/CCL-3, MIP-1β/CCL-4, MIP-2/CXCL-1, and RANTES/CCL-5 mRNA and protein in comparison with mice treated with the vehicle control (Figure 6, A and C). This effect was at least partially chemokine specific because PD did not alter the induction of eotaxin/CCL-11, C10/CCL-6, or MCP-1/CCL-2 (Figure 6, A and C). Similarly, dox-treated IL-13 Tg+/dnMEK1+ mice also had significantly lower levels of MIP-1α/CCL-3, MIP-1β/CCL-4, MIP-2/CXCL-1, and RANTES/CCL-5 and unchanged levels of eotaxin/CCL-11, C10/CCL-6, and MCP-1/CCL-2 mRNA and protein (Figure 6, B and D). These studies demonstrate that IL-13 stimulates pulmonary chemokines via ERK1/2-dependent and -independent pathways.
Figure 6.
Role of ERK1/2 MAPK in IL-13–induced chemokine mRNA and protein expression. (A and B) Whole-lung RNA was used for RT-PCR to detect mRNA for IL-13–regulated chemokines in lungs from IL-13– and IL-13+ mice. Comparisons are made of mice administered PD and vehicle control (A) and mice with and without dnMEK1 (B). β-actin mRNA expression was used as a loading control. (C and D) Levels of the noted chemokines were assessed by ELISA in BAL fluids from IL-13– and IL-13+ mice. Comparisons are made of mice administered PD or vehicle control (C) and mice with and without dnMEK1 (D). Data are expressed as mean ± SEM and represent n = 5 for each group. *P < 0.05 compared with IL-13+.
Role of STAT6 in IL-13–induced chemokine elaboration.
To further define the pathways that IL-13 uses to stimulate chemokine responses, we compared the effects of IL-13 in CC10–rtTA–IL-13 mice with wild-type, ERK1/2-inhibited, and STAT6-null signaling pathways. In these evaluations, STAT6 deficiency caused a significant decrease in IL-13 stimulation of MIP-1α/CCL-3, MIP-1β/CCL-4, and RANTES/CCL-5 (Figure 7, A and B). This inhibition was comparable to that seen in PD-treated or dnMEK1 mice (Figure 6). In contrast to ERK1/2-manipulated animals, however, null mutations of STAT6 also significantly decreased the ability of IL-13 to stimulate eotaxin/CCL-11, MCP-1/CCL-2, and C10/CCL-6 mRNA and protein (Figure 7, A and B). As an additional contrast, null mutations of STAT6 did not alter IL-13 induction of MIP-2/CXCL-1 (Figure 7, A and B) while PD treatment and dnMEK1 significantly decreased MIP-2/CXCL-1 mRNA and protein (Figure 6). These studies demonstrate that IL-13–induced chemokine responses can be comparably dependent on ERK1/2 and STAT6 (MIP-1α/CCL-3, MIP-1β/CCL-4, and RANTES/CCL-5), largely dependent on STAT6 (eotaxin/CCL-11, MCP-1/CCL-2 and C10/CCL-6), or largely dependent on ERK1/2 (MIP-2/CXCL-1).
Figure 7.
Role of STAT6 in IL-13–induced chemokine mRNA and protein expression. IL-13– and CC10–rtTA–IL-13 Tg+ (IL-13+) mice with wild-type or null STAT6 loci were generated. (A) Whole-lung RNA was processed for RT-PCR to detect mRNA for IL-13–regulated chemokines. β-actin mRNA expression was used as a loading control. (B) Levels of the noted chemokines in BAL fluids were evaluated by ELISA. Data are expressed as mean ± SEM and represent n = 4–5 for each group. *P < 0.05 compared with IL-13+/STAT6+/+ mice.
Role of ERK1/2 MAPK activation in IL-13–induced tissue remodeling responses.
IL-13 is a potent stimulator of pulmonary remodeling that induces lung and alveolar enlargement as well as mucus metaplasia (4–6, 26). In order to assess the importance of ERK1/2 activation in IL-13–induced remodeling, we defined the effects of chemical and genetic inhibition of ERK1/2 on these responses. In accord with our previous reports (4, 6, 26), Tg IL-13 caused increases in lung volume, alveolar size, goblet cell number, and Gob-5, Muc-5ac, and Muc-1 gene expression in Tg+ ERK1/2 MAPK-sufficient animals (Figure 8, A–H). Interestingly, PD administration decreased IL-13–induced alveolar remodeling responses. This was readily apparent in comparisons of the histologic appearance, lung volumes, and chord length measurements of IL-13 Tg+ mice treated with PD or vehicle control (Figure 8, A–C). In accord with these findings, the dnMEK1 construct also ameliorated these parameters of alveolar remodeling in IL-13 Tg+ animals when we performed parallel studies in IL-13+/dnMEK1+ mice (Figure 8, D–F). In contrast, neither the chemical nor the genetic manipulations of ERK1/2 altered the IL-13–induced mucus responses (goblet cell hyperplasia, Gob-5, Muc-5ac, and Muc-1 gene expression and mucin secretion; Figure 8, G and H, and data not shown). These studies demonstrate that ERK1/2 signaling plays an important role in the pathogenesis of selected aspects of the IL-13 remodeling response, with alveolar remodeling being at least partially ERK1/2 dependent and mucus metaplasia being largely ERK1/2 independent.
Figure 8.
Role of ERK1/2 MAPK in IL-13–induced tissue remodeling responses. Parameters of remodeling were assessed in IL-13– and CC10–rtTA–IL-13 Tg+ (IL-13+) mice. (A–C) Comparison of (A) alveolar histology (H&E), (B) lung volume, and (C) alveolar chord length in mice treated with PD or vehicle control. Original magnification, ×10. (D–F) Comparison of (D) alveolar histology (H&E), (E) lung volume, and (F) alveolar chord length in mice with and without dnMEK1. Original magnification, ×10. (G) PAS with diastase staining and (H) levels of mucin and Gob-5 mRNA in these animals. β-actin mRNA expression was used as a loading control. Original magnification, ×40. Arrow highlights PAS with diastase staining of epithelial cells in IL-13+/STAT6–/– animals. Values in graphs illustrate the mean ± SEM and represent n = 4–6 for each group. *P < 0.05 compared with IL-13+.
Role of STAT6 in IL-13–induced tissue remodeling.
To gain additional insight into the mechanisms of IL-13–induced alveolar remodeling, the importance of STAT6 in these responses was also evaluated. As seen with the ERK1/2-based interventions, IL-13–induced alveolar remodeling was partially abrogated in the absence of STAT6 signaling (Figure 9, A–C). Interestingly, the amelioration that was seen with null mutations of STAT6 was comparable to that seen with ERK1/2-based interventions. In contrast, the ERK1/2- and STAT6-based interventions played very different roles in the IL-13–induced mucus responses. Unlike the ERK1/2 interventions, which did not alter these responses, IL-13–induced stimulation of goblet cell hyperplasia, Muc-5ac, and Gob-5 gene expression and mucin secretion was reduced in the absence of STAT6 (Figure 9, D–F). In all cases, however, these effects were not complete, with modest IL-13–induced mucus and mucin responses being appreciated in the absence of STAT6 (Figure 9, D–F). These studies demonstrate that IL-13–induced alveolar remodeling is dependent on ERK1/2 and STAT6 signaling. They also demonstrate that IL-13–induced mucus metaplasia is largely STAT6-dependent and ERK1/2-independent.
Figure 9.
Role of STAT6 in IL-13–induced tissue remodeling responses. IL-13– and CC10–rtTA–IL-13 Tg+ (IL-13+) mice with wild-type or null STAT6 loci were generated. Lungs were processed for (A) alveolar histology (H&E), (B) lung volume, (C) alveolar chord length, (D) PAS with diastase staining, (E) mucin gene expression, (F) BAL-fluid Muc-5ac content. Arrow highlights residual PAS+ epithelial cells in IL-13+/STAT6–/– lungs. Original magnification, ×10 (A); ×40 (D). For E, β-actin mRNA expression was used as loading control. Graphical values illustrate the mean ± SEM and represent n = 4–5 for each group. *P < 0.05 compared with IL-13+/STAT6+/+ mice.
Role of ERK1/2 MAPK in IL-13 regulation of proteases and antiproteases.
Our previous studies demonstrated that IL-13 causes MMP- and cathepsin-dependent lung remodeling and inhibits the expression of α1–AT (4). Therefore, studies were undertaken to define the effects of PD and dnMEK1 on MMP, cathepsin, and α1-antitrypsin (α1-AT) expression in our modeling systems. In accord with our prior report (4), the levels of mRNA encoding MMP-2, -9, -12, and -14, cathepsin S, and cathepsin B were increased while the levels of mRNA encoding α1-AT were decreased in dox-treated CC10-rtTA–IL-13 Tg+ mice compared with IL-13 Tg– control animals (Figure 10). PD administration decreased the levels of mRNA encoding each of these MMPs and cathepsin B and increased the levels of mRNA encoding α1-AT in lungs from dox-treated Tg+ mice (Figure 10, A and C). Consistent with these findings, the dnMEK1 construct also decreased the levels of expression of these MMPs and cathepsin B and increased the expression of α1-AT in lungs from Tg+ but not Tg– animals. (Figure 10, B and D). Neither ERK1/2 intervention, however, altered the expression of cathepsin S (Figure 10). Thus, these chemical and genetic studies demonstrate that IL-13 stimulates MMPs and cathepsins and inhibits α1-AT via pathways that are at least partially dependent on ERK1/2 signaling.
Figure 10.
Role of ERK1/2 MAPK in IL-13 regulation of proteases and antiproteases. (A–D) Whole-lung RNA was used for RT-PCR (A and B) and real-time RT-PCR (C and D) to detect mRNA for proteases and anti-proteases in IL-13 (–) and IL-13 (+) mice. Comparisons are made of mice treated with PD or vehicle control (A and C) and mice with and without dnMEK1 (B and C). β-actin mRNA expression was used as a loading control. Data are expressed as mean ± SEM and represent n = 4–5 for each group. *P < 0.05 compared with IL-13+.
Role of STAT6 in IL-13 regulation of proteases and antiproteases.
To clarify the pathways that IL-13 uses to regulate proteases and antiproteases, the importance of STAT6 in these molecular events was also evaluated. In accord with the ERK1/2 interventions, STAT6-null mutations also decreased the levels of mRNA encoding MMP-2, -9, -12, and -14 and cathepsin B and increased the levels of mRNA encoding α1-AT in lungs from dox-treated CC10–rtTA–IL-13 Tg mice (Figure 11, A and B). Interestingly, the contributions of the ERK1/2 and STAT6 pathways to these regulatory events were similar in some circumstances and different in others. Specifically, the ERK1/2- and STAT6-based interventions both decreased IL-13 induction of cathepsin B to a comparable degree (Figures 10 and 11). As a quantitative contrast, PD and dnMEK1 decreased MMP-12 by approximately 50% whereas STAT6-null mutations diminished MMP-12 by approximately 80% as quantified by real-time RT-PCR (Figure 11B). Qualitative differences were also seen, with only STAT6 contributing to IL-13 induction of cathepsin S (Figure 11). Thus, ERK1/2 and STAT6 signaling both play important roles in IL-13 regulation of proteases and antiproteases in the murine lung.
Figure 11.
Role of STAT6 in IL-13 regulation of proteases and antiproteases. IL-13 (–) and CC10–rtTA–IL-13 Tg (+) mice with wild-type or null STAT6 loci were generated. Protease and antiprotease mRNA levels were assessed using RT-PCR (A) and real-time RT-PCR (B). β-actin mRNA expression was used as a loading control. Data are expressed as mean ± SEM and represent n = 4–5 for each group. *P < 0.05 compared with IL-13+/STAT6+/+ mice.
Discussion
IL-13 is a powerful stimulator of inflammation and tissue remodeling and is a key effector cytokine at sites of Th2 inflammation. In these responses, IL-13 is purported to use the canonical STAT6 signaling pathway to mediate its tissue inflammatory and structural effects. This widely held belief is the result of the well-documented ability of IL-13 to activate STAT6 in a variety of tissues (20, 27). In addition, studies from a number of laboratories have demonstrated that many of the effects of intratracheal IL-13 and Tg IL-13 are diminished in STAT6-null mice (7, 13, 14). However, IL-13 can also activate MAPKs in cell lines in vitro (20, 22, 23). These observations led us to hypothesize that IL-13 might also signal in vivo via MAPK-dependent pathways. To address this hypothesis, we took advantage of Tg systems established in our laboratory in which IL-13 can be selectively targeted to the murine lung (4–6). The current studies demonstrate, we believe for the first time, that IL-13 is a potent and early stimulator of ERK1/2 MAPK. Our data also demonstrate that ERK1/2 activation is a proximal event in the IL-13 pathway that occurs in the absence of STAT6. Lastly, we demonstrate, using chemical and genetic approaches, that ERK1/2 MAPK activation plays a critical role in IL-13–induced inflammation and alveolar remodeling.
In accord with the importance of IL-13 in inflammation and disease, many studies have sought to define the mechanisms of IL-13–induced tissue alterations. These studies have demonstrated that the effects of IL-13 are mediated to a great extent by its ability to regulate a number of downstream genes and effector pathways (6, 26, 28–30). Prominent in this regard are studies from our laboratory that demonstrate that IL-13 is a powerful stimulator of macrophage and epithelial chemokines and that chemokine signaling via CC chemokine receptor 2 (CCR2) and CCR1 is required for optimal IL-13–induced inflammation and remodeling (6, 26). Given that we found significant ERK1/2 activation in IL-13 Tg mice, studies were undertaken to define the role(s) of this activation in IL-13–induced inflammation. These studies demonstrate that chemical (PD) and genetic approaches preventing ERK1/2 activation diminish IL-13–induced tissue inflammation. They also provide insight into potential mechanisms of these inflammatory alterations by demonstrating that ERK1/2 activation plays an essential role in IL-13 stimulation of MIP-1α/CCL-3, MIP-1β/CCL-4, and MIP-2/CXCL-1. Interestingly, these effects were chemokine-specific because eotaxin/CCL-11, C10/CCL-6, and MCP-1/CCL-2 were not altered by these interventions.
As noted above, our studies demonstrate that ERK1/2-based interventions decrease tissue and BAL-fluid eosinophilia without altering IL-13 stimulation of eotaxin/CCL-11. This is surprising in light of the well-known ability of eotaxin/CCL-11 to recruit and activate eosinophils (31–33). However, studies from our laboratory and others have demonstrated that IL-13–induced eosinophilia and eotaxin/CCL-11 production can be dissociated. In our experiments, the neutralization of C10/CCL6 and the genetic ablation of its putative receptor, CCR1, both diminished IL-13–induced BAL-fluid and tissue eosinophilia without altering eotaxin/CCL-11 induction (26). Similarly, Yang et al. demonstrated that IL-13 induces eosinophilia via a mechanism that is independent of eotaxin/CCL-11 and IL-5 (13). In evaluating these findings, it is important to appreciate that eosinophil chemotaxis is also regulated by chemokines such as MIP-1α/CCL-3, MIP-1β/CCL-4, and RANTES/CCL-5 (34, 35). In accord with these observations, our ERK1/2-based interventions diminished the production of all 3 of these moieties. When our studies and the literature are viewed in combination, it is clear that eotaxin/CCL-11 is induced by IL-13 via a mechanism that is largely ERK1/2, C10/CCL6, and CCR1 independent. They also demonstrate that chemokines other than eotaxin/CCL-11 are required for maximal IL-13–induced BAL-fluid and tissue eosinophil responses.
Our studies demonstrate that chemical and genetic interventions blocking ERK1/2 MAPK activation ameliorate IL-13–induced alveolar remodeling. These effects could be caused by a decrease in IL-13 production or an alteration in IL-13 effector function(s). Our results suggest that effector mechanisms were altered because similar levels of IL-13 were found in BAL fluids from Tg+ mice treated with PD or its vehicle control and Tg+ mice in which dnMEK1 was or was not expressed. A variety of effector pathways might contribute to these findings, including the diminished production of important proteases or the enhanced production of antiproteases. Our studies demonstrate that the ERK1/2 MAPK pathway plays important roles in the regulation of each of these moieties. Specifically, ERK1/2 activation was required for optimal IL-13 stimulation of MMP-2, -9, -12, and -14, and cathepsin B and IL-13 inhibition of α1-AT. These are, to our knowledge, the first studies to demonstrate the importance of ERK1/2 or any MAPK in the pathogenesis of IL-13–induced tissue remodeling responses. We believe they are also the first to demonstrate that IL-13 stimulates MMPs and cathepsin B and inhibits α1-AT via ERK1/2 MAPK-dependent pathways. These findings have important implications for diseases in which IL-13 dysregulation, protease excess, and tissue destruction coexist. They have particularly intriguing implications for pulmonary emphysema since cigarette smoke exposure has been shown to induce IL-13 elaboration in the murine lung (36), polymorphisms in IL-13 have been associated with emphysema (37), and mice exposed to cigarette smoke as well as lung tissues from patients with emphysema exhibit increased levels of ERK1/2 MAPK activation (38).
To define the role(s) of ERK1/2 MAPK in the pathogenesis of IL-13–induced tissue responses, we initially used the MEK1 inhibitor PD and its corresponding vehicle control. Because the specificity of pharmacologic inhibitors can always be questioned, we also used genetic approaches to address this issue. To accomplish this, we generated IL-13 Tg+ mice that expressed and did not express dnMEK1. As shown in Figure 3, mice that expressed this MEK1 inactivating construct were unable to activate downstream ERK1/2 MAPK in our modeling system. Importantly, identical results were noted with the chemical and genetic approaches. Specifically, both interventions decreased IL-13–induced inflammation, alveolar remodeling, MIP-1α/CCL-3, MIP-1β/CCL-4, RANTES/CCL-5, and MIP-2/CXCL-1 production, and MMP-2, -9, -12, and -14 and cathepsin B mRNA accumulation while increasing α1-AT expression. These mutually supportive studies demonstrate that ERK1/2 MAPK plays critical roles in these responses. Since the dnMEK1 construct is selectively targeted to the epithelial cells in our Tg mice, these studies also suggest that ERK1/2 signaling in lung epithelia is particularly crucial in these responses.
It is important to point out that in most cases, ERK1/2 inhibition decreased but did not totally reverse IL-13–induced tissue and molecular alterations. Given that we show virtually complete ERK1/2 inactivation with PD or dnMEK1 (see Figure 3), we conclude that there are aspects of the IL-13 phenotype that are mediated, at least in part, by alternative signaling pathways. Previous studies from our laboratory and our associates demonstrated that STAT6 also plays an important role in the pathogenesis of selected IL-13–induced tissue events (7, 14). As a result, studies were undertaken to compare the relative contributions of ERK1/2 and STAT6 signaling in the tissue and regulatory effects of IL-13. These studies highlight complex relationships between ERK1/2 and STAT6 in the genesis of these responses. This can be easily seen in IL-13–induced inflammation where ERK1/2 and STAT6 play similar roles in tissue and BAL-fluid eosinophilia but only STAT6 contributes to lymphocyte and neutrophil accumulation. In accord with this finding, IL-13 stimulated pulmonary chemokines via pathways that were equally dependent on ERK1/2 and STAT6, largely dependent on STAT6 and largely dependent on ERK1/2. Similarly, both the ERK1/2 and STAT6 pathways contributed to IL-13–induced alveolar remodeling, with each pathway contributing in specific ways to the regulation of the proteases and antiproteases that mediate these responses. In contrast, STAT6 was the dominant pathway, and ERK1/2 played a minimal role in the pathogenesis of IL-13–induced mucus metaplasia. STAT6 was not, however, the only signaling pathway involved in these responses because null mutations of this gene did not completely abolish IL-13–induced goblet cell hyperplasia or Muc-5ac and Gob-5 expression. These are, to our knowledge, the first studies to appreciate the importance of ERK1/2 MAPK pathways in the pathogenesis of the in vivo effects of IL-13. We believe they are also the first to define the effector repertoires of the ERK1/2 and STAT6 pathways in this setting. Collectively, they demonstrate that the ERK1/2 and STAT6 pathways play similar and dissimilar roles in the pathogenesis of the tissue and molecular effects of IL-13. These observations provide a clear rationale for the use of ERK1/2 MAPK pathway inhibitors, alone or in combination with STAT6 inhibitors, in the treatment of IL-13–induced disorders.
In summary, our studies demonstrate that IL-13 is a potent activator of ERK1/2 MAPK and that this activation takes place even in the absence of STAT6. They also demonstrate that ERK1/2 MAPK activation plays a critical role(s) in the pathogenesis of IL-13–induced inflammation and alveolar remodeling and that the effects of this activation can be similar to and different from those mediated by STAT6. Lastly, they provide mechanistic insights by demonstrating that ERK1/2 MAPK activation is required for optimal IL-13 stimulation of chemokines (MIP-1α/CCL-3, MIP-1β/CCL-4, MIP-2/CXCL-1, and RANTES/CCL-5) and proteases (MMP-2, -9, -12, and -14 and cathepsin B) and IL-13 inhibition of α1-AT. Exaggerated IL-13 production has been implicated in the pathogenesis of a wide variety of disorders, including asthma, chronic obstructive pulmonary disease, pulmonary fibrosis, scleroderma, hepatic fibrosis, and nodular sclerosing Hodgkin disease. The present studies suggest that the effects of IL-13 in these disorders may be beneficially controlled via interventions that control and/or prevent the activation of ERK1/2 MAPK. This establishes the ERK1/2 MAPK pathway as a worthwhile site for future investigation designed to identify therapeutic agents that can be used to treat these and other IL-13–mediated disorders.
Methods
Tg and genetically modified mice.
CC10–rtTA–IL-13 Tg+ and Tg– littermate mice generated in our laboratory were used (4). These are dual Tg+ animals on a C57Bl/6 background in which rtTA drives the expression of the murine IL-13 gene in a lung-specific and externally regulatable fashion. The Tg is activated by adding dox to the animals’ drinking water. The details of both genetic constructs, the methods of microinjection and genotype evaluation, Tg inducibility, and the emphysematous and inflammatory phenotype of CC10–rtTA–IL-13 mice have been described previously (4). CC10-dnMEK1 Tg mice were generated using a dnMEK1 sequence in which serines 218 and 222 had been mutated to valines (39). Eliminating these critical activation loop phosphorylation sites inhibits activation of dnMEK1 by upstream kinases. Lungs, hearts, and livers from Tg-positive founders were screened by RT-PCR to confirm expression of the dnMEK1 Tg selectively in lung (see below for details of the dnMEK1 primer sequences). The dnMEK1 sequence was flanked by a rat CC10 promoter at the 5′ end (kindly provided by Jeffrey Whitsett, Children’s Hospital of Cincinnati, Cincinnati, Ohio, USA; ref. 40) and a human growth hormone polyadenylation sequence at the 3′ end. Mice were bred for 10 or more generations onto the C57BL6 background. CC10–rtTA–IL-13/dnMEK1 Tg mice were generated by breeding the IL-13+ mice with the dnMEK1+ mice. PCR was used to define the Tg status of all offspring using primers that detected rtTA and/or the junction region of murine IL-13–human growth hormone construct for IL-13 mice. For IL-13/dnMEK1 Tg detection, in addition to the primers for rtTA and the IL-13–human growth hormone region, we used the following primers: sense, 5′-TCGGGAGAAGCACAAGAT-3′; antisense, 5′-GAGCTGTTTGTTTTTCTCTCTCC-3′.
IL-13+ mice with wild-type (+/+) or null (–/–) STAT6 loci were generated by breeding the CC10–rtTA–IL-13 or constitutive CC10–IL-13 mice (5) with STAT6-null mutant (STAT6–/–) mice (Jackson Laboratory). Except for experiments shown in Figure 2A, the inducible CC10–rtTA–IL-13 mice were used for these studies. The mice that were used in these experiments were bred for at least 10 generations onto a pure C57Bl/6 background. All experiments were approved by the Institutional Animal Care and Use Committee of Yale University.
Dox water administration.
In experiments performed with CC10–rtTA–IL-13 Tg+ mice and their littermate controls, all mice were maintained on normal water until they were 4–6 weeks old. They were then randomized to receive either normal water or water with dox for the duration of the experiment. Dox was administered at 500 mg/l in 4% sucrose and kept in dark bottles to prevent light-induced degradation.
Administration of PD.
The MEK1 inhibitor PD (5 mg/kg body wt/d) (EMD Biosciences) was administered to IL-13 Tg mice and wild-type littermates by intraperitoneal injection. PD administration started 1 day prior to administration of dox water and was repeated daily.
Lung volume, histologic evaluation, and morphometric analysis.
Lung volume was assessed as previously described (4). In brief, mice were anesthetized, the trachea was cannulated, and the lungs were removed and inflated with PBS at 25 cm H2O. The size of the lung was evaluated by volume displacement. H&E or PAS with diastase staining was performed after pressure fixation with Streck solution (Streck) in the Research Pathology Laboratory at Yale University. Alveolar size was estimated from the mean chord length of the airspace as previously described by our laboratory (4).
Immunohistochemistry.
Formalin-fixed, paraffin-embedded lung tissue sections were deparafinized with xylene, rehydrated gradually with graded alcohol solutions (100%, 95%, and 80%), and then washed with deionized water. For antigen unmasking, sections were incubated with 0.1% trypsin for 10 minutes followed by incubation with 3% H2O2 in methanol for 10 minutes to block endogenous peroxidase activity. Sections were then incubated with a 1:4000 dilution of rabbit anti-MBP antibody (gift from J. Lee, Mayo Clinic, Scottsdale, Arizona, USA) for 1 hour at 37°C. After 3 PBS washes, sections were incubated with anti-rabbit–HRP antibody (ImmunoVision Technologies Co.) at 37°C for 30 minutes. Diaminobenzidine substrate (DakoCytomation) was applied as the chromogen, resulting in a brown reaction product, and the sections were counterstained with Mayer’s hematoxylin. PBS was used instead of the primary antibody as negative control for nonspecific binding. Eosinophil quantification was accomplished by counting the number of MBP+ cells in the tissues in 20 random fields from 10 random sections from each lung at amplification ×400.
Western blot analysis.
Lung protein analyses were performed as previously described with minor modifications (41). Whole-lung lysates were homogenized in 62.5 mM Tris buffer, and the protein concentrations of the lysates were determined by BCA Protein Assay (Pierce Biotechnology Inc.). Samples were electrophoresed in a 12% ready-made Tris-HCl gel (Bio-Rad) and electrophoretically transferred onto a nitrocellulose membrane. The membranes were then incubated overnight with rabbit anti–phospho-p38, phospho-ERK, phospho-JNK, phospho-STAT6 (Tyr651), total p38, total ERK, total JNK, total STAT6, or β-tubulin polyclonal antibody (1:1,000 dilution; Cell Signaling Technology). The membranes were incubated with HRP-conjugated goat anti-rabbit IgG antibody followed by the detection of signal with a chemiluminescence LumiGLO detection kit (Cell Signaling Technology).
Quantification of IL-13 and IL-13–regulated chemokines.
The levels of IL-13, MIP-1α/CCL-3, MIP-1β/CCL-4, MIP-2, eotaxin, MCP-1/CCL-2, and RANTES/CCL-5 in BAL fluids were quantitated using commercial ELISA kits (R&D Systems) according to the manufacturer’s instructions as described previously (6).
Quantification of mucin secretion.
The secretion of mucin protein into BAL fluids was evaluated by quantifying the levels of Muc-5ac in BAL fluids using slot blots and immunostaining, as previously described (42). Briefly, equal volumes of BAL fluids from mice were loaded onto a nitrocellulose membrane within the slot blot device. The membrane was incubated overnight with mouse anti–Muc-5ac antibody (1:500 dilution; Lab Vision Corp.). The membranes were then incubated with HRP-conjugated goat anti-mouse IgG antibody followed by luminol reagent (Santa Cruz Biotechnology Inc.).
mRNA analysis.
mRNA levels were evaluated with RT-PCR evaluations using whole lung RNA and primers as previously described (4). Amplified PCR products were detected using ethidium bromide gel electrophoresis.
In selected experiments, real-time RT-PCR was used. These evaluations were performed using a QuantiTect SYBR Green RT-PCR kit (QIAGEN) according to the instructions provided by the manufacturer. In these evaluations, reactions were made by a combination of 12.5 μl SYBR RT-PCR Master Mix (QIAGEN), 0.25 μl QuantiTect RT Mix (QIAGEN), 1 μl upstream primer, 1 μl downstream primer, 8.75 μl RNase-free water, and 1.5 μl (200 ng/μl) RNA template. A negative control containing no RNA template was introduced in each run. Mouse β-actin gene was amplified as an internal control. RT-PCR was performed using ABI sequence Detection System (Applied Biosystems), in which the mixture was heated to 50°C for 30 minutes for reverse transcription and 95°C for 15 minutes, then cycled 40 times at 94°C for 15 seconds, 60°C for 30 seconds, and 72°C for 30 seconds. The relative quantification values for MMP-12 (5′-CATTTCGCCTCTCTGCTGATG-3′ and 5′-TTGATGGTGGACTGCTAGGTTTT-3′), cathepsin B (5′-GCAGGACTTCCAAAAGAACGA-3′ and 5′-GACGAATGCCTGCCACAAG-3′), cathepsin S (5′-GAGACCCTACCCTGGACTACCA-3′) and β-actin (5′-GTGGGCCGCTCTAGGCACCA-3′ and 5′-TGGCCTTAGGGTTCAGGGGG-3′) gene expression were calculated from the accurate threshold cycle (CT), which is the PCR cycle at which an increase in reporter fluorescence from SYBR green dye can first be detected above a baseline signal. The CT values for β-actin were subtracted from the CT values for MMP-12, cathepsin B, and cathepsin S in each well to calculate ΔCT. The triplicate ΔCT values for each sample were averaged. To calculate the fold induction of MMP-12, cathepsin B, and cathepsin S mRNA over controls (ΔΔCT), the average ΔCT values calculated for control animals were subtracted from ΔCT values calculated for IL-13 Tg mice treated with PD, IL-13 Tg mice treated with dnMEK1, or STAT6–/– animals. Then the fold induction for each well was calculated by using 2–(ΔΔCT) formula. The fold induction values for triplicate wells were averaged, and data were presented as the mean ± SEM of triplicate wells.
Statistics.
Data are expressed as mean ± SEM and were analyzed by 2-tailed Student’s t test. Significance was accepted at P < 0.05.
Acknowledgments
We thank Susan Ardito and Kathy Bertier for their administrative and editorial assistance. P.J. Lee was supported by NIH grant HL04034, an American Heart Association Heritage Affiliate grant, and an American Lung Association Career Investigator Award. Z. Zhu was supported by NIH grant HL074095 and an American Lung Association Grant. The CC10-dnMEK1 mice were created with the support of Linda Weiss, Raymond Robledo, and NIH grant P01-HL67004 (to B.T. Mossman). J.A. Elias was supported by NIH grants HL-64242, HL-78744, HL-66571, and HL-56389.
Footnotes
Nonstandard abbreviations used: α1-AT, α1-antitrypsin; BAL, bronchoalveolar lavage; CC10, Clara cell 10; CCR, CC chemokine receptor; CT, threshold cycle; dnMEK1, dominant-negative MEK1; dox, doxycycline; MBP, major basic protein; MEK1, MAPK/ERK kinase 1; PD, PD98059; rtTA, reverse tetracycline transactivator.
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Elias JA, Lee CG, Zheng T, Shim Y, Zhu Z. Interleukin-13 and leukotrienes: an intersection of pathogenetic schema. Am. J. Respir. Cell Mol. Biol. 2003;28:401–404. doi: 10.1165/rcmb.F264. [DOI] [PubMed] [Google Scholar]
- 2.Wills-Karp M, et al. Interleukin-13: central mediator of allergic asthma. Science. 1998;282:2258–2261. doi: 10.1126/science.282.5397.2258. [DOI] [PubMed] [Google Scholar]
- 3.Grunig G, et al. Requirement for IL-13 independently of IL-4 in experimental asthma. Science. 1998;282:2261–2263. doi: 10.1126/science.282.5397.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng T, et al. Inducible targeting of IL-13 to the adult lung causes matrix metalloproteinase- and cathepsin-dependent emphysema. J. Clin. Invest. 2000;106:1081–1093. doi: 10.1172/JCI10458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu Z, et al. Pulmonary expression of interleukin-13 causes inflammation, mucus hypersecretion, subepithelial fibrosis, physiologic abnormalities, and eotaxin production. J. Clin. Invest. 1999;103:779–788. doi: 10.1172/JCI5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu Z, et al. IL-13-induced chemokine responses in the lung: role of CCR2 in the pathogenesis of IL-13-induced inflammation and remodeling. J. Immunol. 2002;168:2953–2962. doi: 10.4049/jimmunol.168.6.2953. [DOI] [PubMed] [Google Scholar]
- 7.Kuperman DA, et al. Direct effects of interleukin-13 on epithelial cells cause airway hyperreactivity and mucus overproduction in asthma. Nat. Med. 2002;8:885–889. doi: 10.1038/nm734. [DOI] [PubMed] [Google Scholar]
- 8.Blease K, et al. Enhanced pulmonary allergic responses to Aspergillus in CCR2–/– mice. J. Immunol. 2000;165:2603–2611. doi: 10.4049/jimmunol.165.5.2603. [DOI] [PubMed] [Google Scholar]
- 9.Chiaramonte MG, Donaldson DD, Cheever AW, Wynn TA. An IL-13 inhibitor blocks the development of hepatic fibrosis during a T-helper type 2-dominated inflammatory response. J. Clin. Invest. 1999;104:777–785. doi: 10.1172/JCI7325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hancock A, Armstrong L, Gama R, Millar A. Production of interleukin 13 by alveolar macrophages from normal and fibrotic lung. Am. J. Respir. Cell Mol. Biol. 1998;18:60–65. doi: 10.1165/ajrcmb.18.1.2627. [DOI] [PubMed] [Google Scholar]
- 11.Ohshima K, et al. Interleukin-13 and interleukin-13 receptor in Hodgkin’s disease: possible autocrine mechanism and involvement in fibrosis. Histopathology. 2001;38:368–375. doi: 10.1046/j.1365-2559.2001.01083.x. [DOI] [PubMed] [Google Scholar]
- 12.Mattes J, et al. IL-13 induces airways hyperreactivity independently of the IL-4R alpha chain in the allergic lung. J. Immunol. 2001;167:1683–1692. doi: 10.4049/jimmunol.167.3.1683. [DOI] [PubMed] [Google Scholar]
- 13.Yang M, et al. Interleukin-13 mediates airways hyperreactivity through the IL-4 receptor-alpha chain and STAT-6 independently of IL-5 and eotaxin. Am. J. Respir. Cell Mol. Biol. 2001;25:522–530. doi: 10.1165/ajrcmb.25.4.4620. [DOI] [PubMed] [Google Scholar]
- 14.Kuperman D, Schofield B, Wills-Karp M, Grusby MJ. Signal transducer and activator of transcription factor 6 (Stat6)-deficient mice are protected from antigen-induced airway hyperresponsiveness and mucus production. J. Exp. Med. 1998;187:939–948. doi: 10.1084/jem.187.6.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blenis J. Signal transduction via the MAP kinases: proceed at your own RSK. Proc. Natl. Acad. Sci. U. S. A. 1993;90:5889–5892. doi: 10.1073/pnas.90.13.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis RJ. The mitogen-activated protein kinase signal transduction pathway. J. Biol. Chem. 1993;268:14553–14556. [PubMed] [Google Scholar]
- 17.Davis RJ. MAPKs: new JNK expands the group. Trends Biochem. Sci. 1994;19:470–473. doi: 10.1016/0968-0004(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 18.Duan W, Chan JH, Wong CH, Leung BP, Wong WS. Anti-inflammatory effects of mitogen-activated protein kinase kinase inhibitor U0126 in an asthma mouse model. J. Immunol. 2004;172:7053–7059. doi: 10.4049/jimmunol.172.11.7053. [DOI] [PubMed] [Google Scholar]
- 19.Pahl A, Zhang M, Kuss H, Szelenyi I, Brune K. Regulation of IL-13 synthesis in human lymphocytes: implications for asthma therapy. Br. J. Pharmacol. 2002;135:1915–1926. doi: 10.1038/sj.bjp.0704656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.David M, Ford D, Bertoglio J, Maizel AL, Pierre J. Induction of the IL-13 receptor alpha2-chain by IL-4 and IL-13 in human keratinocytes: involvement of STAT6, ERK and p38 MAPK pathways. Oncogene. 2001;20:6660–6668. doi: 10.1038/sj.onc.1204629. [DOI] [PubMed] [Google Scholar]
- 21.Hirst SJ, Hallsworth MP, Peng Q, Lee TH. Selective induction of eotaxin release by interleukin-13 or interleukin-4 in human airway smooth muscle cells is synergistic with interleukin-1beta and is mediated by the interleukin-4 receptor alpha-chain. Am. J. Respir. Crit. Care Med. 2002;165:1161–1171. doi: 10.1164/ajrccm.165.8.2107158. [DOI] [PubMed] [Google Scholar]
- 22.Laporte JC, et al. Direct effects of interleukin-13 on signaling pathways for physiological responses in cultured human airway smooth muscle cells. Am. J. Respir. Crit. Care Med. 2001;164:141–148. doi: 10.1164/ajrccm.164.1.2008060. [DOI] [PubMed] [Google Scholar]
- 23.Moore PE, Church TL, Chism DD, Panettieri RA, Jr, Shore SA. IL-13 and IL-4 cause eotaxin release in human airway smooth muscle cells: a role for ERK. Am. J. Physiol. Lung Cell. Mol. Physiol. 2002;282:L847–L853. doi: 10.1152/ajplung.00245.2001. [DOI] [PubMed] [Google Scholar]
- 24.Peng Q, Matsuda T, Hirst SJ. Signaling pathways regulating interleukin-13-stimulated chemokine release from airway smooth muscle. Am. J. Respir. Crit. Care Med. 2004;169:596–603. doi: 10.1164/rccm.200307-888OC. [DOI] [PubMed] [Google Scholar]
- 25.Xu B, et al. Interleukin-13 induction of 15-lipoxygenase gene expression requires p38 mitogen-activated protein kinase-mediated serine 727 phosphorylation of Stat1 and Stat3. Mol. Cell. Biol. 2003;23:3918–3928. doi: 10.1128/MCB.23.11.3918-3928.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma B, et al. The C10/CCL6 chemokine and CCR1 play critical roles in the pathogenesis of IL-13-induced inflammation and remodeling. J. Immunol. 2004;172:1872–1881. doi: 10.4049/jimmunol.172.3.1872. [DOI] [PubMed] [Google Scholar]
- 27.Nelms K, Keegan AD, Zamorano J, Ryan JJ, Paul WE. The IL-4 receptor: signaling mechanisms and biologic functions. Annu. Rev. Immunol. 1999;17:701–738. doi: 10.1146/annurev.immunol.17.1.701. [DOI] [PubMed] [Google Scholar]
- 28.Blackburn MR, et al. Adenosine mediates IL-13-induced inflammation and remodeling in the lung and interacts in an IL-13-adenosine amplification pathway. J. Clin. Invest. 2003;112:332–344. doi:10.1172/JCI200316815. doi: 10.1172/JCI16815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lanone S, et al. Overlapping and enzyme-specific contributions of matrix metalloproteinases-9 and -12 in IL-13-induced inflammation and remodeling. J. Clin. Invest. 2002;110:463–474. doi:10.1172/JCI200214136. doi: 10.1172/JCI14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee CG, et al. Interleukin-13 induces tissue fibrosis by selectively stimulating and activating transforming growth factor beta(1) J. Exp. Med. 2001;194:809–821. doi: 10.1084/jem.194.6.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mould AW, et al. The effect of IL-5 and eotaxin expression in the lung on eosinophil trafficking and degranulation and the induction of bronchial hyperreactivity. J. Immunol. 2000;164:2142–2150. doi: 10.4049/jimmunol.164.4.2142. [DOI] [PubMed] [Google Scholar]
- 32.Rothenberg ME, MacLean JA, Pearlman E, Luster AD, Leder P. Targeted disruption of the chemokine eotaxin partially reduces antigen-induced tissue eosinophilia. J. Exp. Med. 1997;185:785–790. doi: 10.1084/jem.185.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rothenberg ME, et al. Eotaxin triggers eosinophil-selective chemotaxis and calcium flux via a distinct receptor and induces pulmonary eosinophilia in the presence of interleukin 5 in mice. Mol. Med. 1996;2:334–348. [PMC free article] [PubMed] [Google Scholar]
- 34.Oliveira SH, et al. Increased responsiveness of murine eosinophils to MIP-1beta (CCL4) and TCA-3 (CCL1) is mediated by their specific receptors, CCR5 and CCR8. J. Leukoc. Biol. 2002;71:1019–1025. [PubMed] [Google Scholar]
- 35.Rot A, et al. RANTES and macrophage inflammatory protein 1 alpha induce the migration and activation of normal human eosinophil granulocytes. J. Exp. Med. 1992;176:1489–1495. doi: 10.1084/jem.176.6.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bartalesi B, et al. Different lung responses to cigarette smoke in two strains of mice sensitive to oxidants. Eur. Respir. J. 2005;25:15–22. doi: 10.1183/09031936.04.00067204. [DOI] [PubMed] [Google Scholar]
- 37.van der Pouw Kraan TC, et al. Chronic obstructive pulmonary disease is associated with the -1055 IL-13 promoter polymorphism. Genes Immun. 2002;3:436–439. doi: 10.1038/sj.gene.6363896. [DOI] [PubMed] [Google Scholar]
- 38.Mercer BA, Kolesnikova N, Sonett J, D’Armiento J. ERK MAP kinase is upregulated in pulmonary emphysema and mediates MMP-1 induction by cigarette smoke. J. Biol. Chem. 2004;279:17690–17696. doi: 10.1074/jbc.M313842200. [DOI] [PubMed] [Google Scholar]
- 39.Alessandrini A, Greulich H, Huang W, Erikson RL. Mek1 phosphorylation site mutants activate Raf-1 in NIH 3T3 cells. J. Biol. Chem. 1996;271:31612–31618. doi: 10.1074/jbc.271.49.31612. [DOI] [PubMed] [Google Scholar]
- 40.Stripp BR, et al. cis-acting elements that confer lung epithelial cell expression of the CC10 gene. J. Biol. Chem. 1992;267:14703–14712. [PubMed] [Google Scholar]
- 41.Zhang X, et al. Mitogen-activated protein kinases regulate HO-1 gene transcription after ischemia-reperfusion lung injury. Am. J. Physiol. Lung Cell. Mol. Physiol. 2002;283:L815–L829. doi: 10.1152/ajplung.00485.2001. [DOI] [PubMed] [Google Scholar]
- 42.McGuckin MA, Thornton DJ. Detection and quantitation of mucins using chemical, lectin, and antibody methods. Methods Mol. Biol. 2000;125:45–55. doi: 10.1385/1-59259-048-9:045. [DOI] [PubMed] [Google Scholar]