Abstract
Insulin receptor substrates, including Irs1 and Irs2, integrate insulin and IGF receptor signals with heterologous pathways to coordinate growth and metabolism. Since Irs2 is thought to be especially important in hepatic nutrient homeostasis, we deleted Irs1 from hepatocytes of WT mice (called LKO) or genetically insulin-resistant Irs1–/– mice (called LKO::Irs1–/–). Viable LKO::Irs1–/– mice were 70% smaller than WT or LKO mice, and 40% smaller than Irs1–/– mice. Hepatic insulin receptors were functional in all the mice, but insulin signaling via the Akt—FoxO1 pathway was reduced in Irs1–/– and LKO liver, and undetected in LKO::Irs1–/– liver; however, Gsk3β phosphorylation (Ser9) and hepatic glycogen stores were nearly normal in all of the mice. LKO and Irs1–/– mice developed insulin resistance and glucose intolerance that never progressed to diabetes, whereas LKO::Irs1–/– mice developed hyperglycemia and hyperinsulinemia immediately after birth. Regardless, few hepatic genes changed expression significantly in Irs1–/– or LKO mice, whereas hundreds of genes changed in LKO::Irs1–/– mice — including elevated levels of Pck1, G6pc, Ppargc1, Pparg, and Igfbp1. Thus, signals delivered by Irs1 or Irs2 regulate hepatic gene expression that coordinates glucose homeostasis and systemic growth.
Introduction
The liver is central to nutrient homeostasis, because it stores glucose after a meal as glycogen or converts excess glucose to fatty acids, while oxidizing fatty acids to provide energy for gluconeogenesis during fasting (1). The integration of insulin signals with those generated by counterregulatory hormones and neuronal inputs establishes a dynamic network that coordinates systemic nutrient homeostasis (2–4). Whether the consequences of dysregulated hepatic metabolism associated with the metabolic syndrome and diabetes arise directly from insulin resistance or indirectly from changes in nutrient flux is controversial (5). Acute suppression of hepatic insulin receptors does not alter insulin action on glucose production; this suggests that other mechanisms, including neuronal inputs, provide sufficient metabolic regulation (6). Even genetic deletion of the hepatic insulin receptors (the liver-specific insulin receptor knockout [LIRKO] mouse) does not cause diabetes, as fasting glucose levels are nearly normal — although transient hyperglycemia occurs after meals and liver failure develops in old mice (7). Regardless, systemic insulin signaling is essential, as complete deletion of the insulin receptor in mice causes hyperglycemia and death immediately after birth. These lethal consequences of the systemic insulin receptor deletion can be prevented by transgenic expression of the insulin receptor in liver, brain, and β cells, which promotes compensatory hyperinsulinemia that normalizes hepatic glucose production — while other peripheral tissues remain insensitive to insulin (8, 9). Thus, cellular signals regulated by insulin receptors are central to hepatic nutrient flux (10–12).
Insulin signaling is coordinated with counterregulatory signaling through tyrosine phosphorylation of the insulin receptor substrates (Irs’s) — Irs1, Irs2, Irs3, and Irs4 — and other proteins including Shc, APS, SH2B, Gab1/2, Grb isoforms, Dock1/2, and Cbl (13–21). Although the role of each of these substrates merits attention, work with transgenic mice reveals that many insulin responses, especially those that are associated with somatic growth and carbohydrate metabolism, are mediated largely through Irs1 and Irs2 (22, 23). In mice, the Irs2 branch of the insulin/IGF signaling cascade plays an important role in systemic nutrient homeostasis, as it promotes pancreatic β cell function and survival, and central nutrient sensing (24–26). The conditional knockout of Irs2 in mouse pancreas β cells and parts of the brain, including the hypothalamus, increases appetite, lean and fat body mass, linear growth, and insulin resistance that progresses to diabetes (26); however, diabetes resolves between 6 and 10 months of age when functional β cells expressing Irs2 repopulate the pancreas to restore sufficient β cell function to compensate for insulin resistance in the obese mice. Strategies to enhance the Irs2 branch of the insulin/IGF signaling pathway in β cells and hypothalamus could be a rational approach to prevent obesity and diabetes.
Irs2 is an important regulatory point for the PI3K—Akt cascade in hepatocytes (27, 28). Cell-based experimentation with immortalized hepatocytes suggests that Irs2 is the major effector of both the metabolic and the growth-promoting actions of insulin (27). Moreover, Irs2 signaling is essential for the suppression of gluconeogenesis and apoptosis in immortalized neonatal hepatocytes (29, 30). Importantly, Irs2 is highly regulated through feedback and counterregulatory cascades. During nutrient deprivation, hepatic Irs2 expression is upregulated by nuclear forkhead box O1 (FoxO1) and phosphorylated CREB (28, 31). By contrast, SREBPs, transcriptional regulators of lipid synthesis, suppress Irs2 expression, at least in part by interfering with FoxO1 binding to the Irs2 promoter (28). Thus, the depletion of Irs2 signaling can explain the relation between nutrient excess and dysregulated hepatic metabolism.
Although Irs2 appears to play a critical role in hepatic metabolism, transient suppression of hepatic Irs1 or Irs2 expression by more than 70% has only modest effects on murine nutrient metabolism (32). To establish whether signals from Irs1 and Irs2 contribute to hepatic metabolic regulation in vivo, we disrupted the Irs2 gene in hepatocytes of WT mice (liver-specific knockout [LKO] mice) or in hepatocytes of genetically insulin-resistant Irs1–/– mice (LKO::Irs1–/– mice). Contrary to expectations, the LKO mice displayed mild glucose intolerance, as the hepatic insulin resistance was largely compensated for by increased insulin secretion. However, the LKO::Irs1–/– mice developed diabetes with severe fasting hyperinsulinemia immediately after birth. This work reveals the central role of the Irs1 and Irs2 branches of the insulin signaling cascade in hepatic function and metabolic regulation.
Results
Hepatic disruption of Irs2.
Systemic disruption of Irs2 causes insulin resistance in mice that progresses to glucose intolerance and diabetes, owing to a combination of pancreatic β cell failure, hepatic insulin resistance, and dysregulated hypothalamic signaling (23, 25, 26, 33, 34). By contrast, systemic Irs1–/– mice display peripheral insulin resistance that never progresses to diabetes. Hepatic and hypothalamic Irs2 signaling is robust in lean Irs1–/– mice, and β cell hypertrophy ensures the delivery of sufficient insulin throughout life (35–38). To investigate the role of hepatic Irs2 in nutrient homeostasis of WT and genetically insulin-resistant Irs1–/– mice, we disrupted homozygous floxed Irs2 (fIrs2) alleles in hepatocytes using a cre recombinase transgene under control of the rat albumin promoter (crAlb).
The crAlb transgene progressively deletes floxed genes in hepatocytes until complete deletion is achieved at 6 weeks of age (7, 39). Hepatic specificity was confirmed in our experiments by RT-PCR, as Irs2 mRNA was amplified normally in nonhepatic tissues (data not shown) but was undetected in the liver of fIrs2::crAlb mice (called LKO mice throughout this report) at 6 weeks of age, or at 4, 6, or 11 months of age (Figure 1A). Moreover, Irs2 protein detected by specific immunoblotting was expressed at equal levels in WT and fIrs2 liver, but it was undetected in LKO liver (Figure 1B). Irs2 was always detected in muscle and adipose tissues of these mice (data not shown).
Figure 1.
Deletion of the liver Irs2 gene. (A) Irs1 and Irs2 mRNA from WT or LKO livers was analyzed by RT-PCR using gene-specific primers. (B) Irs1 and Irs2 proteins in liver extracts (1 mg) from WT, fIrs2, and LKO mice were identified by immunoblotting with specific antibodies. (C) Identification of Irs1 and Irs2 mRNA in the LKO::Irs1–/–, Irs1–/–, LKO, and fIrs2 liver extracts by RT-PCR. (D) Irs1 and Irs2 proteins in liver extracts (1 mg) from LKO::Irs1–/–, Irs1–/–, LKO, and fIrs2 mice were identified by immunoblotting with specific antibodies. (E) Irs4 mRNA in the livers of LKO::Irs1–/–, Irs1–/–, LKO, and fIrs2 mice was analyzed by real-time PCR. The mean ± SEM of normalized data is plotted.
Irs1–/– mice lacking hepatic Irs2 (LKO::Irs1–/– mice) were generated by intercrossing of fIrs2::crAlb and fIrs2::Irs1+/– mice. LKO littermates were obtained at the expected frequency of 12.5%, whereas one LKO::Irs1–/– littermate was obtained routinely from 5–6 litters of normal size (2.7% yield). To have these drastic effects on fetal development, crAlb apparently deleted fIrs2 alleles in fetal Irs1–/– mice. Regardless, more than half of the viable LKO::Irs1–/– mice survived beyond 4 weeks and were used in our experiments. RT-PCR analysis and specific immunoblotting confirmed the absence of Irs1 and Irs2 mRNA and protein in LKO::Irs1–/– liver (Figure 1, C and D). Irs1 mRNA and protein were expressed normally in LKO and fIrs2 liver but were undetected in Irs1–/– liver (Figure 1, C and D). Hepatic Irs4 expression increases during liver regeneration, suggesting that it could compensate for systemic metabolic stress (40). Compared with that in control fIrs2 mice, Irs4 mRNA quantified by real-time RT-PCR was slightly reduced in Irs1–/– mice but was normal in LKO and LKO::Irs1–/– mice (Figure 1E).
Hepatic gene expression.
Insulin regulates the expression of various hepatic genes involved in nutrient homeostasis and growth control, and Irs2 is thought to play an important role (28, 31). To investigate the effect of Irs2 deletion on hepatic gene expression, we used Affymetrix Mouse Genome 430 2.0 GeneChips to compare mRNA levels in liver extracts from overnight-fasted WT mice versus fIrs2, LKO, Irs1–/–, or LKO::Irs1–/– mice. Two mice were used in each group, and ANOVA was performed across the groups of replicate samples, without multiple testing corrections, using GeneSpring 7.2. This analysis revealed 1,414 probe sets that were statistically different (P < 0.05) across at least 1 group-pair. Next, a 1-sample Student’s t test was used to determine whether the mean normalized expression for these probes was statistically different from the WT. Only 15 probe sets in the fIrs2 sample were significantly different from the WT, and only 1 increased (probe set 141949_at: defensin 1β) or decreased (probe set 1425521_at: polyadenylate binding protein–interacting protein 1) more than 2-fold (Figure 2A). The similarity between WT and fIrs2 was consistent with the indistinguishable metabolic parameters reported previously (26). By contrast, 197 probe sets changed significantly in the Irs1–/– sample, 23 of them more than 2-fold (Figure 2C); 290 probe sets changed significantly in the LKO sample, 33 of them more than 2-fold (Figure 2D); and 970 probe sets changed significantly in the LKO::Irs1–/– sample, 335 of them more than 2-fold (Figure 2B). After correction for redundant probe sets, 254 genes changed significantly, more than 2-fold, in the LKO::Irs1–/– sample; 14 did so in the Irs1–/– sample; and 24 did so in the LKO sample (Table 1).
Figure 2.
Analysis of liver gene expression on Affymetrix Mouse Genome 430 2.0 GeneChips. The mRNA was isolated from the livers of overnight-fasted WT, fIrs2, LKO, Irs1–/–, and LKO::Irs1–/– mice (2 mice per genotype). Each sample was labeled and analyzed on Affymetrix Mouse Genome 430 2.0 GeneChips at the Harvard Medical School Biopolymers Facility. ANOVA without a multiple testing correction identified 1,414 probe sets that changed significantly (P < 0.05) across all of the groups, and a 1-sample Student’s t test was used to determine whether the mean normalized expression value for each of the 1,414 probes was statistically different from the WT control. (A–D) The P value for each probe set was plotted against its fold change (Δ) fIrs2, LKO::Irs1–/–, Irs1–/–, and LKO. Probe sets falling outside the gray zone (P < 0.05 and at least a 2-fold change) are listed in Table 1. (E) Real-time PCR was used to estimate the expression of several candidate genes known to play a role in hepatic glucose homeostasis — Pck1, Glut2, G6pc, Gck, and Ppargc1 — or lipid homeostasis — Pparg, Srebp1, Acc1, Fasn, carnitine palmitoyltransferase 1a (Cpt1a), and Hmgcs1. Data represent the average of replicate determinations from fasted mouse liver extracts normalized against 18S. Expression relative to the fIrs2 liver is presented by log 2 (fold change). *P < 0.05 (ANOVA).
Table 1.
Genes that changed significantly in liver
The binding of nuclear FoxO1 transcription factor to the canonical insulin response element (G/CAAAAT/CAA) is an important mechanism of gene regulation in the liver (41). However, only 36% of the probe sets that changed significantly in the LKO::Irs1–/– samples contain this element within the first 2,500 bp of the promoter region (Table 1). Three genes in this group are known to be induced in hepatocytes by nuclear FoxO1: IGF-binding protein-1 (Igfbp1, 5-fold change compared with control), glucose-6-phosphatase (G6pc, 2-fold change compared with control), and pyruvate dehydrogenase kinase 4 (Pdk4, 21-fold change compared with control) (42, 43). Other genes in this group are expected to regulate hepatic metabolism: Grb10 (5-fold change compared with control), serum- and glucocorticoid-inducible kinase (Sgk, 5-fold change compared with control), PPARγ coactivator 1 (Ppargc1, 8-fold change compared with control), the regulatory subunit of PI3K (Pik3r1, 0.2-fold change compared with control), and the leptin receptor (Lpr, 150-fold change compared with control) (Table 1). Although a complete analysis of these data is beyond the scope of this report, the results show how the systemic deletion of Irs1 or the liver-specific deletion of Irs2 has relatively small effects on liver gene expression by comparison with the dramatic changes that develop in the fasted LKO::Irs1–/– mice.
Next we used RT-PCR to measure the expression of specific candidate genes known to regulate glucose and lipid metabolism (Figure 2E). The expression of phosphoenolpyruvate carboxykinase 1 (Pck1), glucokinase (Gck), and Pparg increased significantly (approximately 2-fold) in LKO liver. In LKO::Irs1–/– liver, G6pc, Pparg, and Ppargc1 increased significantly, Pck1 increased 5 ± 2–fold (P = 0.07), and Gck became undetectable (Figure 2E). In Irs1–/– liver, gene expression was either unchanged or decreased (Glut2, Srebp1, Fasn, and 3-hydroxy-3-methylglutaryl-CoA synthase 1 [Hmgcs1]) (Figure 2E). Only the increases for G6pc and Ppargc1 in LKO::Irs1–/– liver were confirmed on the GeneChips (Table 1).
Growth of LKO and LKO::Irs1–/– mice.
Insulin and IGF signaling plays an important role in body and brain growth during development and adult life (44). Previous reports show that Irs1 signaling promotes body growth (35, 37, 38). By contrast, Irs2–/– mice are born with a normal body size, but with a small brain, and gain adipose mass until β cell failure causes severe diabetes (38, 45). LKO mice had a normal brain and body size at birth and grew normally between 6 and 12 weeks of age (Figure 3A and data not shown). As expected, systemic Irs1–/– mice were consistently 50% smaller than controls, whereas viable LKO::Irs1–/– mice were 70% smaller than controls, reaching only 10 g by 12 weeks of age and never exceeding 15 g (Figure 3, A and B, and data not shown). Compared with the normal bone density observed in control and LKO mice, bone density was reduced 30% in the Irs1–/– mice, as previously shown (46), and further reduced in the LKO::Irs1–/– mice (Figure 3C). The liver and pancreas of LKO::Irs1–/– mice grew in proportion to the body size, whereas the perigonadal fat mass was disproportionately reduced by 83%, exceeding the 59% reduction in systemic Irs1–/– mice (Figure 3D). Leptin levels were normal in LKO and lean Irs1–/– mice, but leptin decreased 50% in the lipodystrophic LKO::Irs1–/– mice (Figure 3E).
Figure 3.
Body growth, bone mineral density, and adiposity. (A) Growth curves were constructed by plotting of the mean ± SEM body weight of each group. *P < 0.05 for Irs1–/– versus LKO::Irs1–/–; **P < 0.001 for Irs1–/– versus LKO::Irs1–/–. (B) A representative picture of 4-week-old LKO, Irs1–/–, and LKO::Irs1–/– mice. (C) The average ± SEM bone density (g/cm2) determined by dual-energy x-ray absorptiometry scanning. (D) Tissue weight — liver, pancreas, and perigonadal fat pad — normalized to body weight. (E) Average ± SEM serum leptin levels for each group were determined in the random-fed mice using mouse leptin ELISA. (F) Igf1 mRNA levels were determined by real-time PCR in liver extracts from each group. (G) Serum Igf1 levels (average ± SEM, n = 6) in six 8-week-old male mice in each group were determined using a mouse Igf1 ELISA kit.
Changes in hepatic gene expression were consistent with small and lipodystrophic LKO::Irs1–/– mice. RT-PCR revealed a significant reduction in Igf1 mRNA levels that was confirmed on the GeneChips (Figure 3F and Table 1). The serum Igf1 level was also decreased by 64% in the LKO::Irs1–/– mice compared with control fIrs2 mice (Figure 3G). The GeneChips revealed additional expression that was consistent with growth retardation — the growth hormone receptor (Ghr, 0.33-fold change compared with control), the IGF-binding protein–acid labile subunit (Igfals, 0.33-fold change compared with control), and IGF-binding protein-1 (Igfbp1, 5-fold change compared with control) (Table 1). Strategies to normalize these hepatic genes could establish their contribution to the growth of LKO::Irs1–/– mice.
The effect of Irs2 on hepatic glucose homeostasis.
Previous reports suggest that Irs2 plays an important role in hepatic glucose homeostasis (27, 28, 30, 32, 34). Regardless, at 6 weeks of age, blood glucose was normal in both fasting and fed LKO mice (Figure 4, A and B). However, mild glucose intolerance was detected by an i.p. glucose tolerance test at 5 weeks of age (Figure 4C). By 16 weeks the LKO mice developed mild fasting hyperglycemia (Figure 4D and data not shown). By contrast, LKO::Irs1–/– mice were severely hyperglycemic when the first measurements were conducted at 2 weeks of age (blood glucose: LKO::Irs1–/–, 267 ± 20 mg/dl; fIrs2, 144 ± 10 mg/dl). Hyperglycemia occurred in viable fasted and nonfasted LKO::Irs1–/– mice at 8 weeks of age and persisted until the experiments were terminated at 6 months (Figure 4, E and F). Glucose tolerance was severely impaired in 6-week-old LKO::Irs1–/– mice, in striking distinction to the LKO and Irs1–/– mice (Figure 4G). Intraperitoneal injections of insulin had no effects on hyperglycemia of the LKO::Irs1–/– mice, whereas they reduced blood glucose normally in the LKO mice and nearly normally in the Irs1–/– mice (Figure 4H). Thus, genetic deletion of Irs2 in the liver of Irs1–/– mice completely abolished insulin response and glucose tolerance in the LKO::Irs1–/– mice.
Figure 4.
Glucose homeostasis. (A and B) Nonfasting (A) and fasting (B) blood glucose (average ± SEM) was measured in at least 8 male mice (6 weeks old) per group. (C) Glucose tolerance tests were performed at 5 weeks of age in male mice fasted for 16 hours (at least 8 mice per genotype). The average ± SEM blood glucose was plotted at the indicated time intervals after i.p. injection of 2 g D-glucose/kg body weight. *P < 0.05, LKO vs. control. (D) Fasting blood glucose levels (average ± SEM, n = 8) were measured in 16-week-old male mice in each group. (E and F) Blood glucose levels (average ± SEM, n = 8) were measured in fasted and nonfasted male mice of each group at 8 weeks of age. (G) Glucose tolerance tests were conducted on at least 6 male mice in each group (6 weeks old) after a 16-hour overnight fast. Blood glucose levels (average ± SEM) were determined at the indicated time points after i.p. injection of 2 g D-glucose/kg body weight. (H) Insulin tolerance tests were performed in nonfasted 8-week-old male mice in each group. Blood glucose (average ± SEM, n = 6) was plotted against the time after the i.p. insulin injection.
In addition to the increased expression of Pck1 and G6pc and decreased expression of Gck that promoted gluconeogenesis (Figure 2E), the GeneChips revealed that expression of 3 other genes involved in glycolysis/gluconeogenesis increased significantly in the fasted LKO::Irs1–/– liver: aldolase (Aldo1, 3-fold change compared with control), lactate dehydrogenase (Ldh2, 12-fold change compared with control), and pyruvate dehydrogenase kinase 4 (Pdk4, 21-fold change compared with control) (Table 1). Pdk4 inhibits the mitochondrial pyruvate dehydrogenase complex to preserve pyruvate for gluconeogenesis (47). The importance of Pdk4 for dysregulated gluconeogenesis awaits more direct experiments with the LKO::Irs1–/– liver.
Insulin secretion and β cell morphology.
Consistent with the severe insulin resistance that developed in the LKO::Irs1–/– mice, circulating insulin levels were elevated 10-fold compared with those in the control fIrs2 mice at 8 weeks of age (Figure 5B). By contrast, insulin levels were increased 2-fold in the Irs1–/– and LKO mice, revealing compensation for the less severe insulin resistance (Figure 5B). Compensation for systemic insulin resistance ordinarily includes expansion of pancreatic β cell mass (48). Compared with the controls, the β cell area in the LKO pancreas increased slightly, whereas it increased 3-fold in 8-week-old Irs1–/– mice (Figure 5C). LKO::Irs1–/– islet area also increased 3-fold, even though these mice were significantly more resistant to insulin (Figure 5C). At 3 months of age, when the Irs1–/– islets had a normal appearance by immunostaining with antibodies against insulin, the LKO::Irs1–/– islets displayed a disordered morphology with weak and inconsistent insulin staining and misplaced α cells (Figure 5A). Thus, compensatory islet hypertrophy did not progress in the LKO::Irs1–/– mice beyond that observed typically in the systemic Irs1–/– mice. In addition, the number of α cells increased significantly and the glucagon staining was strong in the LKO::Irs1–/– mice, suggesting that glucagon levels could be elevated in these mice to promote hyperglycemia (Figure 5, A and D).
Figure 5.
Islet growth and β cell size. (A) Representative pancreas sections from 9-week-old nonfasted male mice of each group were immunostained with antibodies against insulin (green) or glucagon (red). Magnification, ×100. (B) Plasma insulin levels (average ± SEM) were measured in 8-week-old male mice (at least 8 per genotype) after a 16-hour overnight fast. Morphometric analysis was performed on 20 immunostained pancreatic sections from WT, LKO, Irs1–/–, and LKO::Irs1–/– mice. (C) The β cell area is the mean ± SEM percentage of the total pancreatic area surveyed. (D) The β/α cell ratio represents the total β cell number divided by the α cell number in at least 5 pancreatic sections surveyed. (E) Islet size is expressed as the average ± SEM in square micrometers.
The role of Irs2 in hepatic insulin signaling.
To establish whether Irs1 and Irs2 are the central mediators of insulin signaling in hepatocytes, fasted mice were treated i.v. with regular human insulin before the liver was dissected 4 minutes later and frozen in liquid nitrogen. Portions of the liver were homogenized, and signaling events were assessed in specific immunoprecipitates by immunoblotting. As expected, insulin stimulated tyrosine phosphorylation of the insulin receptor β subunit in all of the extracts (Figure 6A). Tyrosine autophosphorylation occurred normally in Irs1–/– liver; however, compared with the controls, receptor autophosphorylation — normalized to β subunit protein levels — was stronger in LKO::Irs1–/– and strongest in LKO liver, suggesting that Irs2 might mediate an inhibitory feedback to the insulin receptor (Figure 6A).
Figure 6.
Insulin signaling in liver, muscle, and white adipose tissue. Mice in each experimental group were fasted overnight and stimulated with insulin (Ins) by i.v. injection. Four minutes after injection, the liver, muscle, and perigonadal white adipose tissue were isolated and frozen immediately in liquid nitrogen. (A) Insulin receptor level and tyrosine phosphorylation in liver extracts. Each sample was immunoprecipitated with antibodies against the insulin receptor (αIR), Irs1, or Irs2. After SDS-PAGE, the samples were immunoblotted with αIR, or with antibodies against phosphotyrosine (αpY). (B) Erk1 and Erk2 and their phosphorylated forms (phospho-Thr202/Tyr204) were analyzed with specific antibodies in the liver lysates of each experimental group. (C) PI3K activity was assayed in vitro using the liver extracts from each group after immunoprecipitation with anti-phosphotyrosine antibodies. The data represent the percentage of PI3K activity normalized against that measured for the fIrs2 mice. PIP3, 32P-labeled phosphatidylinositol-3,4,5-triphosphate. (D) The phosphorylation of Akt, FoxO1, and Prkaa1 was determined in the liver lysates of each experimental group. After SDS-PAGE, the samples were immunoblotted with phosphospecific antibodies against Akt (αpAkt, phospho-Ser473), FoxO1 (αpFoxO1, phospho-Ser256), or Prkaa1 (αpPrkaa1, phospho-Thr172). The expressions of Akt and Prkaa1 were also determined in the liver extracts by immunoblotting with specific antibodies. (E and F) Phosphorylation of Akt and Erk1/2 was analyzed by immunoblotting as described for liver using tissue lysates from epididymal fat or hind-limb skeletal muscle of LKO::Irs1–/–, Irs1–/–, LKO, and control fIrs2 mice.
Insulin increased tyrosine phosphorylation of Irs1 and Irs2 in fIrs2 liver extracts. Irs1 phosphorylation was not detected in the Irs1–/– liver, Irs2 phosphorylation was not detected in LKO liver, and neither phosphoprotein was detected in LKO::Irs1–/– liver (Figure 6A). Similar to previous results in systemic Irs2–/– mice (23), basal Irs1 tyrosine phosphorylation was increased in the LKO mice, whereas basal Irs2 phosphorylation was not elevated in the Irs1–/– mice (Figure 6A).
The effect of insulin on the MAPK cascade in untreated and insulin-treated liver extracts was examined with phosphospecific antibodies against Erk1 and Erk2. Erk1 and Erk2 protein levels were identical in the liver extracts of all the mice examined (Figure 6B). Insulin stimulated the phosphorylation of Erk2 more strongly than that of Erk1 in control liver and in Irs1–/– liver. Erk1 phosphorylation was undetected in the LKO and LKO::Irs1–/– liver, whereas insulin weakly stimulated Erk2 phosphorylation (Figure 6B). Additional work is required to establish whether Gab1, Shc, or Irs4 mediates Erk2 phosphorylation in the LKO::Irs1–/– liver (49–51).
Irs1 and Irs2 play an important role in the activation of the hepatic PI3K—Akt cascade, which controls glucose homeostasis through direct effects on FoxO1 phosphorylation (12, 13). As expected, insulin stimulated PI3K in the control fIrs2 liver; however, this effect was reduced by 25% in the Irs1–/– and LKO liver extracts, and by 70% in LKO::Irs1–/– liver (Figure 6C). Whether the insulin receptor directly activates PI3K in LKO::Irs1–/– liver — or whether Irs4 or other scaffold proteins are involved — remains to be established (40, 52).
Phospholipids produced by PI3K during insulin stimulation promote the phosphorylation of Akt and FoxO1 (53). As expected, insulin strongly stimulated the phosphorylation of Akt (Ser473) and FoxO1 (Ser256) in the control fIrs2 liver. Akt and FoxO1 phosphorylation was reduced in Irs1–/– and LKO liver and was undetected in LKO::Irs1–/– liver (Figure 6D). The complete reduction of the Akt—FoxO1 cascade was most likely related to a signaling defect rather than depletion of hepatic ATP levels, as the phosphorylation of AMP-activated protein kinase, catalytic subunit α1 (Prkaa1; Thr172), was normal in the Irs1–/– and LKO liver extracts and reduced 15% in the LKO::Irs1–/– liver (Figure 6D) (54). The loss of FoxO1 phosphorylation in the LKO::Irs1–/– liver can explain the increased expression of several hepatic genes that promote hepatic glucose production, including G6pc, Pck1, Pdk4, and Ppargc1 (Figure 2E and Table 1).
Finally, we measured insulin-stimulated phosphorylation of Akt and Erk1/2 in adipose tissue and skeletal muscle of control (fIrs2), Irs1–/–, LKO, and LKO::Irs1–/– mice. Akt was stimulated strongly after the insulin injection in epididymal fat and in muscle from each group, including the LKO::Irs1–/– mice (Figure 6, E and F). Erk phosphorylation was unchanged in adipose tissues but barely increased above the basal by insulin (Figure 6E). By contrast, insulin strongly stimulated Erk1 and Erk2 phosphorylation in fIrs2 and Irs1–/– muscle, whereas Erk1/2 phosphorylation was reduced in LKO and LKO::Irs1–/– muscle (Figure 6F). Regardless of these partial changes, insulin signaling in muscle and fat was never completely lost in the LKO::Irs1–/– mice, suggesting that these peripheral tissues continue to respond to insulin.
The regulation of glycogen synthesis.
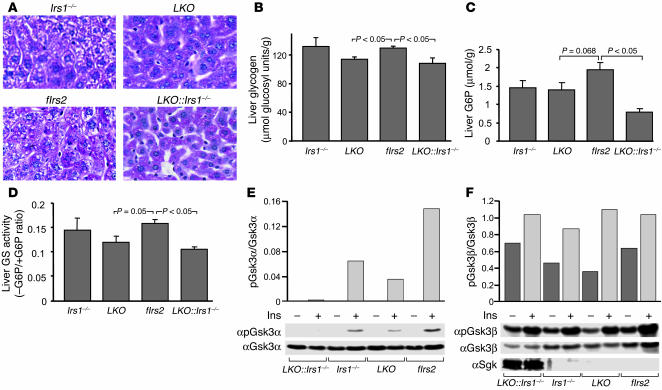
Liver sections stained with periodate suggested that hepatic glycogen content was nearly unchanged in the 4 experimental groups of mice — normal in Irs1–/– liver, but reduced 12% in LKO liver and 17% in LKO::Irs1–/– liver (Figure 7, A and B). The presence of glycogen in the LKO::Irs1–/– liver was unexpected owing to the absence of insulin-stimulated Akt signaling and the extreme hyperglycemia of this mouse. Reduced hepatic glucose-6-phosphate (G6P) might contribute to reduced glycogen content, as glycogen synthase activity is stimulated allosterically by G6P (55). However, the hepatic G6P level correlated poorly with glycogen content, as G6P was decreased about 27% in the liver of Irs1–/– or LKO mice, and 60% in LKO::Irs1–/– liver (Figure 7C). By comparison, glycogen synthase activity was reduced 24% and 34% in the liver of LKO and LKO::Irs1–/– mice, respectively, which correlated more closely to the hepatic glycogen levels (Figure 7D).
Figure 7.
Glycogen synthesis in the LKO::Irs1–/– liver. Liver extracts were prepared from 6 nonfasted mice in each experimental group at 6 weeks of age. (A) Liver sections from all the mice were stained with PAS reagent, and representative sections are shown. Magnification, ×200. (B) Liver glycogen contents (average ± SEM, n = 6 mice per group) were measured in the liver lysates. (C) G6P contents (average ± SEM, n = 6) were determined in the liver extracts. (D) Liver glycogen synthase (GS) activity is reported as the average ± SEM of the ratio of the enzymatic activity in the absence of G6P over that in the presence of G6P. (E and F) Liver extracts described in Figure 6 were used to measure the expression and phosphorylation of Gsk3α (Ser21) and Gsk3β (Ser9). The intensity of each relevant band was determined using ImageQuant to analyze the scanned autograph. The ratio of phosphorylated to total protein was plotted. One of 2 individual experiments is shown. Sgk was also identified by immunoblotting (F).
During insulin stimulation, glycogen synthase is activated by dephosphorylation, which is mediated at least in part when glycogen synthase kinase 3β (Gsk3β) is inhibited by Akt-mediated phosphorylation (56). Since Akt was not activated in LKO::Irs1–/– mice, we were surprised that glycogen levels were nearly normal. Consistent with the level of Akt phosphorylation, insulin-stimulated phosphorylation of Gsk3βSer9 was normal in Irs1–/– and LKO mice (Figure 7F). Moreover, insulin-stimulated phosphorylation of Gsk3αSer21, a closely related kinase, was also detected in these mice (Figure 7E). However, Gsk3αSer21 phosphorylation correlated with Akt phosphorylation and was barely detected in LKO::Irs1–/– liver (Figure 7E). By contrast, basal and insulin-stimulated phosphorylation of Gsk3βSer9 was comparable to that in control fIrs2 liver, although the protein levels were decreased (Figure 7, E and F). Thus, a normal level of phosphorylation of Gsk3βSer9 could promote glycogen synthase activity needed to preserve the hepatic glycogen levels in LKO::Irs1–/– mice.
GeneChip analysis revealed some explanations for the storage of glycogen in LKO::Irs1–/– liver. One possibility was the increased expression of Sgk (5.4-fold change compared with control), an Akt homolog, in the LKO::Irs1–/– liver (Table 1). Immunoblotting confirmed that Sgk protein was increased and migrated as a doublet during SDS-PAGE (Figure 7F). The upper band, believed to be the hyperphosphorylated form, can phosphorylate and inactivate Gsk3β to promote glycogen synthesis (57). Another possibility was the increased expression of UDP-glucose pyrophosphorylase (Ugp2, 3.3-fold change compared with control), one of the key enzymes in glycogen synthesis (Table 1).
Lipid metabolism.
Type 2 diabetes is associated with hepatic steatosis owing to decreased fatty acid oxidation and increased lipogenesis (7, 32). Histological sections obtained at 4 months of age revealed increased triglyceride in the liver of LKO mice, but normal levels in the LKO::Irs1–/– liver (Figure 8A). Biochemical analysis confirmed that triglycerides were normal in the Irs1–/– liver, elevated slightly in the LKO liver, but decreased in the LKO::Irs1–/– liver (Figure 8B). By comparison with the control, circulating FFA levels were normal in LKO mice, but reduced 50% in Irs1–/– and LKO::Irs1–/– mice (Figure 8C). Apparently the Irs1 branch of the insulin signaling system controls systemic circulating FFA levels. Serum triglyceride levels decreased significantly in all the experimental groups, the largest decrease occurring in Irs1–/– and LKO::Irs1–/– mice (Figure 8D).
Figure 8.
Lipid metabolism in the LKO::Irs1–/– mice. (A) Liver morphology was analyzed by H&E staining in the liver sections from the experimental groups (total magnification, ×100). (B) Liver triglyceride (TG) contents were determined in liver extracts and presented as the average ± SEM of 5 overnight-fasted mice (8 weeks old) per genotype. (C and D) Serum FFAs (C) and triglycerides (D) were measured in overnight-fasted 8-week-old mice.
Discussion
Our results show that hepatic Irs2 is required for efficient insulin action in the liver, as its deletion from WT mice (LKO mice) causes mild hyperinsulinemia and glucose intolerance that is detected by a glucose tolerance test by 5–6 weeks of age. Since LKO mice retain Irs2 expression in β cells and the hypothalamus, central nutrient homeostasis and β cell function remain intact to provide the needed compensation under ordinary conditions. This result stands in striking contrast to the deletion of Irs2 from β cells — and partially from the brain (hypothalamus) — which causes obesity and severe diabetes between 6 and 8 weeks of age; however, glucose homeostasis is gradually normalized as functional β cells with intact Irs2 alleles regenerate in the pancreas between 4 and 10 months of age (24, 26). By contrast, LKO::Irs1–/– mice develop severe hyperglycemia immediately after birth that persists for at least 6 months, when all our experiments were terminated. Moreover, LKO::Irs1–/– mice are unresponsive to compensatory hyperinsulinemia or exogenous insulin injections. Thus, Irs1 or Irs2 can mediate sufficient hepatic insulin action to maintain systemic glucose tolerance — at least while β cell function can compensate for insulin resistance. Recent reports reveal some effect of neuronal inputs on hepatic glucose production, even when insulin receptors are ablated (4, 9). Regardless, without signals from Irs1 and Irs2, hepatic glucose production is largely uncontrolled.
Irs proteins coordinate various signals that regulate cell growth, survival, and metabolism, including activation of the PI3K—Akt and the ras—Erk cascades (11). The PI3K—Akt cascade is one of the best-studied pathways in hepatocytes (12, 58–60). One of the consequences of Akt stimulation is the phosphorylation and nuclear exclusion of FoxO1. FoxO1, a member of the O class of forkhead/winged helix transcription factor family, promotes the expression of genes that contain a canonical insulin response element [(G/C)AAAA(T/C)AA] in their promoters. In hepatocytes the best-characterized examples include Pck1 and G6pc, which strongly promote gluconeogenesis. However, their expression is suppressed when FoxO1 is excluded from the nucleus by Akt-mediated phosphorylation (12). Signaling experiments in liver extracts from control or insulin-stimulated LKO, Irs1–/–, or LKO::Irs1–/– mice show that either Irs1 or Irs2 can promote sufficient FoxO1 phosphorylation to suppress the expression of these genes, at least during compensatory hyperinsulinemia. However, Akt and FoxO1 phosphorylation is undetectable in LKO::Irs1–/– liver, and enzymes promoting hepatic glucose production are dysregulated even during hyperinsulinemia.
All insulin signals are not completely lost in the LKO::Irs1–/– liver. The β subunit of the insulin receptor is rapidly phosphorylated after insulin injection; PI3K is activated to about 30% of WT levels; and some Erk2 phosphorylation occurs. Moreover, Gsk3βSer9 is hyperphosphorylated during starvation, and the stimulation by insulin is detectable. Understanding how these signaling cascades are regulated without activating Akt through Irs1 or Irs2 will require additional work focusing on the role of Gab1, Shc, or Irs4, or the direct association of PI3K with the phosphorylated insulin receptor β subunit (40, 52).
Previous studies suggest that PI3K-dependent activation of Akt involves phosphorylation of Thr308 by 3-phosphoinositide–dependent protein kinase 1 (Pdk1) and of Ser473 by a second kinase, possibly the mTOR:Rictor:GβL complex (61–63). Deletion of hepatic Pdk1 prevents insulin-stimulated Akt (Thr308) phosphorylation and Gsk3β (Ser9) phosphorylation; however, basal Ser473 phosphorylation increases in the Pdk1–/– liver (64). Unexpectedly, liver-specific Pdk1–/– mice display normal serum glucose and insulin levels under ordinary experimental conditions, whereas liver glycogen is depleted (64). These results contrast with those in the LKO::Irs1–/– mice, which are hyperglycemic with nearly normal hepatic glycogen levels. Akt (Ser473) phosphorylation is completely lost in the LKO::Irs1–/– liver, suggesting that the phosphorylation of the hydrophobic motif in Akt might be essential to control hepatic gluconeogenesis (64).
Insulin-stimulated phosphorylation of Gsk3 is an important step in the stimulation of glycogen synthase. Gsk3 is generally thought to be a direct Akt substrate, and, as expected, Gsk3α and Gsk3β phosphorylation is strongly decreased in Pdk1–/– liver (64). Consistent with the reduced activation of Akt, Gsk3β phosphorylation decreases in LKO and Irs1–/– liver and is barely detected in LKO::Irs1–/– liver. By contrast, Gsk3α phosphorylation is normal in LKO, Irs1–/–, and LKO::Irs1–/– liver, suggesting that signals independent of the Pdk1—Akt pathway might be involved. The 6-fold induction in LKO::Irs1–/– liver of Sgk, an Akt homolog that can phosphorylate Gsk3β, might promote glycogen synthase activity in hepatocytes even when G6P is low (65).
Conservative estimates reveal well over 200 genes that change significantly, more than 2-fold, in the fasted LKO::Irs1–/– liver. By comparison, 10-fold fewer changes occur in the fasted LKO or Irs1–/– liver extracts, which is consistent with mild glucose intolerance observed in these mice. At least some of these genes contain an insulin response element [(G/C)AAAA(T/C)AA] that could bind nuclear FoxO1 transcription factors — especially G6pc, Igfbp1, Pdk4, and Ppargc1 (41, 66, 67). However, the regulation of gene expression in the LKO::Irs1–/– mice is likely to reflect the interaction of many factors, some of which are regulated by hyperglycemia or other metabolic sequelae. The activity of Gsk3α or Gsk3β can also play a role in the regulation of hepatic gene expression, in some cases influencing genes expected to be controlled by FoxO1 (67). Thus the differential regulation of Gsk3α and Gsk3β can complicate the interpretation of gene expression in the LKO::Irs1–/– mice (67). Regardless, future experiments with LKO::Irs1–/– mice together with the hepatic-specific deletion of Irs1 and Irs2 can provide a model to uncover new mechanisms of insulin-regulated gene expression. Although beyond the scope of this report, detailed informatics informed by pathway interactions might reveal the importance of other significant changes that do not exceed 2-fold.
Viable LKO::Irs1–/– mice are 40% smaller than Irs1–/– mice, reaching only 25–30% of the size of control fIrs2 or WT mice. Moreover, the systemic growth retardation is accompanied by severe lipoatrophy, particularly in perigonadal fat depots. This phenotype might be caused by the intrinsic growth retardation of systemic Irs1–/– deficiency exacerbated by reduced Igf1 signaling. Although overall body growth is not reduced upon deletion of the hepatic Igf1 gene, hepatic Igf1 knockout reduces systemic fat production (68). Moreover, the reduced growth of the Irs1–/– mice is likely to be exacerbated in the LKO::Irs1–/– mice by the elevated expression of Igfbp1 (5-fold change compared with control), and the reduced expression of Igfals and Ghr (both reduced 3-fold compared with control) (Table 1).
Changes in hepatic gene expression strongly favor hepatic glucose production in the LKO::Irs1–/– mice, including increased G6pc and Pck1, and decreased Gck. Deletion of the hepatic insulin receptor also induces these gene expression changes, but fasting glucose levels are barely elevated at 2 months of age and reduced by 6 months. Thus other gene expression changes are likely to contribute to the extreme fasting hyperglycemia of LKO::Irs1–/– mice. The transcription coactivator Ppargc1 is strongly increased in LKO::Irs1–/– liver; this can promote gluconeogenesis and fatty acid oxidation through the association of Ppargc1 with the nuclear hormone receptors HNF4 and PPARα, respectively (69). The 21-fold increase of Pdk4, a direct target of nuclear FoxO1, is especially conspicuous. Pdk4 phosphorylates the E2 subunit of the pyruvate dehydrogenase complex, inhibiting pyruvate oxidation and diverting 3 carbon precursors (pyruvate, lactate, etc.) away from fatty acid synthesis and toward glucose production. These results suggest that a signal from Irs1 or Irs2, however minimal, is essential to maintain the flux of carbon from glucose or its metabolites toward fatty acids. During ordinary type 2 diabetes, the oxidation of pyruvate is never completely blocked so the liver can accumulate FFAs and triglycerides. Moreover, hepatic steatosis accompanies hyperglycemia, because compensatory hyperinsulinemia inhibits fatty acid oxidation more readily than it inhibits gluconeogenesis (1, 2).
Pparg is considered to be a master regulator of adipocyte differentiation and is elevated in the livers of animals that develop fatty livers (70). Consistent with this conclusion, Pparg increases 2-fold in LKO liver, which parallels the mild hepatic steatosis. However, Pparg increases 10-fold in LKO::Irs1–/– liver, so its expression alone cannot predict the hepatic steatosis. Pparg induces the expression of several lipogenic genes in hepatocytes, including adipose differentiation–related protein (Adrp), adipocyte fatty acid–binding protein 4 (Fabp4), sterol regulatory element–binding protein-1 (Srebp1), fatty acid synthase (Fasn), and acetyl-CoA carboxylase (Acc1) (70). Two of these genes, Srebp1 and Fasn, decreased significantly, 2-fold, in the Irs1–/– liver but were not changed in the LKO or LKO::Irs1–/– liver (Figure 2E); a 3-fold increase of Acc1 in LKO::Irs1–/– liver was not significant (P = 0.11). Thus, hepatic gene expression is difficult to predict upon deletion of Irs1 and Irs2.
GeneChip analysis reveals many significant changes that might contribute to reduced circulating triglycerides and FFAs in the LKO::Irs1–/– mice. CD36/FAT, which can be induced by Pparg, was increased 7-fold in LKO::Irs1–/– liver (Table 1). CD36/FAT is a cell surface receptor responsible for the uptake of oxidized LDL (in macrophages) and FFAs (in muscle) (71). In the LKO::Irs1–/– liver, CD36 might facilitate oxidation of circulating lipids. Lipoprotein lipase (Lpl), a key enzyme ordinarily expressed in adipose tissues and muscle, but not in the liver, was increased 16-fold in LKO::Irs1–/– liver (Table 1). Lpl promotes clearance of triglyceride-associated fatty acids from the circulation. Thus, triglyceride uptake into the LKO::Irs1–/– liver can be significantly increased, explaining in part the reduced circulating triglycerides. Together, the gene expression changes in LKO::Irs1–/– liver are consistent with increased production of glucose and clearance of triglyceride.
Unlike the LKO::Irs1–/– mice, LIRKO mice are glucose intolerant but not diabetic and develop mild hepatic steatosis (7). Since Irs1 and Irs2 are expressed in the livers of LIRKO mice, at least a basal signal should be produced to suppress some FoxO1-mediated gene expression and avoid the dramatic gene expression changes that completely dysregulate hepatic metabolism. Clearly, Irs1 and Irs2 signaling is very sensitive in the liver, as partial knock down of Irs1 and Irs2 by short hairpin RNA adenovirus (Irs1 by 80% and Irs2 by 70%) produces a measurable but relatively mild phenotype (32). The dominant effect of Irs1 and Irs2 signaling might explain why acute knock down of the insulin receptor in liver by antisense oligodeoxynucleotide does not dysregulate glucose production (6). The ability of Irs1 and Irs2 to mediate signals without the usual activation by insulin-stimulated tyrosine phosphorylation can explain why liver insulin receptor knockout or knock down from hepatocytes displays milder glucose intolerance with hepatic steatosis.
A direct comparison between LKO::Irs1–/– and LIRKO mice is probably premature, because LKO::Irs1–/– mice have a systemic Irs1 deletion that causes systemic insulin resistance. However, severe hyperglycemia that develops in systemic insulin receptor knockout mice is normalized upon transgenic expression of the insulin receptor in liver, β cells, and brain (6, 7, 9). Thus, insulin signaling in muscle and fat does not appear to contribute significantly to the regulation of hepatic glucose production. Since insulin signaling to Akt and Erk1/2 is at least partially retained in the muscle and adipose tissue of LKO::Irs1–/– mice, we conclude provisionally that the absence of hepatic Irs1 and Irs2 contributes significantly to the dysregulated glucose metabolism. However, liver-specific Irs1 and Irs2 double knockout will need to be investigated to determine directly whether dysregulated hepatic signaling is the major cause of diabetes in the LKO::Irs1–/– mice.
LKO::Irs1–/– mice develop more severe insulin resistance than Irs1–/– mice; however, pancreatic islet compensation — increased β cell area, islet size, and density — is approximately equal. In other backgrounds of persistent insulin resistance, such as the ob/ob mouse, islet expansion progresses unabated. What restrains the expansion of β cell mass in the LKO::Irs1–/– mice to the level obtained in the Irs1–/– mice needs to be investigated. The deterioration of β cells in the LKO::Irs1–/– mice could be caused by defects in the communication between liver and pancreatic β cells (72).
In summary, without Irs2 in the hepatocytes, Irs1 signaling mediates a sufficient insulin response to maintain nearly normal hepatic glucose homeostasis in LKO mice, at least while the β cells can produce sufficient insulin to compensate for the reduced signaling, and the muscle and fat can still respond to insulin. By contrast, functional redundancy between Irs2 and Irs1 in hepatocytes is not observed in either pancreatic β cells or certain hypothalamic nuclei (26). Unlike liver, Irs2 has a predominant role in β cell proliferation and hypothalamic nutrient sensing, as its deficiency in these tissues causes obesity that progresses to diabetes owing to β cell failure (24–26). Additional work might reveal a unique role for hepatic Irs2 during physiological stress and obesity, since Irs2 is regulated by nutrient status through several key transcription factors — FoxO1, SREBPs, and CREB (28, 31). Particularly during the fasting state, the increased expression of Irs2 is expected to generate, without the need for elevated circulating insulin, basal signals that counterregulate the activity of glucagon. Moreover, elevated hepatic Irs2 can prime the liver to respond rapidly to secreted insulin immediately upon feeding, which can maximize the effect of first-phase insulin secretion and reduce the need for prolonged insulin secretion. However, when nutrients are abundant, Irs2 can be inhibited or degraded, and its expression can be suppressed by nuclear SREBPs. Under such conditions, triglyceride synthesis can be sustained by hyperinsulinemia through Irs1 signaling (2). Understanding the balance between Irs1 signaling and Irs2 signaling and their integration with neuronal signals will provide a platform to understand metabolic disease and its progression to diabetes.
Methods
Animal production and genotyping.
The creation of floxed Irs2 (fIrs2) mice has been previously described (26). Albumin-cre (crAlb) mice were purchased from the Jackson Laboratory. To generate liver-specific Irs2 knockout mice, fIrs2 mice were crossed with crAlb mice, and then the generated fIrs2+/–:crAlb mice were crossed with fIrs2+/– mice. To create double-knockout (LKO::Irs1–/–) mice, previously generated Irs1 heterozygous (Irs1+/–) mice (38) were crossed with mice carrying either fIrs2+/–:crAlb or fIrs2+/+. After another round of intercrosses, LKO::Irs1–/– mice were generated from the crosses of fIrs2+/+:crAlb:Irs1+/– and fIrs2+/+:Irs1+/– mice. All the mouse lines were maintained on a mixed background derived from C57BL/6J and 129/Sv. The genotyping was performed as previously described (26, 38). All animal procedures were approved by the Institutional Animal Care and Use Committee of Harvard School of Public Health (Boston, Massachusetts, USA).
Metabolic analyses.
Glucose levels were measured on random-fed (ad libitum) or overnight-fasted mice using a glucometer (Elite XL; Bayer). Serum insulin levels were measured using rat insulin ELISA (Crystal Chem Inc.). Leptin levels were analyzed in the random-fed serum by ELISA (Crystal Chem Inc.). Serum IGF-I levels were analyzed using a Mouse IGF-I EIA kit according to the manufacturer’s instructions (Diagnostic Systems Laboratories Inc.). Plasma FFAs and triglycerides were measured in the serum from overnight-fasted mice using the NEFA and L-Type triglyceride kits from Wako Diagnostics. Glucose tolerance tests were performed on overnight-fasted mice. Animals were injected i.p. with D-glucose (2 g/kg), and blood glucose levels were measured at indicated time points. For insulin tolerance tests, mice were fed ad libitum, and diluted insulin (regular human insulin from Eli Lilly and Co.; 1 U/kg) was injected i.p. Blood glucose concentrations were measured at 0, 15, 30, and 60 minutes after insulin injection.
Islet morphology, immunohistochemistry, and morphometry.
Pancreata were fixed for 16 hours in 4% paraformaldehyde at 4°C and then embedded in paraffin as described previously (73). For general islet histology, paraffin sections (5 μm) were stained by standard H&E staining procedure (Rodent Histopathology Core Facility, Harvard Medical School). To stain pancreatic sections for insulin and glucagons, the sections were deparaffinized, rehydrated, and permeabilized with 1% Triton X-100. Then the sections were incubated with guinea pig anti-insulin and rabbit anti-glucagon antibodies (Zymed Laboratories Inc.) overnight. After 3 washes with PBS buffer, the sections were incubated with FITC-conjugated goat anti–guinea pig and rhodamine-labeled donkey anti-rabbit secondary antibodies (Jackson ImmunoResearch Laboratories Inc.) for 1 hour. The images were captured using a Zeiss Axiovert S100 TV microscope and processed with Openlab image analysis software (Improvision). Islet morphometric analysis was performed as previously described using the Openlab density slice software (74).
Body weight, tissue weight, and bone mineral density.
Mice were fasted overnight before they were weighed. To measure the weights of liver, pancreas, and perigonadal fat pads, animals were anesthetized with Avertin (0.02 ml/g body weight) by i.p. injection, and then the organ/tissue was dissected and immediately weighed. For comparison, the tissue weight was normalized to body weight. Bone mineral density was determined using dual-energy x-ray absorptiometry (Lunar PIXImus II mouse densitometer; GE LUNAR Corp.) as described by the manufacturer. The machine was calibrated daily using a phantom supplied by the manufacturer. Mice were anesthetized using Avertin (0.02 ml/g body weight) by i.p. injection before they were scanned.
Liver histology and apoptosis detection.
To study the liver histology, mice were anesthetized with Avertin (0.02 ml/g body weight), and liver was dissected and fixed in the buffered neutral formalin (10%). The fixed tissue blocks were embedded in paraffin. The paraffin sections (5 μm) were stained by standard H&E staining procedure. The apoptosis assays were carried out on the deparaffinized liver sections using ApopTag Plus Peroxidase In Situ Apoptosis Detection Kit (Chemicon International). The stained sections were viewed and recorded using a Zeiss Axiovert S100 TV microscope and Openlab image analysis software (version 3.5).
Liver glycogen and triglyceride content analysis.
The procedure of glycogen content assay was described previously (75). Briefly, approximately 50 mg of liver samples (in triplicates) was weighed and boiled in 400 μl of 2N HCl for 2 hours. Then 400 μl of 2N NaOH and 22 μl of 1 M Tris-HCl (pH 7.4) were added to neutralize the acid. The glucosyl units from the above glycogen breakdown were measured using Amplex Red Glucose Assay Kit (Invitrogen Corp.). The liver triglyceride assay was performed as previously described (76). Briefly, approximately 100 mg of liver tissue was weighed and digested with 350 μl of ethanolic KOH at 55°C overnight. Then the digest was extracted with 50% ethanol twice. The lysate was finally mixed with an equal volume of 1 M MgCl2 and incubated on ice for 10 minutes. After the precipitate was removed by centrifugation, the supernatant was used for triglyceride assay using the L-Type triglyceride kit from Wako Diagnostics.
Assays for glycogen synthase activity and G6P content.
Liver tissue from random-fed mice was homogenized in buffer consisting of 50 mM Tris, 5 mM EDTA, and 100 mM NaF. The homogenate was used for the glycogen synthase assay as previously described (77). The glycogen synthase activity was represented by the ratio of [14C-UDP-glucose] incorporation in the absence of G6P to that in the presence of G6P. For G6P assay, frozen liver tissue was ground in a porcelain mortar and deproteinized with perchloric acid. The cleared supernatant was assayed according to the method of Lang and Michal (78).
Immunoprecipitation, immunoblotting analyses, and PI3K assay.
Mice were fasted overnight before they were anesthetized with Avertin (0.02 ml/g body weight) by i.p. injection. For insulin stimulation, human insulin (5 U) was injected through the inferior vena cava. Liver, epididymal fat, and hind-limb muscle were dissected and immediately frozen in liquid nitrogen after 4, 4.5, and 5 minutes of insulin stimulation, respectively. To prepare tissue lysates, frozen tissue was homogenized in buffer A (25 mM Tris-HCl, pH 7.4, 50 mM sodium pyrophosphate, 100 mM sodium fluoride, 10 mM EDTA, 1% NP-40, 1 mM PMSF, 1 mM sodium orthovanadate, 10 μg/ml aprotinin, and 10 μg/ml leupeptin). For immunoprecipitation of Irs1, Irs2, and the insulin receptor, 1 mg of liver extracts were incubated with rabbit polyclonal antibodies against Irs1, Irs2, and the insulin receptor for 3 hours at 4°C on a rocker. Then protein A-Sepharose was added and incubated for 1 hour at 4°C. After washing 4 times with buffer A, the immunocomplexes were resolved on 7.5% SDS-PAGE. Phosphorylated or total protein was analyzed by immunoblotting with specific antibodies against Irs1, Irs2, the insulin receptor, and phosphotyrosine (Transduction Laboratories). Phosphorylated or total protein kinase B, Gsk3, FoxO1, Sgk, Prkaa1, and Erk1/2 were analyzed by immunoblotting with specific antibodies (Cell Signaling Technology) after tissue lysates were resolved on SDS-PAGE and transferred to nitrocellulose membrane. Bound antibodies were detected with HRP-conjugated secondary antibodies using ECL detection system (PerkinElmer). For PI3K assay, 1 mg of liver extract was immunoprecipitated with anti-phosphotyrosine antibodies (Transduction Laboratories), and the immune complexes were assayed for PI3K activity using the phosphatidylinositol (Avanti Polar Lipids Inc.) as substrate. The procedure was performed as previously described (23). The kinase activity was quantitated with ImageQuant TL software (Amersham Biosciences).
RT-PCR and real-time PCR.
Total RNA was isolated from livers of overnight-fasted mice using TRIzol (Invitrogen Corp.). cDNA synthesis was performed with the RETROscript kit (Ambion) using random decamers. The real-time RT-PCR was carried out using the QuantiTect SYBR Green PCR kit (QIAGEN) on an iCycler PCR instrument (Bio-Rad Laboratories) using gene-specific primers according to the manufacturer’s instructions. The relative quantitation analysis was performed by calculation of the ratio of the mRNA amount of the gene of interest over the amount of internal control 18S rRNA. For regular RT-PCR analysis of gene expression of Irs1 and Irs2, PCR amplification was carried out under the following conditions (hot start): 94°C for 15 minutes; 26 cycles of 94°C for 30 seconds, 60°C for 30 seconds, and 72°C for 30 seconds; followed by 72°C for 5 minutes. The PCR products were resolved on 2% agarose gel and stained with ethidium bromide. The gel image was captured using a Gel Logic 100 Imaging System (Eastman Kodak Co.).
Affymetrix GeneChip analysis.
Total RNA was isolated from livers of overnight-fasted 6-week-old mice in each experimental group using TRIzol (Invitrogen Corp.). Analysis of mRNA expression was performed on duplicate samples of each group using Mouse Genome 430 2.0 GeneChips (Affymetrix). RNA labeling and microarray hybridizations were performed at the Harvard Medical School Biopolymers Facility, which operates a full-service unit to support Affymetrix microarray analysis using the model 450 Fluidics station and model 3000 Scanner with autoloader. Affymetrix Microarray Suite 5.0 was used to generate cell intensity (CEL) files that were analyzed by Robust Multichip Average Analysis (RMA) implemented in GeneSpring 7.2 (Agilent Technologies). The data were further normalized by gene polishing to center the data on unity. The Cross-Gene Error Model implemented in GeneSpring 7.2 was applied to the replicate samples. ANOVA was performed using the variances estimated by the Cross-Gene Error Model in GeneSpring 7.2, but without a multiple testing correction. Next, a 1-sample, 2-tailed Student’s t test was calculated upon the set of significant probes to determine whether the mean normalized expression values for each probe were statistically different from 1.0. Probes that were significantly different (P < 0.05) were further analyzed using GeneSpring 7.2 to determine the location of the canonical FoxO1 transcriptional regulation element (G/CAAAAT/CAA) between –10 and –2,500 as defined in the Ensembl database build NCBI 32 (http://www.ensembl.org/index.html; data from February 2004, the most recent build at the time of analysis).
Statistics.
All data are presented as mean ± SEM. For analyses of statistical significance, the unpaired Student’s t test (2-tailed) was used, except in the GeneChip analysis described above. P values less than 0.05 were considered significant.
Acknowledgments
This work was supported by NIH grants DK55236 and DK43808. We thank Jake Kushner for the technical suggestions and members of the White laboratory for the helpful discussions.
Footnotes
Nonstandard abbreviations used: crAlb, albumin-cre; Fasn, fatty acid synthase; fIrs2, floxed Irs2; FoxO1, forkhead box O1; Gck, glucokinase; Ghr, growth hormone receptor; G6P, glucose-6-phosphate; G6pc, glucose-6-phosphatase; Gsk3, glycogen synthase kinase 3; Igfals, IGF-binding protein–acid labile subunit; Igfbp1, IGF-binding protein-1; Irs, insulin receptor substrate; LIRKO, liver-specific insulin receptor knockout; LKO, liver-specific knockout; Lpl, lipoprotein lipase; Pck1, phosphoenolpyruvate carboxykinase 1; Pdk1, 3-phosphoinositide–dependent protein kinase 1; Pdk4, pyruvate dehydrogenase kinase 4; Ppargc1, PPARγ coactivator 1; Prkaa1, AMP-activated protein kinase, catalytic subunit α1; Sgk, serum- and glucocorticoid-inducible kinase.
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Gribble FM. Metabolism: a higher power for insulin. Nature. 2005;434:965–966. doi: 10.1038/434965a. [DOI] [PubMed] [Google Scholar]
- 2.Wolfrum C, Asilmaz E, Luca E, Friedman JM, Stoffel M. Foxa2 regulates lipid metabolism and ketogenesis in the liver during fasting and in diabetes. Nature. 2004;432:1027–1032. doi: 10.1038/nature03047. [DOI] [PubMed] [Google Scholar]
- 3.Montminy M, Koo SH. Diabetes: outfoxing insulin resistance? Nature. 2004;432:958–959. doi: 10.1038/432958a. [DOI] [PubMed] [Google Scholar]
- 4.Pocai A, et al. Hypothalamic K(ATP) channels control hepatic glucose production. Nature. 2005;434:1026–1031. doi: 10.1038/nature03439. [DOI] [PubMed] [Google Scholar]
- 5.Barrett EJ. Insulin’s effect on glucose production: direct or indirect? J. Clin. Invest. 2003;111:434–435. doi:10.1172/JCI200317881. doi: 10.1172/JCI17881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buettner C, et al. Severe impairment in liver insulin signaling fails to alter hepatic insulin action in conscious mice. J. Clin. Invest. 2005;115:1306–1313. doi:10.1172/JCI200523109. doi: 10.1172/JCI23109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michael MD, et al. Loss of insulin signaling in hepatocytes leads to severe insulin resistance and progressive hepatic dysfunction. Mol. Cell. 2000;6:87–97. [PubMed] [Google Scholar]
- 8.Okamoto H, et al. Transgenic rescue of insulin receptor-deficient mice. J. Clin. Invest. 2004;114:214–223. doi:10.1172/JCI200421645. doi: 10.1172/JCI21645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okamoto H, Obici S, Accili D, Rossetti L. Restoration of liver insulin signaling in Insr knockout mice fails to normalize hepatic insulin action. J. Clin. Invest. 2005;115:1314–1322. doi:10.1172/JCI200523096. doi: 10.1172/JCI23096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Accili D. Lilly Lecture 2003. The struggle for mastery in insulin action: from triumvirate to republic. Diabetes. 2004;53:1633–1642. doi: 10.2337/diabetes.53.7.1633. [DOI] [PubMed] [Google Scholar]
- 11.Rhodes CJ, White MF. Molecular insights into insulin action and secretion. Eur. J. Clin. Invest. 2002;32(Suppl. 3):3–13. doi: 10.1046/j.1365-2362.32.s3.2.x. [DOI] [PubMed] [Google Scholar]
- 12.Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414:799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- 13.White MF. Insulin signaling in health and disease. Science. 2003;302:1710–1711. doi: 10.1126/science.1092952. [DOI] [PubMed] [Google Scholar]
- 14.Nelms K, et al. Alternative splicing, gene localization, and binding of SH2-B to the insulin receptor kinase domain. Mamm. Genome. 1999;10:1160–1167. doi: 10.1007/s003359901183. [DOI] [PubMed] [Google Scholar]
- 15.Yenush L, White MF. The IRS-signaling system during insulin and cytokine action [review] Bioessays. 1997;19:491–500. doi: 10.1002/bies.950190608. [DOI] [PubMed] [Google Scholar]
- 16.Pawson T, Scott JD. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 17.Kotani K, Wilden P, Pillay TS. SH2-Balpha is an insulin-receptor adapter protein and substrate that interacts with the activation loop of the insulin-receptor kinase. Biochem. J. 1998;335:103–109. doi: 10.1042/bj3350103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lock P, Casagranda F, Dunn AR. Independent SH2-binding sites mediate interaction of Dok-related protein with RasGTPase-activating protein and Nck. J. Biol. Chem. 1999;274:22775–22784. doi: 10.1074/jbc.274.32.22775. [DOI] [PubMed] [Google Scholar]
- 19.Noguchi T, et al. Tyrosine phosphorylation of p62(Dok) induced by cell adhesion and insulin: possible role in cell migration. EMBO J. 1999;18:1748–1760. doi: 10.1093/emboj/18.7.1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiang SH, et al. Insulin-stimulated GLUT4 translocation requires the CAP-dependent activation of TC10. Nature. 2001;410:944–948. doi: 10.1038/35073608. [DOI] [PubMed] [Google Scholar]
- 21.Baumann CA, et al. CAP defines a second signalling pathway required for insulin-stimulated glucose transport. Nature. 2000;407:202–207. doi: 10.1038/35025089. [DOI] [PubMed] [Google Scholar]
- 22.Kido Y, et al. Tissue-specific insulin resistance in mice with mutations in the insulin receptor, IRS-1, and IRS-2. J. Clin. Invest. 2000;105:199–205. doi: 10.1172/JCI7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Withers DJ, et al. Disruption of IRS-2 causes type 2 diabetes in mice. Nature. 1998;391:900–904. doi: 10.1038/36116. [DOI] [PubMed] [Google Scholar]
- 24.Choudhury AI, et al. The role of insulin receptor substrate 2 in hypothalamic and β cell function. J. Clin. Invest. 2005;115:940–950. doi:10.1172/JCI200524445. doi: 10.1172/JCI24445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kubota N, et al. Insulin receptor substrate 2 plays a crucial role in β cells and the hypothalamus. J. Clin. Invest. 2004;114:917–927. doi:10.1172/JCI200421484. doi: 10.1172/JCI21484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin X, et al. Dysregulation of insulin receptor substrate 2 in β cells and brain causes obesity and diabetes. J. Clin. Invest. 2004;114:908–916. doi:10.1172/JCI200422217. doi: 10.1172/JCI22217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rother KI, et al. Evidence that IRS-2 phosphorylation is required for insulin action in hepatocytes. J. Biol. Chem. 1998;273:17491–17497. doi: 10.1074/jbc.273.28.17491. [DOI] [PubMed] [Google Scholar]
- 28.Ide T, et al. SREBPs suppress IRS-2-mediated insulin signalling in the liver. Nat. Cell Biol. 2004;6:351–357. doi: 10.1038/ncb1111. [DOI] [PubMed] [Google Scholar]
- 29.Valverde AM, Fabregat I, Burks DJ, White MF, Benito M. IRS-2 mediates the antiapoptotic effect of insulin in neonatal hepatocytes. Hepatology. 2004;40:1285–1294. doi: 10.1002/hep.20485. [DOI] [PubMed] [Google Scholar]
- 30.Valverde AM, et al. Molecular mechanisms of insulin resistance in IRS-2-deficient hepatocytes. Diabetes. 2003;52:2239–2248. doi: 10.2337/diabetes.52.9.2239. [DOI] [PubMed] [Google Scholar]
- 31.Jhala US, et al. cAMP promotes pancreatic beta-cell survival via CREB-mediated induction of IRS2. Genes Dev. 2003;17:1575–1580. doi: 10.1101/gad.1097103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taniguchi CM, Ueki K, Kahn R. Complementary roles of IRS-1 and IRS-2 in the hepatic regulation of metabolism. J. Clin. Invest. 2005;115:718–727. doi:10.1172/JCI200523187. doi: 10.1172/JCI23187. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Burks DJ, et al. IRS-2 pathways integrate female reproduction and energy homeostasis. Nature. 2000;407:377–382. doi: 10.1038/35030105. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki R, et al. Both insulin signaling defects in the liver and obesity contribute to insulin resistance and cause diabetes in Irs2(–/–) mice. J. Biol. Chem. 2004;279:25039–25049. doi: 10.1074/jbc.M311956200. [DOI] [PubMed] [Google Scholar]
- 35.Araki E, et al. Alternative pathway of insulin signalling in mice with targeted disruption of the IRS-1 gene. Nature. 1994;372:186–190. doi: 10.1038/372186a0. [DOI] [PubMed] [Google Scholar]
- 36.Previs SF, Withers DJ, Ren JM, White MF, Shulman GI. Contrasting effects of IRS-1 vs IRS-2 gene disruption on carbohydrate and lipid metabolism in vivo. J. Biol. Chem. 2000;275:38990–38994. doi: 10.1074/jbc.M006490200. [DOI] [PubMed] [Google Scholar]
- 37.Tamemoto H, et al. Insulin resistance and growth retardation in mice lacking insulin receptor substrate-1. Nature. 1994;372:182–186. doi: 10.1038/372182a0. [DOI] [PubMed] [Google Scholar]
- 38.Withers DJ, et al. Irs-2 coordinates Igf-1 receptor-mediated beta-cell development and peripheral insulin signalling. Nat. Genet. 1999;23:32–40. doi: 10.1038/12631. [DOI] [PubMed] [Google Scholar]
- 39.Postic C, Magnuson MA. DNA excision in liver by an albumin-Cre transgene occurs progressively with age. Genesis. 2000;26:149–150. doi: 10.1002/(sici)1526-968x(200002)26:2<149::aid-gene16>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 40.Escribano O, et al. Insulin receptor substrate-4 signaling in quiescent rat hepatocytes and in regenerating rat liver. Hepatology. 2003;37:1461–1469. doi: 10.1053/jhep.2003.50245. [DOI] [PubMed] [Google Scholar]
- 41.O’Brien RM, Lucas PC, Forest CD, Magnuson MA, Granner DK. Identification of a sequence in the PEPCK gene that mediates a negative effect of insulin on transcription. Science. 1990;249:533–537. doi: 10.1126/science.2166335. [DOI] [PubMed] [Google Scholar]
- 42.Guo S, et al. Phosphorylation of serine 256 by protein kinase B disrupts transactivation by FKHR and mediates effects of insulin on insulin-like growth factor-binding protein-1 promoter activity through a conserved insulin response sequence. J. Biol. Chem. 1999;274:17184–17192. doi: 10.1074/jbc.274.24.17184. [DOI] [PubMed] [Google Scholar]
- 43.Kwon HS, Huang B, Unterman TG, Harris RA. Protein kinase B-alpha inhibits human pyruvate dehydrogenase kinase-4 gene induction by dexamethasone through inactivation of FOXO transcription factors. Diabetes. 2004;53:899–910. doi: 10.2337/diabetes.53.4.899. [DOI] [PubMed] [Google Scholar]
- 44.Accili D, Nakae J, Kim JJ, Park BC, Rother KI. Targeted gene mutations define the roles of insulin and IGF-I receptors in mouse embryonic development. J. Pediatr. Endocrinol. Metab. 1999;12:475–485. doi: 10.1515/jpem.1999.12.4.475. [DOI] [PubMed] [Google Scholar]
- 45.Schubert M, et al. Insulin receptor substrate-2 deficiency impairs brain growth and promotes tau phosphorylation. J. Neurosci. 2003;23:7084–7092. doi: 10.1523/JNEUROSCI.23-18-07084.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ogata N, et al. Insulin receptor substrate-1 in osteoblast is indispensable for maintaining bone turnover. J. Clin. Invest. 2000;105:935–943. doi: 10.1172/JCI9017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harris RA, Bowker-Kinley MM, Huang B, Wu P. Regulation of the activity of the pyruvate dehydrogenase complex. Adv. Enzyme Regul. 2002;42:249–259. doi: 10.1016/s0065-2571(01)00061-9. [DOI] [PubMed] [Google Scholar]
- 48.Rhodes CJ. Type 2 diabetes: a matter of beta-cell life and death? Science. 2005;307:380–384. doi: 10.1126/science.1104345. [DOI] [PubMed] [Google Scholar]
- 49.Bard-Chapeau EA, et al. Deletion of Gab1 in the liver leads to enhanced glucose tolerance and improved hepatic insulin action. Nat. Med. 2005;11:567–571. doi: 10.1038/nm1227. [DOI] [PubMed] [Google Scholar]
- 50.Fantin VR, Wang Q, Lienhard GE, Keller SR. Mice lacking insulin receptor substrate 4 exhibit mild defects in growth, reproduction, and glucose homeostasis. Am. J. Physiol. Endocrinol. Metab. 2000;278:E127–E133. doi: 10.1152/ajpendo.2000.278.1.E127. [DOI] [PubMed] [Google Scholar]
- 51.Sasaoka T, Kobayashi M. The functional significance of Shc in insulin signaling as a substrate of the insulin receptor. Endocr. J. 2000;47:373–381. doi: 10.1507/endocrj.47.373. [DOI] [PubMed] [Google Scholar]
- 52.Van Horn DJ, Myers MG, Jr, Backer JM. Direct activation of the phosphatidylinositol 3′-kinase by the insulin receptor. J. Biochem. 1994;269:29–32. [PubMed] [Google Scholar]
- 53.Lawlor MA, Alessi DR. PKB/Akt: a key mediator of cell proliferation, survival and insulin responses? J. Cell Sci. 2001;114:2903–2910. doi: 10.1242/jcs.114.16.2903. [DOI] [PubMed] [Google Scholar]
- 54.Hardie DG. Minireview. The AMP-activated protein kinase cascade: the key sensor of cellular energy status. Endocrinology. 2003;144:5179–5183. doi: 10.1210/en.2003-0982. [DOI] [PubMed] [Google Scholar]
- 55.Ferrer JC, et al. Control of glycogen deposition. Growth Regul. 2003;546:127–132. doi: 10.1016/s0014-5793(03)00565-9. [DOI] [PubMed] [Google Scholar]
- 56.Frame S, Cohen P. GSK3 takes centre stage more than 20 years after its discovery. Biochem. J. 2001;359:1–16. doi: 10.1042/0264-6021:3590001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park J, et al. Serum and glucocorticoid-inducible kinase (SGK) is a target of the PI 3-kinase-stimulated signaling pathway. EMBO J. 1999;18:3024–3033. doi: 10.1093/emboj/18.11.3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ueki K, et al. Positive and negative roles of p85 alpha and p85 beta regulatory subunits of phosphoinositide 3-kinase in insulin signaling. J. Biol. Chem. 2003;278:48453–48466. doi: 10.1074/jbc.M305602200. [DOI] [PubMed] [Google Scholar]
- 59.Ueki K, et al. Increased insulin sensitivity in mice lacking p85beta subunit of phosphoinositide 3-kinase. Proc. Natl. Acad. Sci. U. S. A. 2002;99:419–424. doi: 10.1073/pnas.012581799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ueki K, et al. Molecular balance between the regulatory and catalytic subunits of phosphoinositide 3-kinase regulates cell signaling and survival. Mol. Cell. Biol. 2002;22:965–977. doi: 10.1128/MCB.22.3.965-977.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alessi DR, et al. 3-Phosphoinositide-dependent protein kinase-1 (PDK1): structural and functional homology with the Drosophila DSTPK61 kinase. Curr. Biol. 1997;7:776–789. doi: 10.1016/s0960-9822(06)00336-8. [DOI] [PubMed] [Google Scholar]
- 62.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 63.Stokoe D, et al. Dual role of phosphatidylinositol-3,4,5-triphosphate in the activation of protein kinase B. Science. 1997;277:567–570. doi: 10.1126/science.277.5325.567. [DOI] [PubMed] [Google Scholar]
- 64.Mora A, Lipina C, Tronche F, Sutherland C, Alessi DR. Deficiency of PDK1 in liver results in glucose intolerance, impairment of insulin-regulated gene expression and liver failure. Biochem. J. 2005;385:639–648. doi: 10.1042/BJ20041782. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 65.Sakoda H, et al. Differing roles of Akt and serum- and glucocorticoid-regulated kinase in glucose metabolism, DNA synthesis, and oncogenic activity. J. Biol. Chem. 2003;278:25802–25807. doi: 10.1074/jbc.M301127200. [DOI] [PubMed] [Google Scholar]
- 66.Daitoku H, Yamagata K, Matsuzaki H, Hatta M, Fukamizu A. Regulation of PGC-1 promoter activity by protein kinase B and the forkhead transcription factor FKHR. Diabetes. 2003;52:642–649. doi: 10.2337/diabetes.52.3.642. [DOI] [PubMed] [Google Scholar]
- 67.Finlay D, et al. Glycogen synthase kinase-3 regulates IGFBP-1 gene transcription through the thymine-rich insulin response element. BMC Mol. Biol. 2004;5:15. doi: 10.1186/1471-2199-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sjogren K, et al. Liver-derived IGF-I is of importance for normal carbohydrate and lipid metabolism. Diabetes. 2001;50:1539–1545. doi: 10.2337/diabetes.50.7.1539. [DOI] [PubMed] [Google Scholar]
- 69.Koo SH, et al. PGC-1 promotes insulin resistance in liver through PPAR-alpha-dependent induction of TRB-3. Nat. Med. 2004;10:530–534. doi: 10.1038/nm1044. [DOI] [PubMed] [Google Scholar]
- 70.Schadinger SE, Bucher NL, Schreiber BM, Farmer SR. PPARgamma2 regulates lipogenesis and lipid accumulation in steatotic hepatocytes. Am. J. Physiol. Endocrinol. Metab. 2005;288:E1195–E1205. doi: 10.1152/ajpendo.00513.2004. [DOI] [PubMed] [Google Scholar]
- 71.Sato O, Kuriki C, Fukui Y, Motojima K. Dual promoter structure of mouse and human fatty acid translocase/CD36 genes and unique transcriptional activation by peroxisome proliferator-activated receptor alpha and gamma ligands. J. Biol. Chem. 2002;277:15703–15711. doi: 10.1074/jbc.M110158200. [DOI] [PubMed] [Google Scholar]
- 72.Flier SN, Kulkarni RN, Kahn CR. Evidence for a circulating islet cell growth factor in insulin-resistant states. Proc. Natl. Acad. Sci. U. S. A. 2001;98:7475–7480. doi: 10.1073/pnas.131192998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kushner JA, et al. Pdx1 restores β cell function in Irs2 knockout mice. J. Clin. Invest. 2002;109:1193–1201. doi:10.1172/JCI200214439. doi: 10.1172/JCI14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kushner JA, et al. Islet-sparing effects of protein tyrosine phosphatase-1b deficiency delays onset of diabetes in IRS2 knockout mice. Diabetes. 2004;53:61–66. doi: 10.2337/diabetes.53.1.61. [DOI] [PubMed] [Google Scholar]
- 75.Howlett KF, et al. Insulin signaling after exercise in insulin receptor substrate-2-deficient mice. Diabetes. 2002;51:479–483. doi: 10.2337/diabetes.51.2.479. [DOI] [PubMed] [Google Scholar]
- 76.Laustsen PG, et al. Lipoatrophic diabetes in Irs1(–/–)/Irs3(–/–) double knockout mice. Genes Dev. 2002;16:3213–3222. doi: 10.1101/gad.1034802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fujii N, et al. Overexpression or ablation of JNK in skeletal muscle has no effect on glycogen synthase activity. Am. J. Physiol. Cell Physiol. 2004;287:C200–C208. doi: 10.1152/ajpcell.00415.2003. [DOI] [PubMed] [Google Scholar]
- 78.Lang, G., and Michal, G. 1974. D-glucose-6-phosphate and D-fructose-6-phosphate. In Methods of enzymatic analysis. Academic. New York, New York, USA. 1238–1242.