Abstract
The bulged insertions of (R)-1-O-(pyren-1-ylmethyl)glycerol (monomer P) in two complementary 8mer DNA strands (intercalating nucleic acids) opposite to each other resulted in the formation of an easily denaturing duplex, which had lower thermal stability (21.0°C) than the wild-type double-stranded DNA (dsDNA, 26.0°C), but both modified oligodeoxynucleotides had increased binding affinity toward complementary single-stranded DNA (ssDNA) (41.5 and 39.0°C). Zipping of pyrene moieties in an easily denaturing duplex gave formation of a strong excimer band at 480 nm upon excitation at 343 nm in the steady-state fluorescence spectra. The excimer band disappeared upon addition of a similar short dsDNA, but remained when adding a 128mer dsDNA containing the same sequence. When P was inserted into 2′-OMe-RNA strands, the duplex with zipping P was found to be more stable (42.0°C) than duplexes with the complementary ssDNAs (31.5 and 19.5°C). The excimer band observed in the ds2′-OMe-RNA with zipping P had marginal changes upon addition of both 8 and 128mer dsDNA. Synthesized oligonucleotides were tested in a transcriptional inhibition assay for targeting of the open complex formed by Escherichia coli RNA polymerase with the lac UV-5 promoter using the above mentioned 128mer dsDNA. Inhibition of transcription was observed for 8mer DNAs possessing pyrene intercalators and designed to target both template and non-template DNA strands within the open complex. The observed inhibition was partly a result of unspecific binding of the modified DNAs to the RNA polymerase. Furthermore, the addition of 8mer DNA with three bulged insertions of P designed to be complementary to the template strand at the +36 to +43 position downstream of the transcription start resulted in a specific halt of transcription producing a truncated RNA transcript. This is to our knowledge the first report of an RNA elongation stop mediated by a small DNA sequence possessing intercalators. The insertions of P opposite to each other in ds2′-OMe-RNA showed inhibition efficiency of 96% compared with 25% for unmodified ds2′-OMe-RNA.
INTRODUCTION
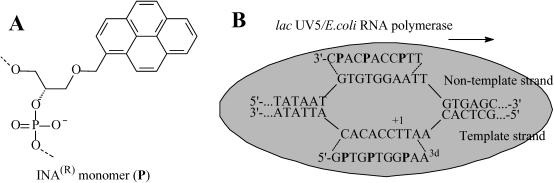
The sequence-specific recognition of double-stranded DNA (dsDNA) using chemically modified oligonucleotides (ONs) is of considerable interest in the development of ON-based tools in molecular biology, therapeutics and bionanotechnology. A competing duplex formation using a pair of modified ONs to disrupt and hybridize to dsDNA through Watson–Crick base pairing is considered to be a more attractive approach than formation of triple helixes through Hoogsteen base pairing and has been termed double-duplex invasion (1) or dual duplex invasion (2). This method may improve detection and sequence-selective treatment of dsDNA for mutagenesis or recombination of genes. For this purpose, invading chemically modified nucleic acids with better binding affinity toward native DNA than with each other is needed. Efficient hybridization affinity of modified ONs toward single-stranded DNA (ssDNA) or ssRNA usually results in even stronger binding to a complementary sequence of their own type, as observed for Peptide Nucleic Acids (PNAs) (3) and Locked Nucleic Acids (LNAs) (4). There have also been several reports about pseudo-complementary nucleic acids having modified nucleic bases which formed considerably less stable base pairs than natural bases (5,6). Pseudo-complementary PNAs (7) have been used for targeting breathed plasmid DNA using non-physiological salt concentration (8). Recently, we have presented the design of easily denaturing nucleic acids constructed from DNA- and RNA-discriminating molecules (9). LNA binds better to ssRNA than to ssDNA (10,11). Contrary to the LNA case, the bulge insertions of (R)-1-O-(pyren-1-ylmethyl)glycerol (monomer P, Figure 1A) into oligodeoxynucleotides [intercalating nucleic acids (INAs®)] induce an increased affinity for complementary ssDNA (ΔTm = +1.0 to +11.0°C), but reduce the affinity for ssRNA (ΔTm = −2.0 to −7.0°C) (12). (INA® is commercially available from Human Genetic Signatures Pty Ltd, Sydney, Australia.) Placement of LNA as an RNA-mimic molecule and INA as a DNA-like molecule opposite each other in complementary regions has been used to make targeting of native nucleic acids more favorable than ‘quenching’ of LNA-rich sites (9).
Figure 1.
(A) Structure of INA monomer. (B) Schematic presentation of the open complex formed by dsDNA and RNA polymerase. A proposed targeting of the template and non-template strand by an easily denaturing duplex at transcription start site (+1) is shown. A3d denotes 3′-deoxyadenosine, P denotesINA monomer.
Recently, we observed another unique feature of INA: the insertion of (R)-1-O-(pyren-1-ylmethyl)glycerol (monomer P) as a bulge in two complementary oligodeoxynucleotide strands opposite to each other resulted in a decreased thermal stability of the INA/INA duplex (Tm = 46.5°C) compared with the corresponding INA/DNA duplexes (Tm = 52.5 and 54.5°C) (13,14). According to a NMR structure of the INA/INA duplex, the two pyrene moieties were zipping in the center of the helix and were stacking with the neighboring nucleobases (13). Therefore, the INA/INA complex can also be considered as an example of easily denaturing nucleic acids. Coaxial stacking of two pyrene residues in a helix usually results in the formation of an excimer band at 480 nm upon excitation at 343 nm in steady-state fluorescence spectra (12,15,16). Therefore, formation and disappearance of this excimer band can be used to monitor the binding/release of pyrene-containing ONs with itself and with complementary DNA. Recently, interstrand communication of pyrene labeled nucleic acids has been reported (15,16) and the disappearance of the pyrene excimer band has been exploited in monitoring of a dual duplex recognition of dsDNA of the same length as the probes (2).
The challenge of targeting and sequence-specific recognition of long relaxed dsDNA under physiological conditions using short invaders has not been met (8,17). It has been shown that short oligoribonucleotides could bind to the template strand during the local opening of the dsDNA by Escherichia coli RNA polymerase and inhibit start of transcription (18–22). It was concluded that the inhibitor should be an RNA mimic possessing a non-extendable terminal 3′-deoxyribonucleotide, an intercalating moiety and should hybridize to position −5 to +2 of the template strand (18–22). We have used a combination of INA and LNA monomers with 2′-OMe-ribonucleotides in pentanucleotidic inhibitors of E.coli RNA polymerase and obtained >95% inhibitory efficiency at 16 µM compared with 60% for unmodified 2′-OMe-RNA in the same assay (23). A considerable drop in inhibition efficiency has been observed for a single mis-matched pentamer with P insertion compared with the corresponding perfectly matched pentamer. We also observed that low hybridization affinity of pentanucleotides to the template strand correlated with low inhibition. However, in some cases high thermal stability did not improve inhibition. In two very recent publications, transfected 19mer PNA and 21mer duplex RNA were reported to inhibit the transcription start site for the human progesterone receptor in T47D breast cancer cells (24,25). Interestingly, despite a very high thermal stability (Tm = 64–84°C), the duplex antigene RNAs showed good inhibition efficiency (25). We anticipate that targeting of an open complex with easily denaturing nucleic acids can improve inhibition of RNA transcription start (Figure 1B) and it may be considered as a next step in the development of sequence-specific molecular recognition and targeting of biologically important dsDNA.
In this paper, we present the design and fluorescence properties of easily denaturing nucleic acids derived from (R)-1-O-(pyren-1-ylmethyl)glycerol (P) inserted as a bulge into either 2′-deoxyoligonucleotides or 2′-OMe-oligonucleotides. These ONs were used as invaders in a competing duplex formation targeting dsDNA of the same size which was followed by fluorescence measurements. Furthermore, the synthesized ONs were evaluated in a transcriptional inhibition assay where they targeted the open complex formed by E.coli RNA polymerase with the lac UV-5 promoter. The insertion of P into ONs led to increased inhibition efficiency compared with unmodified ONs. The effect is explained by a combination of targeting of melted dsDNA within the open complex and by unspecific binding to RNA polymerase. Adding an 8mer DNA with three bulged P as a complement to the template strand at positions +36 to +43 led to the formation of a truncated RNA in the transcription assay. The complement to this ON that was complementary to the non-template strand did not result in the formation of truncated RNAs but the inhibition efficiency for the 61mer RNA was at the same level as for other 8mer DNAs with Ps. The insertions of P opposite to each other into ds2′-OMe-RNA gave inhibition efficiency of 96% compared with 25% for unmodified ds2′-OMe-RNA.
MATERIALS AND METHODS
Oligonucleotide synthesis using 2′-O-methylated-RNA and INA® monomers
Oligonucleotides were synthesized on a 0.2 µmol scale using Expedite™ Nucleic Acid Synthesis System Model 8909 from Applied Biosystems. 2′-O-Methylated-RNA phosphoramidites and A3′deoxy succinyl CPG support were purchased from Glen Research; an universal CPG support 500 was from BioGenex. INA phosphoramidite was prepared as described previously (12). The 5′-DMT-on ONs were cleaved off the solid support (rt, 3h) and deprotected (55°C, overnight) using 32% aqueous ammonia for succinyl CPG supports or a solution of 2% LiCl in 32% aqueous ammonia (w/w) when using universal CPG support 500 (BioGenex). Purification of 5′-O-DMT-on ONs was carried out using a Waters Delta Prep 4000 Preparative Chromatography System on a Waters Xterra™ MS C18 column and dual-wavelength (254 and 343 nm) detection. The ONs were DMT deprotected in 100 µl of 80% aq. acetic acid, diluted with 100 µl H2O, followed by 1 M aq. NaOAc (150 µl) and precipitated from ethanol (550 µl). Matrix-assisted laser desorption ionization time-of-flight analysis was performed on a Voyager Elite Biospectrometry Research Station from PerSeptive Biosystems. All ONs gave satisfactory composition with a purity above 90% that was verified by ion-exchange chromatography using LaChrom system from Merck Hitachi on GenPak-Fax column (Waters). ON concentrations were determined by absorbance at 260 nm and the calculated single-strand extinction coefficients were based on a nearest neighbor model [the extinction coefficient for P monomer is 22 400 (260 nm) and 43 000 (343 nm)].
Preparation of dsDNA transcription template with a lac UV5 promoter
The lac promoter in pUC18 was mutagenized by changing position −8A and −9C to −8T and −9T (numbering according to transcription start) to create the so-called UV-5 promoter in a 128 nt long dsDNA. The following primers were used: 5′-TGAGTTAGCTCACTCATTAGG-3′ and 5′-CTCGAATTCGTAATCATGGTC-3′ as the ends of the UV5-template and 5′-ACACATTATACGAGCCGGAAG-3′ and 5′-CTTCCGGCTCGTATAATGTGT-3′ to insert the internal TT/AA mutations. The first PCR created a 128mer product around the lac promoter in pUC18. The second and third PCRs created shorter versions with the T/A mutations included and each with one end of the 128mer dsDNA. The fourth PCR with the product from the second and third PCR extended the mutated product to the original 128mer length including the two mutations.
In vitro transcription assay
The above described dsDNA were used as transcription template for a 61mer RNA product. The transcription assay was performed as follows: 10 µl (40 mM Tris–HCl, 50 mM KCl, 10 mM MgCl2, 0.1 mM DTT, 100 µg/ml BSA and 5% glycerol) containing 1 µl of 0.5 µM template fragment, 5′-32P-labeled 30mer DNA control oligo, 1 µl of 0.5 U/µl E.coli RNA polymerase (Sigma) and the inhibitor to be analyzed were preincubated for 10 min at 37°C. The reactions were started by adding an rNTP mixture consisting of 1 µl of 1 mM ATP, GTP, UTP plus 0.2 µl 32P-αCTP (3000 Ci/mmol). The reactions were stopped by adding 5 µl cold formamide loading buffer after 4 min from the transcription initiation and frozen for later electrophoresis or loaded on acrylamide gels. A longer reaction time resulted in some degradation of the RNA product, as observed in transcription experiments without any inhibitor. The samples were heated for 2 min at 80°C before loading on 13% denaturing acrylamide gels. The gels were dried after electrophoresis and analyzed by a Phosphor Imager whereby the amount of transcript could be quantified relative to the added control ON. The effect of the added ONs was assessed by comparison with values for the transcript obtained without any inhibitor. The amount of CTP used was optimized by varying the concentrations of rNTPs. The ON:template ratio was 200:1 at 10 µM of each oligo. All experiments were repeated 2–3 times and average inhibition efficiencies are presented in Table 1.
Table 1.
Thermal stability (Tm) and inhibition efficiency of ONs synthesizeda
| Sequence | ON6 | ON7 | ON8 | ON9 | ON10 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3′-cPaccPu | 3′-cPacPaccPuu | 3′-CPACPACCPTT | 3′-cacaccuu | 3′-CACACCTT | ||||||||
| 0%b | 80% | 56% | 0% | |||||||||
| ON1 | 5′-gPuggPA3d | 18.0c | 29.5 | <5.0 | 21.0 | 13.5 | ||||||
| 42% | 72% | 46% | ||||||||||
| ON2 | 5′-gPugPuggPaA3d | 26.0 | 42.0 | 21.5 | 35.1 | 31.5 | ||||||
| 90% | 94% | 96% | 96% | 92% | ||||||||
| ON3 | 5′-GPTGPTGGPAA | <5.0 | <5.0 | 21.0 | <5.0 | 41.5 | ||||||
| 67% | 72% | 88% | 84% | 77% | ||||||||
| ON4 | 5′-guguggaA3d | 25.5 | 37.5 | 24.0 | 55.5 | 36.5 | ||||||
| 19% | 25% | 83% | 62% | 25% | ||||||||
| ON5 | 5′-GTGTGGAA | <5.0 | 19.5 | 39.0 | 21.0 | 26.0 | ||||||
aP denotes INA monomer; A3d denotes 3′-deoxyadenosine; g, u, a, c denote 2′-O-methylribonucleotides; G, T, A, C denote 2′-deoxyribonucleotides. ON1-5 are complements to the template strand; ON6-10 are complements to the non-template strand.
bInhibition efficiencies are showed in the right lower corner of the cell in italic and defined as percentage decrease of transcription relative to noninhibited RNA transcription at 10 µM concentration of inhibitor.
cTms (°C), which are showed in the left upper corner of the cell, were determined at 260 nm as the maximum of the first derivative plots of the melting curves obtained by measuring absorbance at 260 nm against increasing temperature (1.0°C min−1) on equimolar mixtures (1.0 µM in each strand) of ON1-10 in a hybridization buffer (40 mM Tris–HCl, 100 mM KCl, 10 mM MgCl2, pH 7.9). The bold Tm values represent easily denaturing nucleic acids. The underlined values of Tm represent the expanded list of easily denaturing nucleic acids (see text for details).
Measurement of transition temperatures
The melting temperature measurements were performed using a Perkin-Elmer Lambda 50 UV-visible spectrophotometer fitted with a PTP-6 (Peltier Temperature Programmer) device. The duplexes were formed by mixing the two strands, each at a concentration of 1.0 µM in the hybridization buffer (40 mM Tris–HCl, 100 mM KCl, 10 mM MgCl2, pH 7.9). The solutions were heated at 70°C for 5 min and then cooled to room temperature during 10 min before measuring the melting curves. The melting temperature (Tm, °C) was determined as the maximum of the first derivative plots of the melting curves obtained by measuring absorbance at 260 nm against increasing temperature (1.0°C min−1) from 5.0 to 60.0°C. The control experiments were also performed at 343 nm for P containing sequences. All melting temperatures are reported within an uncertainty ±0.5°C, as determined from multiple experiments.
Fluorescence measurements
Fluorescence measurements were performed on a Perkin-Elmer luminescence spectrometer LS-55 fitted with a Julabo F25 temperature controller. Steady-state spectra were recorded at 10°C upon excitation at 343 nm in 1 ml cuvette; 1 µM of all strands possessing P and 1.5 µM of ON5, ON10 or 128mer dsDNA were in the same buffer as used for the hybridization studies; duplexes were formed by heating to 70°C and cooling to 10°C.
RESULTS AND DISCUSSION
We suppose that the discrimination properties of INA with ssDNA relative to ssRNA and INA can be a result of a unique ability of a B-type duplex to adopt the bulged insertions of (R)-1-O-(pyren-1-ylmethyl)glycerol (P). Incorporation of P as a bulge in a sequence leads to different hybridization properties depending on its incorporation into A- or B-type duplexes. The influence of P incorporation into RNA-like sequences on thermal stability has not been studied in detail. Some preliminary results have been obtained using 2′-OMe-pentaribonucleotides with P for improving transcription inhibition (23). In extension of this work, we synthesized 5 and 8mer 2′-OMe-ribonucleotides with P insertions designed to target either template or non-template strand of the open complex formed by E.coli RNA polymerase with the lac UV-5 promoter. It was expected that an 8mer sequence will give better sequence-selective binding to ssDNA than pentamers and that it can still target the open transcription initiation complex (20). The 2′-OMe-sequences targeting the template strand contain a terminal 3′-deoxynucleotide to prevent elongation, while there is no need for this for DNA sequences or ONs targeting the non-template strand. For comparison, the similar 8mer DNA sequences with P insertions were also synthesized (ON3 and ON8). To unveil the relationship between inhibition and binding affinity of ONs toward ssDNA and toward a complementary sequence with P insertions, we performed thermal denaturation (Table 1) and fluorescence studies (Figure 2) in a buffer similar to the one used for the RNA transcription assay. Finally, all the synthesized ONs, except pure DNAs, were checked in an in vitro transcription assay at 10 µM (Table 1). The inhibition efficiencies were quantified and compared with the melting and fluorescence data.
Figure 2.
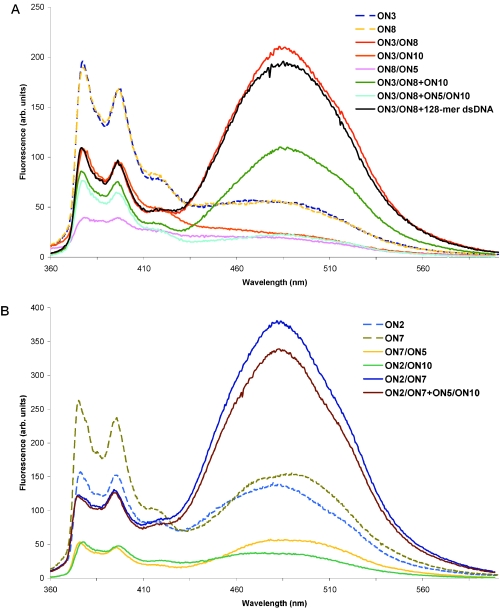
(A) Fluorescence spectra of ON3 and ON8 and their combinations with ON5, ON10 and 128mer dsDNA. (B) Fluorescence spectra of ON2 and ON7 and their combinations with ON5 and ON10. Spectra recorded at 10°C upon excitation at 343 nm in 1 ml cuvette; 1 µM of all strands possessing P and 1.5 µM of ON5, ON10 or 128mer dsDNA in the same buffer as in the hybridization studies; duplexes were preformed by heating to 70°C and cooling to 10°C. Plus sign indicates that the latter mentioned ON or preformed duplex were added to a solution of the first mentioned duplex.
Easily denaturing duplexes
As the Tm value of the wild-type dsDNA (ON5/ON10, Table 1) was 26.0°C, those pairs of ONs with lower thermal stability were considered easily denaturing nucleic acids if both ONs had higher Tm toward their complementary ssDNA. Although a variety in thermal stability of complementary chemically modified ONs were observed, three pairs of ONs were found that were regarded as easily denaturing nucleic acids (Tms marked in bold in Table 1).
As it was expected (13,14) the incorporation of three Ps in the 8mer dsDNA opposite each other resulted in the formation of easily denaturing duplex: ONs had increased binding affinity toward ssDNA (ON3/ON10, ON5/ON8, Table 1) and INA/INA duplex (ON3/ON8) had lower thermal stability when compared with the wild-type dsDNA (ON5/ON10). Different effect was observed for ds2′-OMe-RNA, which is believed to be an A-type duplex. Unmodified ds2′-OMe-RNA (ON4/ON9) was the most stable duplex and insertions of P into 2′-OMe-RNA sequence destabilized duplexes in all cases. Placing of the pyrene units opposite to each other in the ds2′-OMe-RNA led to a zipping duplex (ON2/ON7) with higher thermal stability than those duplexes possessing P in one of the strands (ON2/ON9 and ON4/ON7) and it was the least destabilized duplex among all the 2′-OMe-RNA duplexes studied here with P insertions. This 2′-OMe-RNA duplex with zipping pyrenes cannot, in contrary to INA/INA duplex, be called an easily denaturing duplex.
It was also observed that the insertions of P into either 2′-OMe-RNA or DNA strands of 2′-OMe-RNA/DNA duplexes (ON2/ON10, ON4/ON8 and ON5/ON7, ON3/ON9) reduced binding affinity as compared with the corresponding unmodified 2′-OMe-RNA/DNA duplexes (ON4/ON10 and ON5/ON9, respectively). However, when compared with wild-type dsDNA, increased binding affinity could be obtained upon insertion of P into 2′-OMe-RNA strands as observed in the case of ON2/ON10 (Tm = 31.5°C). This makes it possible to use ON2 in combination with ON8 as an easily denaturing duplex. Incorporation of P into 2′-OMe-RNA also considerably reduces the stability of ds2′-OMe-RNA with little influence on the stability toward ssDNA. This suggests that INA monomers can be applied to destabilize self-hybridization of 2′-OMe-RNA probes.
For ssONs (ON2, ON3, ON7 and ON8, Figure 2A and B), we observed the formation of a monomer fluorescence at 380 and 405 nm and an excimer band at 480 nm upon excitation at 343 nm. The appearance of the excimer band for ssONs could be explained by folding of the single strand or by communication of the pyrenes separated by 2 bp. Another possibility is intermolecular communication of pyrenes in different single strands. These intra- or intermolecular structures are believed to be unstable because they do not interfere in the annealing with the complementary strands as deduced from thermal stability experiments.
As expected for duplexes with zipping pyrenes ON3/ON8 and ON2/ON7 (Figure 2A and B, respectively), they formed a strong excimer band at 480 nm in the steady-state fluorescence spectra upon excitation at 343 nm. This can indicate duplex formation where the signals appear as a result from coaxial stacking of two pyrene moieties positioned inside the nucleic acids helix. To ascertain the formation of the duplexes, all fluorescence experiments were measured at 10°C. The intensity of the excimer band at 480 nm for the corresponding ssONs (ON2, ON3, ON7 and ON8) was considerably lower than the intensity of the excimer band for the above mentioned duplexes. Lowering of the monomer fluorescence at 380 and 405 nm in these duplexes compared with the spectra of the single strands also supports a placement of pyrenes inside the helixes which allows their interstrand communication.
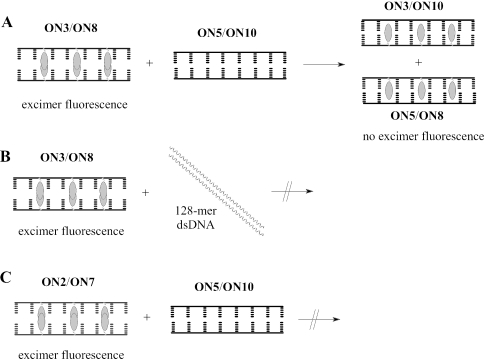
The excimer band of the easily denaturing duplex ON3/ON8 was partially quenched upon addition of one of the complementary ssDNAs (ON3/ON8+ON10). The complete disappearance of the excimer band was observed when the preformed wild-type dsDNA (ON5/ON10, 1.5 eqv) of the same size as the probes was added to the preformed dsINA (ON3/ON8). There is evidence for the formation of two duplexes ON3/ON10 and ON5/ON8, because no excimer bands were observed for these duplexes when studied separately. The fluorescence spectra are in agreement with the thermal stability studies for this pairs of ONs. Therefore, the process of dual duplex invasion using easily denaturing duplex ON3/ON8 can be represented as shown in Figure 3A. The strong excimer band was still observed upon addition of non-complementary and single mis-matched dsDNA to dsINA (ON3/ON8) showing that dual duplex invasion is sequence-selective (spectra not shown). Importantly, this strand invasion experiments were performed under medium salt conditions and with dsDNA of the same size as the INA probes. When longer linear dsDNA (128mer) was added to the probes (ON3/ON8+long dsDNA), the fluorescence spectra showed marginal changes. The long dsDNA had the same sequence as the dsDNA used in the transcription assay with the target site located in the middle of the sequence. The spectrum of INA probes (ON3/ON8) and 128mer dsDNA did not change after 5 h incubation at 10°C or after heating samples at 90°C followed by cooling to 10°C (spectra not showed). From these data, we concluded that there was no strand invasion of the short probes into the middle of the 128mer dsDNA (Figure 3B). This difference in fluorescence spectra of short probes upon targeting of short (8mer) and long dsDNA (128mer) reflects the general problem of invasion into longer dsDNA. We did not study plasmid DNA, in which DNA strands may be accessible for strand invasion targeting because of dsDNA breathing of the supercoiled structures. Instead we focused on targets participating in events with partially denaturation or destabilization of relaxed dsDNA under physiological conditions using formation of the open complex during the transcription start as an example described in the next section.
Figure 3.
Illustration of the strand invasion experiments. (A) Easily denaturing duplex ON3/ON8 with dsDNA of the same size (ON5/ON10). (B) Easily denaturing duplex ON3/ON8 with 128mer dsDNA. (C) Non-easily denaturing duplex ON2/ON7 with dsDNA of the same size (ON5/ON10). The ovals denote INA monomer (P) inserted as a bulge into ONs.
Another purpose of constructing easily denaturing duplexes is their possible use as helical-structured probes. The advantage of such a probe is a better mismatch distinction and a higher affinity toward the DNA target compared with unstructured probes as it has already been presented (26,27). The benefit of structured probes over linear nucleic acids has been used for real-time PCR detection (28), specific DNA triplex formation (29) and DNA mis-match detection (30). Easily denaturing duplexes are also participating in the so-called strand-exchange reactions between dsDNA probes and ss target DNA (30,31) as we saw for the transition ON3/ON8 → ON3/ON8+ON10 in the fluorescence spectra. The change in the excimer intensity at 480 nm can be used to monitor this process. Alternatively, easily denaturing duplexes can be labeled with a fluorophore at the 3′ end of one of the strands and with a quencher at the 5′ end in the complement. In such case, a double check is possible: (i) excimer band at 480 nm and no fluorescence of the fluorophore in the easily denaturing duplex form and (ii) considerable lowering of the excimer band and high fluorescence signal after the strand exchange reactions with targets. These features can be explored in the design of probes for fluorescent in situ hybridization (FISH) where the detection of single copy sequence tracts and good discrimination between matched and single mis-matched targets are still a challenge (32).
Contrary to the fluorescence spectra of intercalating dsDNA (ON3/ON8) with addition of short native dsDNA, a very small decrease in the excimer band was detected on mixing of intercalating ds2′-OMe-RNA (ON2/ON7) with the native 8mer dsDNA (1.5 eqv) compared with the duplex alone, even after heating up and cooling down (Figure 2B). Since the fluorescence spectra of the corresponding duplexes (ON2/ON10 and ON7/ON5) had lower intensity of monomeric and excimer bands compared with the ssON2 and ssON10, one could conclude that the duplex ON2/ON7 remained untouched after addition of the short dsDNA (Figure 3C). This also correlates with considerably higher thermal stability of ON2/ON7 than those for the corresponding duplexes with unmodified ssDNAs.
In biological assays, those pairs of modified complementary ONs with lower Tm values than for native dsDNA could be considered easily denaturing nucleic acids even though only one of the modified strands has a higher Tm value with the complementary ssDNA than Tm of the dsDNA. The weakly binding strand may also have enhanced affinity to a complementary DNA strand due to concomitant binding to proteins. If these are included, the list of pairs of modified ONs, which are able to target dsDNA, can be expanded (underlined values of Tm in Table 1) with for example ON3/ON7. The thermal stability of one of the P possessing strands toward ssDNA (ON7/ON5, Tm = 19.5°C) is lower than the Tm value for the native dsDNA (26°C), while another strand has higher Tm (ON3/ON10, Tm = 41.5°C).
Inhibition of transcription start
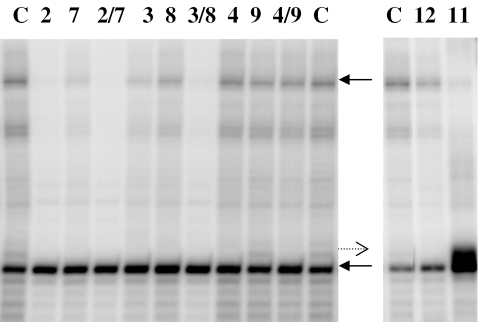
We studied the synthesized ONs alone and in pairs with the complementary sequences targeting the template and/or the non-template strand of the open complex formed by E.coli RNA polymerase with the lac UV-5 promoter (Table 1 and Figure 4). As expected, the 8mer 2′-OMe-RNA possessing P (ON2) targeting the template strand was inhibiting more efficiently (90%) than the pentamer ON1 (42%), which is probably a result of increased binding affinity. Interestingly, the inhibition by the non-intercalating 8mer 2′-OMe-RNA (ON4) was the lowest of all ONs complementary to the template strand. This is despite the high binding affinity of ON4 toward ssDNA (36.5°C). All other tested ONs complementary to the template strand had insertions of P. A good inhibition (67%) was observed for the DNA sequence ON3 possessing three Ps in a DNA sequence. This sequence had also the highest Tm value toward ssDNA (ON3/ON10) and it is the first example of a DNA-like molecule exhibiting inhibition in the RNA polymerase assay, as DNA sequences have never showed any substantial inhibition earlier (21,25). From our thermal stability studies, it was concluded that a B-type helix could adopt the bulged P insertions in a thermodynamically more favorable fashion than an A-type helix. Therefore, the DNA sequence with P insertions (ON3) in comparison with ON1 and ON2 provide evidence that the design of short ONs targeting the template strand of the open complex should not be restricted to RNA-like molecules. Recently, PNA designed to target the template strand has showed efficient inhibition of transcription in mammalian cells (24) despite earlier observations of very low inhibition efficiency for PNA in the same assay as used here (21). We have previously mentioned that stacking interactions of P with aromatic amino acids in the RNA polymerase could be an alternative explanation of enhanced inhibition efficiency (23). Removal of water molecules by an intercalator and complex stabilization should also be taken into account. An efficient inhibition of 80 and 56% was detected for 8mer ssONs with P insertions (ON7 and ON8, respectively), which were designed to target the non-template strand, whereas the 2′-OMe-RNA (ON9) without insertions of intercalators was inactive (Table 1). The non-template targeting sequence does not complement to any part of the template strand. ON12, which was designed to be a complement to the non-template strand at the +36 to +43 position of the dsDNA sequence and to be a scrambled ON for the open complex, showed inhibition efficiency at the same level (62%, Figure 4) as other 8mer DNAs with P insertions. Therefore, it is believed that the observed inhibition efficiency for DNA sequences with P insertions is a result of sequence unspecific binding within the open complex. However, a truncated RNA transcript (ca. 35mer) was observed when using the ON ON11 with three P monomers, that is a complement to the template strand at positions +36 to +43 (Figure 4). This is the first example of stop of the transcription using a sequence derived from DNA and INA monomers targeting the template strand in the middle of the dsDNA downstream from the transcription starting site. We believe that the targeting occurs by local melting of dsDNA by the RNA polymerase. Therefore, the observed inhibition efficiency for ON3 can still be ascribed as binding to the template strand within the open complex. The pentamer ON6 targeting the non-template strand did not show any activity. This proves sequence specificity for inhibition by 2′-OMe-RNA pentamers targeting the template strand, which showed 42% inhibition efficiency (ON1) in analogy with previous results (23).
Figure 4.
Denaturing gel electrophoresis of RNA transcription using E.coli RNA polymerase in the presence of ON 2, 3, 4, 7–9, ON2/ON7 (2/7), ON3/ON8 (3/8), ON4/ON9 (4/9), ON 11, 12 and in the absence of inhibitors (C). ON11 is 5′-GPCTPATGPAC (complement to the template strand at +36 to +43); ON12 is 3′-GTPCATPAGPC (complement to the non-template strand at +36 to +43). The upper arrow points to the 61mer RNA transcript, and the lower arrow to a 30mer DNA quantification marker. The dashed arrow points to an ∼35mer RNA product. The concentration of inhibitors is 10 µM.
The use of two complementary INA sequences (ON3/ON8), which were considered as easily denaturing duplex, gave a better inhibition than using them separately. The same effect was observed for duplexes ON2/ON8 and ON1/ON8. Slightly increased inhibition of complexes compared with the ssONs was detected for ON4/ON8, ON3/ON6, ON2/ON6 and ON3/ON9. Owing to the unspecific transcription inhibition by ONs designed for the non-template strand, the increased inhibition efficiency observed for easily denaturing nucleic acids should most likely not be explained by simultaneous targeting of template and non-template strands. In a previous study, Milne et al. (20) considered this possibility unlikely based on results from Marr and Roberts (33) who showed interaction of Sigma factor 70 with the non-template DNA strand in transcription initiation. Their investigation was concentrated on the −10 region. However, new X-ray crystal structures of the initiation complex [reviewed in (34)] do not exclude that non-template DNA at the transcription start site in the open complex form may be accessible for hybridization.
As indicated by the thermal denaturation and fluorescence studies, the duplexes ON2/ON7 and ON2/ON9 are too stable to be termed easily denaturing nucleic acids and are therefore expected to be inefficient as transcription start inhibitors. However, an efficient inhibition of transcription start was observed with these two duplexes. The melting temperatures of these complexes were very close to the temperature of the RNA polymerase assay (37°C). It is therefore speculated that either partial melting of these duplexes at 37°C or unspecific interactions in the RNA polymerase/dsDNA/inhibitors complex led to increased inhibition efficiency. We observed that insertion of an intercalator and thereby a decreased duplex thermal stability led to a considerable enhancement of inhibition from 25% for ds2′-OMe-RNA (ON4/ON9) to 96% for intercalating ds2′-OMe-RNA (ON2/ON7). We do not believe that targeting with these ONs as duplexes is likely. It is possible to envision that melting of ds2′-OMe-RNA with P insertions might also be mediated by the open complex. In order to evaluate unspecific interactions of pyrene bearing ONs with polymerase we performed fluorescence experiments with ON2, ON7, ON3, ON8, 128mer dsDNA and with addition of E.coli polymerase at 37°C. Unfortunately, the results were unsuccessful so assessment about interactions and positions of intercalators covalently attached to ONs inside of the open complex needs more detailed investigations using for example crystallographic analysis. It should also be mentioned that depending on the nature of the RNA polymerase and open complex (34) the effect of unspecific binding driven by intercalating units might differ. The use of either single-stranded or double-stranded ONs according to the size of the open complex and derivatization of ONs to make additional interactions with the RNA polymerase widens the possibilities of creating sequence-specific and promoter-specific inhibitors of RNA transcription. The recent evidence of gene expression inhibition at transcription start sites on mammalian cells (24,25) is a promising indication of further development of this approach. The screening of ON inhibitors for different polymerases should be more effective if using a larger library of ONs. Therefore, the synthesis of ON library with the possibilities for post-synthetic modifications is highly desirable as an alternative to the routine and time-consuming oligosynthesis performed nowadays.
In conclusion, we have synthesized 5 and 8mer ONs comprising INA monomer (P) in a combination with 2′-OMe-RNA and DNA and evaluated their ability to target both 8mer dsDNA and 128mer dsDNA. Using fluorescence and thermal stability studies, a number of easily denaturing nucleic acids were found that were able to bind sequence-selectively to dsDNA of the same size. Using E.coli RNA polymerase and a lac UV-5 promoter the inhibition of transcription start was evaluated for the synthesized ONs on 128mer dsDNA. For the first time an inhibition of transcription start was observed for sequences based on DNA structures possessing several pyrene units. We also found that insertion of the intercalator P in ds2′-OMe-RNA was a prerequisite to exhibit high transcription inhibition efficiency. When the 8mer DNA sequence with three bulged insertions of P and complementary to the positions +36 to +43 of the template strand, a truncated RNA transcript (ca. 35mer) was observed as the main product. The latter finding encourages the performance of further studies devoted to the use of INA in RNA transcription assays for the development of new therapeutic systems.
Acknowledgments
Mrs Kirsten Østergaard and Ms Suzy W. Lena are thanked for the assistance. The Nucleic Acid Center is funded by The Danish National Research Foundation for studies on nucleic acid chemical biology. Funding to pay the Open Access publication charges for this article was provided by The Danish National Research Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lohse J., Dahl O., Nielsen P.E. Double duplex invasion by peptide nucleic acid: a general principle for sequence-specific targeting of double-stranded DNA. Proc. Natl Acad. Sci. USA. 1999;96:11804–11808. doi: 10.1073/pnas.96.21.11804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bryld T., Højland T., Wengel J. DNA-selective hybridization and dual strand invasion of short double-stranded DNA using pyren-1-ylcarbonyl-functionalized 4′-C-piperazinomethyl-DNA. Chem. Commun. (Camb) 2004:1064–1065. doi: 10.1039/b402414a. [DOI] [PubMed] [Google Scholar]
- 3.Wittung P., Nielsen P.E., Buchardt O., Egholm M., Norden B. DNA-like double helix formed by peptide nucleic acid. Nature. 1994;368:561–563. doi: 10.1038/368561a0. [DOI] [PubMed] [Google Scholar]
- 4.Koshkin A.A., Nielsen P., Meldgaard M., Rajwanshi V.K., Singh S.K., Wengel J. LNA (Locked Nucleic Acid): An RNA Mimic Forming Exceedingly Stable LNA:LNA Duplexes. J. Am. Chem. Soc. 1998;120:13252–13253. [Google Scholar]
- 5.Kutyavin I.V., Rhinebart R.L., Lukhtanov E.A., Gorn V.V., Meyer R.B., Jr, Gamper H.B., Jr Oligonucleotides containing 2-aminoadenine and 2-thiothymine act as selectively binding complementary agents. Biochemistry. 1996;35:11170–11176. doi: 10.1021/bi960626v. [DOI] [PubMed] [Google Scholar]
- 6.Nguyen H.-K., Southern E.M. Minimising the secondary structure of DNA targets by incorporation of a modified deoxynucleoside: implications for nucleic acid analysis by hybridization. Nucleic Acids Res. 2000;28:3904–3909. doi: 10.1093/nar/28.20.3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demidov V.V., Protozanova E., Izvolsky K.I., Price C., Nielsen P.E., Frank-Kamenetskii M.D. Kinetics and mechanism of the DNA double helix invasion by pseudocomplementary peptide nucleic acids. Proc. Natl Acad. Sci. USA. 2002;99:5953–5958. doi: 10.1073/pnas.092127999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaihatsu K., Janowski B.A., Corey D.R. Recognition of chromosomal DNA by PNAs. Chem. Biol. 2004;11:749–758. doi: 10.1016/j.chembiol.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 9.Filichev V.V., Christensen U.B., Pedersen E.B., Babu B.R., Wengel J. Locked nucleic acids and intercalating nucleic acids in the design of easily denaturing nucleic acids: thermal stability studies. Chembiochem. 2004;5:1673–1679. doi: 10.1002/cbic.200400222. [DOI] [PubMed] [Google Scholar]
- 10.Singh S.K., Nielsen P., Koshkin A.A., Wengel J. LNA (locked nucleic acids): synthesis and high-affinity nucleic acid recognition. Chem. Commun. 1998:455–456. [Google Scholar]
- 11.Obika S., Nanbu D., Hari Y., Andoh J., Morio K., Doi T., Imanishi T. Stability and structural features of the duplexes containing nucleoside analogues with a fixed N-type conformation, 2′-O,4′-C-methyleneribonucleosides. Tetrahedron Lett. 1998;39:5401–5404. [Google Scholar]
- 12.Christensen U.B., Pedersen E.B. Intercalating nucleic acids containing insertions of 1-O-(1-pyrenylmethyl)glycerol: stabilisation of dsDNA and discrimination of DNA over RNA. Nucleic Acids Res. 2002;30:4918–4925. doi: 10.1093/nar/gkf624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nielsen C.B., Petersen M., Pedersen E.B., Hansen P.E., Christensen U.B. NMR structure determination of a modified DNA oligonucleotide containing a new intercalating nucleic acid. Bioconjug. Chem. 2004;15:260–269. doi: 10.1021/bc0341932. [DOI] [PubMed] [Google Scholar]
- 14.Filichev V.V., Hilmy K.M.H., Christensen U.B., Pedersen E.B. Intercalating nucleic acids: The inversion of the stereocenter in 1-O-(pyren-1-ylmethyl)glycerol from R to S. Thermal stability towards ssDNA, ssRNA and its own type of oligodeoxynucleotides. Tetrahedron Lett. 2004;45:4907–4910. [Google Scholar]
- 15.Hrdlicka P.J., Babu B.R., Sørensen M.D., Wengel J. Interstrand communication between 2′-N-(pyren-1-yl)methyl-2′-amino-LNA monomers in nucleic acid duplexes: directional control and signalling of full complementarity. Chem. Commun. (Camb) 2004:1478–1479. doi: 10.1039/b404446k. [DOI] [PubMed] [Google Scholar]
- 16.Langenegger S.M., Haner R. Excimer formation by interstrand stacked pyrenes. Chem. Commun. (Camb) 2004:2792–2793. doi: 10.1039/b412831a. [DOI] [PubMed] [Google Scholar]
- 17.Ishihara T., Corey D.R. Rules for strand invasion by chemically modified oligonucleotides. J. Am. Chem. Soc. 1999;121:2012–2020. [Google Scholar]
- 18.Perrin D.M., Mazumder A., Sadeghi F., Sigman D.S. Hybridization of a complementary ribooligonucleotide to the transcription start site of the lacUV-5-Escherichia coli RNA polymerase open complex. Potential for gene-specific inactivation reagents. Biochemistry. 1994;33:3848–3854. doi: 10.1021/bi00179a008. [DOI] [PubMed] [Google Scholar]
- 19.Perrin D.M., Chen C.-H.B., Xu Y., Pearson L., Sigman D.S. Gene-specific transcription inhibitors. Pentanucleotides complementary to the template strand of transcription start sites. J. Am. Chem. Soc. 1997;119:5746–5747. [Google Scholar]
- 20.Milne L., Xu Y., Perrin D.M., Sigman D.S. An approach to gene-specific transcription inhibition using oligonucleotides complementary to the template strand of the open complex. Proc. Natl Acad. Sci. USA. 2000;97:3136–3141. doi: 10.1073/pnas.050544597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Milne L., Perrin D.M., Sigman D.S. Oligoribonucleotide-based gene-specific transcription inhibitors that target the open complex. Methods. 2001;23:160–168. doi: 10.1006/meth.2000.1117. [DOI] [PubMed] [Google Scholar]
- 22.Hwang J.-T., Baltasar F.E., Cole D.L., Sigman D.S., Chen C.-H., Greenberg M.M. Transcription inhibition using modified pentanucleotides. Bioorg. Med. Chem. 2003;11:2321–2328. doi: 10.1016/s0968-0896(03)00071-3. [DOI] [PubMed] [Google Scholar]
- 23.Filichev V.V., Vester B., Hansen L.H., Abdel Aal M.T., Babu B.R., Wengel J., Pedersen E.B. Enhanced inhibition of transcription start by targeting with 2′-OMe pentaribonucleotides comprising locked nucleic acids and intercalating nucleic acids. Chembiochem. 2005;6:1181–1184. doi: 10.1002/cbic.200400457. [DOI] [PubMed] [Google Scholar]
- 24.Janowski B.A., Kaihatsu K., Huffman K.E., Schwartz J.C., Ram R., Hardy D., Mendelson C.R., Corey D.R. Inhibiting transcription of chromosomal DNA with antigene peptide nucleic acids. Nature Chem. Biol. 2005;1:210–215. doi: 10.1038/nchembio724. [DOI] [PubMed] [Google Scholar]
- 25.Janowski B.A., Huffman K.E., Schwartz J.C., Ram R., Hardy D., Shames D.S., Minna J.D., Corey D.R. Inhibiting gene expression at transcription start sites in chromosomal DNA with antigene RNAs. Nature Chem. Biol. 2005;1:216–222. doi: 10.1038/nchembio725. [DOI] [PubMed] [Google Scholar]
- 26.Demidov V.V., Frank-Kamenetskii M.D. Two sides of the coin: affinity and specificity of nucleic acid interactions. Trends Biochem. Sciences. 2004;29:62–71. doi: 10.1016/j.tibs.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 27.Bonnet G., Tyagi S., Libchaber A., Kramer F.R. Thermodynamic basis of the enhanced specificity of structured DNA probes. Proc. Natl Acad. Sci. USA. 1999;96:6171–6176. doi: 10.1073/pnas.96.11.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kong D.-M., Huang Y.-P., Zhang X.-B., Yang W.-H., Shen H.-X., Mi H.-F. Duplex probes: a new approach for the detection of specific nucleic acids in homogenous assays. Anal. Chim. Acta. 2003;491:135–143. [Google Scholar]
- 29.Roberts R.W., Crothers D.M. Specificity and stringency in DNA triplex formation. Proc. Natl Acad. Sci. USA. 1991;88:9397–9401. doi: 10.1073/pnas.88.21.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Q., Luan G., Guo Q., Liang J. A new class of homogeneous nucleic acid probes based on specific displacement hybridization. Nucleic Acids Res. 2002;30:E5. doi: 10.1093/nar/30.2.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim W.J., Akaike T., Maruyama A. DNA strand exchange stimulated by spontaneous complex formation with cationic comb-type copolymer. J. Am. Chem. Soc. 2002;124:12676–12677. doi: 10.1021/ja0272080. [DOI] [PubMed] [Google Scholar]
- 32.Hacia J.G., Novotny E.A., Mayer R.A., Woski S.A., Ashlock M.A., Collins F.S. Design of modified oligodeoxyribonucleotide probes to detect telomere repeat sequences in FISH assays. Nucleic Acids Res. 1999;27:4034–4039. doi: 10.1093/nar/27.20.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marr M.T., Roberts J.W. Promoter recognition as measured by binding of polymerase to nontemplate strand oligonucleotide. Science. 1997;276:1258–1260. doi: 10.1126/science.276.5316.1258. [DOI] [PubMed] [Google Scholar]
- 34.Murakami K.S., Darst S.A. Bacterial RNA polymerases: the wholo story. Curr. Opin. Struct. Biol. 2003;13:31–39. doi: 10.1016/s0959-440x(02)00005-2. [DOI] [PubMed] [Google Scholar]