Abstract
Aptamers are nucleic acids developed by in vitro evolution techniques that bind to specific ligands with high affinity and selectivity. Despite such high affinity and selectivity, however, in vitro evolution does not necessarily reveal the minimum structure of the nucleic acid required for selective ligand binding. Here, we show that a 35mer RNA aptamer for the cofactor flavin mononucleotide (FMN) identified by in vitro evolution can be computationally evolved to a mere 14mer structure containing the original binding pocket and eight scaffolding nucleotides while maintaining its ability to bind in vitro selectively to FMN. Using experimental and computational methodologies, we found that the 14mer binds with higher affinity to FMN (KD ∼ 4 µM) than to flavin adenine dinucleotide (KD ∼ 12 µM) or to riboflavin (KD ∼ 13 µM),despite the negative charge of FMN. Different hydrogen-bond strengths resulting from differing ring-system electron densities associated with the aliphatic-chain charges appear to contribute to the selectivity observed for the binding of the 14mer to FMN and riboflavin. Our results suggest that high affinity and selectivity in ligand binding is not restricted to large RNAs, but can also be a property of extraordinarily short RNAs.
INTRODUCTION
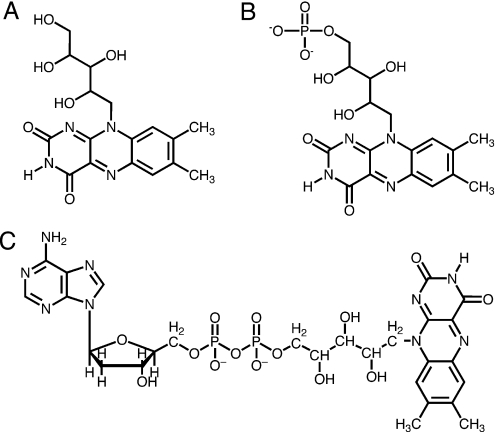
Aptamers can be exploited for their selective ligand binding. Among the numerous types (1) of ligands for which RNA aptamers have been selected are flat, aromatic compounds, including flavin mononucleotide (FMN) (2,3). Aptamers are not useful merely as passive ligand binders, but can play active roles in gene regulation. Riboswitches, for example, are elements in certain mRNAs that regulate gene expression by acting as receptors for specific metabolites (4–6). Riboswitches regulate metabolic pathways for the biosynthesis of a variety of important molecules (7,8), including the cofactor FMN (4,5,9), which mediates oxidation-reduction reactions when bound to flavoproteins as a prosthetic group. Riboswitches usually contain a ligand-binding (aptamer) domain and a downstream region whose conformation is able to permit or repress gene expression (10,11). The aptamer domains of riboswitches are similar to aptamers developed by in vitro techniques (12–16) in that they bind their ligands with exquisite affinity and selectivity (7,8). For instance, the FMN riboswitch (4,5,9) is able to discriminate against riboflavin (Figure 1A) by three orders of magnitude with respect to FMN (Figure 1B), despite the negative charge of FMN (5). In addition, recent studies have demonstrated that synthetic aptamers for small molecules can also be inserted into the 5′-UTR region of mRNA in vitro and in vivo, significantly reducing mRNA translation upon ligand addition (17,18). Other examples of aptamer uses for gene regulation include engineered RNA switch systems that act as ligand-responsive riboregulators (19–21), which have recently been reviewed (22).
Figure 1.
Ligand structures for FMN aptamer and riboswitch. (A) Riboflavin. (B) FMN. (C) FAD.
Determining the minimum sequence requirements of aptamers for selective ligand binding can allow the design of structures that are less likely to get trapped in energetically favorable inactive conformations (23,24). The notion of ‘conformational hell’ can be an issue of concern, as, according to theoretical models, both the mean and maximum barrier heights for a given RNA sequence scale with sequence length N roughly as N1/2 (25). Moreover, the number of competing minima, in which the RNA can form inactive metastable structures, swells with increasing sequence size (26,27).
From this standpoint, it may be fortunate, then, that the final aptamer nucleotide sequences that are obtained from in vitro evolution techniques are not necessarily the minimum sequences necessary for the aptamers to perform their functions (13,28). The in vitro selection methodology requires the use of many randomized nucleotide positions in the primary structures of nucleic acid libraries, and consequently the final evolved structures usually contain unnecessary stretches of nucleotides. At present, there are several experimental protocols for determining the simplest binding site of an aptamer. These protocols include (i) decreasing the size of the randomized region until there remains no more affinity selection (28), (ii) random terminal truncation experiments (29), (iii) minimization using random internal deletions (30) and (iv) selection from remutagenized pools (31). We have recently predicted computationally and shown experimentally (32) that a 33mer RNA aptamer for theophylline (33) can be truncated to a 13mer structure containing only seven binding-pocket and six scaffolding nucleotides from the original 33mer while maintaining its ability to bind selectively to theophylline. This finding suggests that, in certain cases, large stretches of primary structure may potentially be removed from aptamers without compromising the aptamers' functions.
We have developed an iterative computational approach to determine the shortest truncated structure of the FMN-binding 35mer RNA (2,3). This approach may serve as a supplement to the extensive experimental protocols that are currently relied upon for identifying minimum structure requirements for selective ligand binding. The 35mer which we used as a model for our methodology binds to FMN with KD = 500 nMand to flavin adenine dinucleotide (FAD) (Figure 1C) with KD = 700 nM (2). We have applied our computational methodology to show that a 14mer RNA containing only six binding-pocket nucleotides and eight additional scaffolding nucleotides from the original 35mer is able to bind FMN. The 14mer is able to discriminate against the structurally similar compound riboflavin and also against FAD. In addition, we have investigated the preference of the polyanionic RNA molecule for negatively charged FMN over neutral riboflavin using density functional theory (DFT) calculations. We observed that significantly different hydrogen-bonding strengths of FMN and riboflavin complexed with an adenine residue in the binding pocket are consistent with the difference between binding energies and with the aptamer's preferential binding to FMN.
MATERIALS AND METHODS
In vitro nucleic acids
RNA and DNA oligomers were purchased from Integrated DNA Technologies (Coralville, IA). The original 35mer RNA aptamer sequence was transcribed from a 52 nt antisense DNA containing a 17 nt promoter using T7 polymerase.
Computational design of 14mer RNA
The NMR structure of the 35mer RNA complexed with FMN was loaded from the PDB (PDB code 1FMN) into the MacroModel version 7.0 software package (34). The first NMR structure was selected. Nucleotide pairs that appeared qualitatively to be unnecessary for FMN binding were deleted from the RNA, beginning at the 5′ and 3′ ends. The farther a nucleotide pair was located from the binding pocket, the more suitable it was considered for deletion. The first deletion consisted of nucleotides G1 to G6 and A31 to C35. An energy minimization was performed on the modified structure using 500 steps of steepest descent followed by Polak-Ribiere conjugate gradient minimization until convergence of the gradient (0.01 kJ/mol Å) was reached using the AMBER force field (35) and a generalized Born/solvent-accessible surface area (GB/SA) implicit water solvent model (36) with a dielectric constant of 78. A 500 ps molecular dynamics (MD) simulation at 300 K using the same force field and implicit-solvent model and 1 fs time steps was then performed on the minimized structure. The FMN was predicted to remain bound and the binding-pocket structure was not perturbed. Individual base pairs were subsequently excised one by one from the newest 5′ and 3′ ends resulting from the previous truncations, with each new structure undergoing energy minimization and 500 ps of molecular dynamics as described above. This stepwise removal of base pairs was continued until the remaining RNA structure was sufficiently short that the binding pocket was shown to open up and become entirely solvent exposed during the course of the MD simulation. The 14mer was the shortest structure that maintained integrity of the binding pocket and a tight ligand-pocket fit. Deletions of either the A4-G11 base pair or the C5-G10 base pair of the 14mer resulted in RNA structures that lost intrastrand stabilization and significant disruption of the binding pocket's structure.
Explicit-solvent MD simulation of 14mer:FMN complex
The 14mer RNA structure identified above was loaded into the AMBER 7 suite of programs [(37) AMBER 7, University of California, San Francisco] and solvated in a 54 Å × 63 Å × 54 Å orthorhombic box containing ∼4250 TIP3P (38) water molecules. Twelve Na+ counterions were added automatically to regions of greatest electrostatic potential in order to achieve a neutral system charge. Employing the ff02 force field (39) for RNA and the gaff force field (40) for FMN, the water molecules and solute were relaxed with five separate energy-minimization runs, each of which consisted of 500 steepest-descent steps followed by 2000 conjugate-gradient steps. Harmonic restraints on all solute (RNA and FMN) atoms decreased gradually from 500 kcal mol−1 Å−2 to zero over the five separate minimizations. The system was then equilibrated to 300 K and 1 g/ml density by heating from 100 to 300 K over the course of 60 ps using 2 fs time steps, an 11 Å electrostatic cutoff and the particle-mesh Ewald (41) treatment of long-range electrostatic interactions. All covalent bonds containing hydrogen were constrained by the SHAKE algorithm (42), and the system temperature was controlled by coupling to an external bath using the Berendsen method (43). Following equilibration, a production run of 3 ns was performed at 300 K using the same simulation conditions.
In vitro binding assays
All fluorescence measurements (λexcitation = 445 nm, λemission = 525 nm) were performed on a Hitachi F-3010 fluorimeter at 295 K. For both the 14mer, the 35mer and the first non-binding RNA (12mer) aptamers, a solution of 1 µM FMN was prepared in a buffer containing 100 mM HEPES, 50 mM NaCl and 10 mM MgCl2. In the case of FAD and riboflavin binding, solutions of 1 µM FAD and 1 µM riboflavin in the same HEPES buffer were used. Binding constants were measured by observing the quenching of ligand fluorescence by bound RNA. Aliquots of each RNA were successively added to the FMN, FAD and riboflavin solutions to yield increasing total RNA concentrations, and the decreases in fluorescence intensity as a function of RNA concentration were measured. The binding isotherms are consistent with the formation of 1:1 complexes. Fluorescence data, after correction for dilution, were fit to the isotherm (44)
using the software package GraphPad Prism4 to yield the dissociation constant Kd, where ΔF is the change in FMN, riboflavin or FAD fluorescence upon addition of RNA, [L] is the RNA concentration at each addition, and Fo is the original FMN, riboflavin or FAD solution fluorescence.
Ab initio calculations
All calculations were performed using the Gaussian 98 software package (45). The riboflavin molecule, FMN molecule and the adenine base were each optimized separately in vacuo using B3LYP DFT and the 6-31G basis set. In order to make the optimization of FMN and riboflavin more tractable, the three main dihedral angles of the aliphatic chain in both molecules were constrained. The optimized FMN and riboflavin structures were each subsequently complexed with the optimized adenine base, and the complexes optimized using B3LYP/6-31G and applying the same chain dihedral-angle constraints as in the optimizations of the separate molecules. Single-point energy calculations on the optimized complex structures were performed using B3LYP/6-31+G(p,d).
RESULTS AND DISCUSSION
Determining shortest RNA that binds FMN
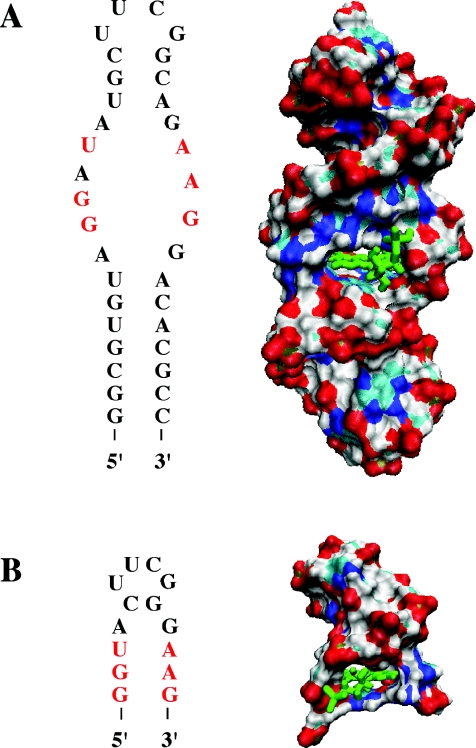
An iterative computational approach was employed to determine the shortest truncated structure of the original FMN-binding 35mer RNA that can still bind FMN (Figure 2A). From the original NMR structure of the 35mer, base pairs were deleted computationally one or two pairs at a time, beginning at the 5′ and 3′ ends. Each separate deletion was followed by a 500 ps implicit-solvent molecular dynamics (MD) simulation. This process was repeated until simulations no longer predicted binding of the ligand. Finally, a 14mer, predicted as the shortest sequence capable of binding FMN, was simulated in its complex with FMN using a 3 ns explicit-solvent molecular dynamics simulation. The FMN remained bound in the binding pocket over the course of the whole MD trajectory. The 14mer contains only the original binding pocket's 6 nt and eight additional nucleotides holding the binding pocket in place (Figure 2B).
Figure 2.
Secondary structures and surface renderings of original and truncated FMN-binding aptamers. Nucleotides of binding pocket are highlighted in red letters and FMN is green. (A) 35mer Aptamer. (B) 14mer Truncation of 35mer, retaining six binding-pocket nucleotides (red) and eight additional scaffolding nucleotides, including the UUCG loop.
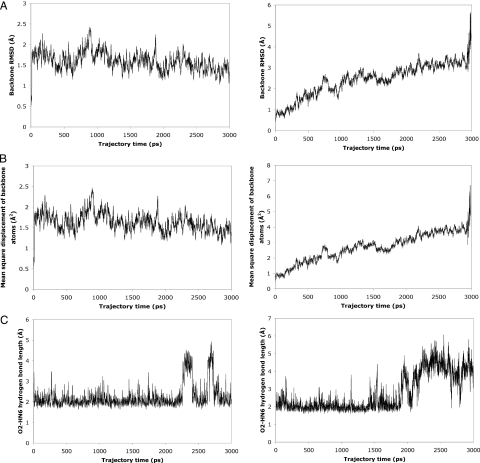
MD simulations of sequences containing fewer than eight scaffolding nucleotides and the six binding-pocket nucleotides indicate a perturbation of the binding-pocket and overall RNA structures. The 5′ and 3′ ends of the RNA dissociate, allowing increased solvent exposure to FMN. For the 12mer formed by removal of the A4-G11 base pair of the 14mer, for example, the backbone stability of the whole RNA is reduced, as shown by a steadily increasing root-mean-square deviation (RMSD) of the RNA backbone atoms (C3′, C4′, C5′, O3′, O5′ and P) relative to their starting positions as a function of time (Figure 3A). Similarly, the diffusional mean square displacement of the backbone atoms of the 12mer reaches >5 Å2 within 3 ns, while that of the 14mer has an average value of only 1.6 Å2 and never exceeds 2.5 Å2 (Figure 3B). Figure 3A and B also illustrates how the 12mer begins to unfold at ∼2.9 ns. Furthermore, at ∼2.2 ns in the 12mer trajectory a significant disruption of the hydrogen bond between the N6 hydrogen atom of A13 and the O2 carbonyl oxygen atom of FMN is observed (Figure 3C). Similarly, simulations of 13mers that contain a single terminal guanine residue at either the 5′ or 3′ end likewise predict a loss of backbone stability (data not shown). The 3D structures of the stable 14mer and the unstable 12mer as it begins to unfold are presented in Figure 4. These observations taken together suggested that the 14mer is the minimum structure that binds FMN.
Figure 3.
Molecular dynamics data suggest stable binding of FMN to 14mer and 14mer RNA stability (left-hand column); 12mer shows structural instability and inability to bind FMN (right-hand column). See text for details. (A) RMSD values of backbone atom (C3′, C4′, C5′, O3′, O5′, P) positions relative to starting structure. (B) Average mean-square displacements of backbone atoms. These values reflect the overall distances over which backbone atoms diffuse. (C) Hydrogen-bond length (distance between hydrogen atom and acceptor atom) for hydrogen bond between N6 of adenine 13 and carbonyl oxygen atom O2 of FMN.
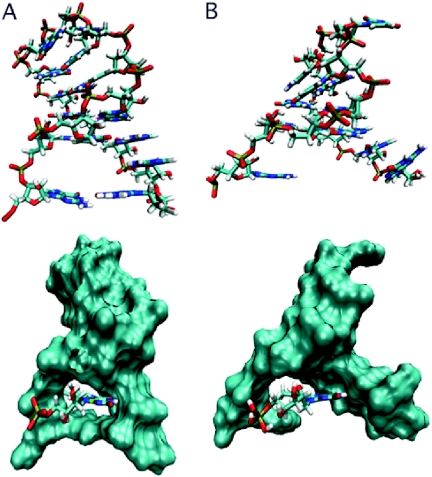
Figure 4.
Comparison of predicted 12 and 14mer RNA complex structures. Structures are averages obtained from MD simulations. The top panels show the RNAs without FMN; the bottom panels show the RNAs as surface representations with the ligand as a tube structure. (A) The 14mer forms a binding pocket that encapsulates FMN tightly and shields its flavin ring from contact with solvent water molecules. (B) The 12mer loses stability that had been conferred by the three hydrogen bonds formed by the sheared A4·G11 N7-amino, amino-N3 non-Watson–Crick base pair of the 14mer. The 5′ and 3′ ends of the RNA become separated, leaving FMN with no hydrogen bonds to the adenine residue A11 (corresponding to A13 of 14mer) and exposed to solvent water molecules.
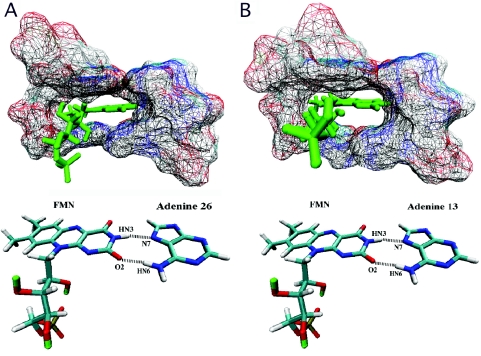
The orientation of the FMN ligand in the binding pocket of the 35mer (Figure 5A) as determined by NMR (3) and that predicted for the 14mer (Figure 5B) are similar. Both binding pockets encapsulate the ligand to roughly the same extent. The NMR structure of the 35mer aptamer shows two hydrogen bonds formed between the FMN molecule and A26. The hydrogen-bond lengths and angles for the 14mer complexed with FMN were calculated at each 1 ps interval of the MD trajectory. The 14mer is predicted to maintain the same two hydrogen bonds with FMN over the course of its whole trajectory (Table 1 and Figure 5B). Moreover, the 14mer RNA exhibits a similar base–ligand–base stacking geometry in the binding pocket as that in the original aptamer. In the experimental 35mer structure the FMN molecule is sandwiched between G10, U12 and A25 above and G9 and G27 below, with ∼3.4 Å separating the planes of each layer's aromatic system. In the 14mer average simulation structure, FMN is sandwiched ∼4.4 Å above and ∼4.0 Å below by the aromatic systems of the same set of bases.
Figure 5.
Binding pockets and hydrogen bonding in the 35 and 14mer RNA aptamers. (A) 35mer Binding pocket complexed with FMN. (B) 14mer Binding pocket complexed with FMN as predicted by a 3 ns molecular dynamics simulation.
Table 1.
Hydrogen-bonding pairs observed in both 35 and 14mer RNA aptamers complexed with FMN
| Simulation | ||
|---|---|---|
| Hydrogen bond | Average length (Å) | Average angle (°) |
| 1 | 1.96 | 163 |
| 2 | 2.23 | 162 |
| Experimental (NMR) | ||
| 1 | 2.11 | 160 |
| 2 | 2.14 | 136 |
Hydrogen bonding data for the 35mer are based on the NMR structure (PDB code 1FMN, first structure). Hydrogen-bonding analysis for the 14mer was performed using the PTRAJ program of AMBER 7. Cutoff distance between heavy atoms is 4.0 Å. Cutoff donor-hydrogen-acceptor angle is 120°. In both cases, hydrogen bond 1 corresponds to that between atom HN3 of FMN and atom N7 of adenine 13 (adenine 26 in the 35mer); hydrogen bond 2 corresponds to that between atom O2 of FMN and atom HN6 of adenine 13 (adenine 26 in the 35mer).
In vitro study of FMN binding
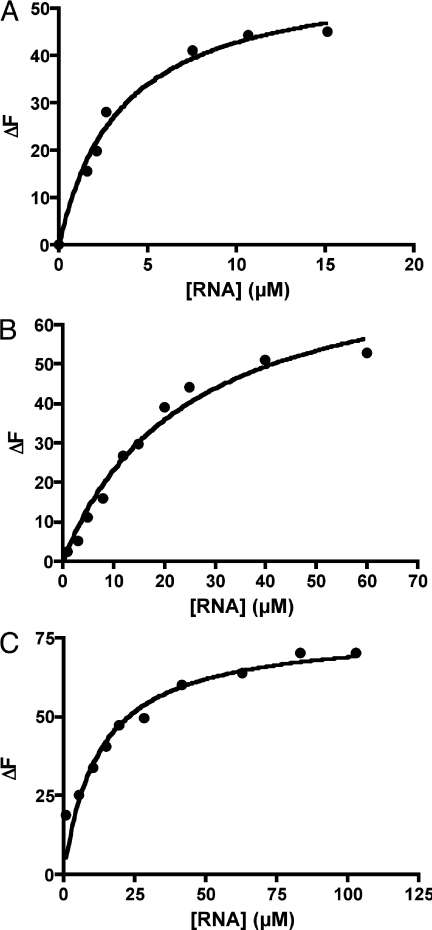
In order to verify our prediction that the 14mer remains bound to FMN, we performed an in vitro binding assay of the 14mer RNA interacting with FMN at 295 K. Fluorescence quenching of FMN was monitored as a function of RNA concentration in order to determine the amount of bound FMN. Non-linear regression of the binding data (Figure 6A) indicates a KD value of 4.1 ± 1.2 µM, corresponding to a ΔGassociation of −31 ± 1 kJ mol−1. The original 35mer binds FMN with a KD of ∼500 nM (2), which corresponds to a ΔGassociation of −36 kJ mol−1. Thus, roughly an 8-fold reduction in binding affinity to FMN occurs in going from the 35mer to the 14mer. However, the 12mer formed by removal of the A4-G11 base pair from the 14mer structure showed no fluorescence quenching even at RNA concentrations as high as 100 µM (data not shown). This finding agrees with the prediction by MD simulations that a major loss of the binding pocket's structural integrity accompanies the elimination of the A4-G11 base pair of the 14mer. The experimental observation of a discontinuity in the relation between sequence length and binding energy when making the 14 to 12 nt transition, and the abrupt fall in binding energy for RNA sequences shorter than the 14mer suggest that the 14mer is the minimum sequence necessary for FMN binding.
Figure 6.
Binding curves for 14mer RNA aptamer. (A) Fluorescence quenching (ΔF) of a solution containing 1 µM FMN as a function of 14mer RNA concentration. (B) Fluorescence quenching (ΔF) of a solution containing 1 µM FAD as a function of 14mer RNA concentration. (C) Fluorescence quenching (ΔF) of a solution containing 1 µM riboflavin as a function of 14mer RNA concentration.
A binding assay was also performed on the original 35mer RNA to verify our fluorescence assay methodology (data not shown). The resulting binding curve indicates a ΔGassociation of −37 ± 1 kJ mol−1, which agrees well with the reported value of −36 kJ mol−1.
Selectivity studies
The FMN-binding 35mer aptamer is able to bind FMN (KD = 500 nM) while discriminating against FAD (KD = 700 nM). In order to evaluate the selectivity of the 14mer truncation for FAD, a binding assay using the same methodology as for the FMN binding assay was performed. The binding curve (Figure 6B) and non-linear regression yield a KD value of 12.0 ± 2.0 µM, which corresponds to a ΔGassociation of −28 ± 1 kJ mol−1. Thus, FAD binds to the 14mer roughly three times more weakly than does FMN.
We next investigated whether the 14mer is able to bind more tightly to FMN than to riboflavin, despite the anionic character of both FMN and RNA. The binding curve (Figure 6C) and non-linear regression indicate a KD value of 12.5 ± 2.7 µM, corresponding to a ΔGassociation of −28 ± 1 kJ mol−1. Thus, riboflavin binds to the 14mer with ∼3 kJ mol−1 less energy than does FMN and with the same affinity as that of FAD. This is not surprising, as the binding of either FAD or riboflavin to the designed 14mer is based only on the interactions of the flavin ring with the RNA bases. The observed selectivity is modest, however, compared with the ∼15 kJ mol−1 energy difference between FMN and riboflavin associating with the FMN-binding riboswitch (5). The selectivity between FMN and related, neutral flavins is surprising considering that a negatively charged molecule such as FMN should, in principle, bind less to the polyanionic RNA than should the corresponding, structurally similar, neutral molecule riboflavin. It has been suggested that the selectivity of the natural riboswitch may be due to the close proximity between a cation complexed in the riboswitch and the phosphate group of FMN (5). However, this possibility has to be excluded in the case of the artificial 14mer aptamer shown here. This short RNA does not have enough nucleotides to complex both FMN and a cation such as divalent magnesium. In the absence of additional interactions that can stabilize the FMN complex compared with that formed by neutral riboflavin, the selectivity observed for FMN has to be the result of the effect of the charge of the phosphate ion on the electronic structure of FMN. In particular, we hypothesized that the electronic density on the phosphate group could be partially delocalized on the flavin ring and used to strengthen both the hydrogen bond between carbonyl oxygen O2 (FMN/riboflavin) and the hydrogen on N6 (Adenine 13) (Figure 5) and π-stacking with the nucleic bases. The former interaction was estimated with ab initio studies. Density functional ab initio calculations performed on the complexes FMN/adenine and riboflavin/adenine [B3LYP/6-31G//B3LYP/6-31+G(p,d)] indicate that in the gas phase FMN binds adenine more favorably than riboflavin (−62 kJ mol−1 versus −44 kJ mol−1). While the level and basis sets used in this calculation reflect the large size of the system and the absolute values of the calculated energies can be improved, the trend of adenine binding FMN more favorably than the neutral riboflavin is consistent with our experimental observations. Thus, different hydrogen-bond strengths resulting from differing ring-system electron densities associated with the aliphatic-chain charges appear to contribute to the selectivity observed for the binding of the 14mer to FMN and riboflavin.
We have demonstrated that a 35mer RNA aptamer for FMN can be truncated to a 14mer, a 60% reduction in sequence size, while losing only 5 kJ mol−1 of binding energy upon complex formation. The 14mer contains no more than 6 nt of the original binding pocket of the 35mer and eight additional scaffolding nucleotides that are retained from the 35mer. Removal of merely one additional base pair to form a 12mer, however, eliminated binding of FMN altogether, indicating that the 14mer contains no unnecessary structural elements. Presumably, this ability to maintain binding would hold true for other aptamer systems involving small, planar aromatic ligands that are capable of forming stacking interactions with nucleic acid bases. These findings suggest that, in principle, the aptamer region of both natural and artificially evolved RNAs can be extraordinarily short while maintaining high affinity and selectivity. It is interesting to note that the aptamer binding theophylline—another small, planar aromatic ligand—which we have studied previously (32) can be truncated to a 13mer and maintain selective ligand binding, and that any sequences shorter than 13 nt in length abolish binding. It would appear, then, that 13 to 14 nt may constitute a sequence-length threshold for short RNAs that selectively bind and recognize small, planar aromatic molecules. This size threshold is likely attributable to destabilization of the folded RNA structure once too few stabilizing intrastrand hydrogen bonds remain. Furthermore, the fact that the ability of the original 35mer to bind FMN selectively can be emulated by a 14mer RNA implies that very short RNAs may also be able to perform specific functions in a cellular environment.
The methodology used in the design of the FMN-binding 14mer RNA aptamer can easily be extended to the search for the minimum sequences of other RNAs that bind to small molecules. Availability of structural data of the complex between the small molecule and a larger RNA aptamer is needed as the starting point of the sequence search. In vitro evolution of RNA and X-ray- or NMR-based RNA structures are then indispensable tools for the identification of very short active RNAs.
Acknowledgments
The authors thank Prof. Charles Lauhon for useful discussions and help in preparing the 1FMN RNA aptamer and the University of Wisconsin-Madison and the WARF Foundation for funding. Funding to pay the Open Access publication charges for this article was provided by the University of Wisconsin-Madison.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hermann T., Patel D.J. Adaptive recognition by nucleic acid aptamers. Science. 2000;287:820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- 2.Burgstaller P., Famulok M. Isolation of RNA aptamers for biological cofactors by in vitro selection. Angew. Chem. Int. Ed. Engl. 1994;33:1084–1087. [Google Scholar]
- 3.Fan P., Suri A.K., Fiala R., Live D., Patel D.J. Molecular recognition in the FMN-RNA aptamer complex. J. Mol. Biol. 1996;258:480–500. doi: 10.1006/jmbi.1996.0263. [DOI] [PubMed] [Google Scholar]
- 4.Mironov A.S., Gusarov I., Rafikov R., Lopez L.E., Shatalin K., Kreneva R.A., Perumov D.A., Nudler E. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell. 2002;111:747–756. doi: 10.1016/s0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- 5.Winkler W., Cohen-Chalamish S., Breaker R.R. An mRNA structure that controls gene expression by binding FMN. Proc. Natl Acad. Sci. USA. 2002;99:15908–15913. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Winkler W., Nahvi A., Breaker R.R. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 7.Mandal M., Breaker R.R. Gene regulation by riboswitches. Nature Rev. Mol. Cell. Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- 8.Soukup J.K., Soukup G.A. Riboswitches exert genetic control through metabolite-induced conformational change. Curr. Opin. Struct. Biol. 2004;14:344–349. doi: 10.1016/j.sbi.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 9.Vitreschak A.G., Rodionov D.A., Mironov A.A., Gelfand M.S. Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res. 2002;30:3141–3151. doi: 10.1093/nar/gkf433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winkler W., Breaker R.R. Genetic control by metabolite-binding riboswitches. ChemBioChem. 2003;4:1024–1032. doi: 10.1002/cbic.200300685. [DOI] [PubMed] [Google Scholar]
- 11.Barrick J.E., Corbino K.A., Winkler W.C., Nahvi A., Mandal M., Collins J., Lee M., Roth A., Sudarsan N., Jona I., et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc. Natl Acad. Sci. USA. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 13.Gold L., Polisky B., Uhlenbeck O., Yarus M. Diversity of oligonucleotide functions. Annu. Rev. Biochem. 1995;64:763–797. doi: 10.1146/annurev.bi.64.070195.003555. [DOI] [PubMed] [Google Scholar]
- 14.Wilson D.S., Szostak J.W. In vitro selection of functional nucleic acids. Annu. Rev. Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 15.Joyce G.F. In vitro evolution of nucleic acids. Curr. Opin. Struct. Biol. 1994;4:331–336. doi: 10.1016/s0959-440x(94)90100-7. [DOI] [PubMed] [Google Scholar]
- 16.Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 17.Werstuck G., Green M.R. Controlling gene expression in living cells through small molecule-RNA interactions. Science. 1998;282:296–298. doi: 10.1126/science.282.5387.296. [DOI] [PubMed] [Google Scholar]
- 18.Hanson S., Bauer G., Fink B., Suess B. Molecular analysis of a synthetic tetracycline-binding riboswitch. RNA. 2005;11:503–511. doi: 10.1261/rna.7251305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Isaacs F.J., Collins J.J. Plug-and-play with RNA. Nat. Biotechnol. 2005;23:306–307. doi: 10.1038/nbt0305-306. [DOI] [PubMed] [Google Scholar]
- 20.Bayer T.S., Smolke C.D. Programmable ligand-controlled riboregulators of eukaryotic gene expression. Nat. Biotechnol. 2005;23:337–343. doi: 10.1038/nbt1069. [DOI] [PubMed] [Google Scholar]
- 21.Buskirk A.R., Landrigan A., Liu D.R. Engineering a ligand-dependent RNA transcriptional activator. Chem. Biol. 2004;11:1157–1163. doi: 10.1016/j.chembiol.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 22.Rimmele M. Nucleic acid aptamers as tools and drugs: recent developments. ChemBioChem. 2003;4:963–971. doi: 10.1002/cbic.200300648. [DOI] [PubMed] [Google Scholar]
- 23.Uhlenbeck O.C. Keeping RNA happy. RNA. 1995;1:4–6. [PMC free article] [PubMed] [Google Scholar]
- 24.Higgs P.G. RNA secondary structure: physical and computational aspects. Q. Rev. Biophys. 2000;33:199–253. doi: 10.1017/s0033583500003620. [DOI] [PubMed] [Google Scholar]
- 25.Morgan S.R., Higgs P.G. Barrier heights between ground states in a model of RNA secondary structure. J. Phys. A: Math. Gen. 1998;31:3153–3170. [Google Scholar]
- 26.Higgs P.G. Thermodynamic properties of transfer RNA: a computational study. J. Chem. Soc. Faraday T. 1995;91:2531–2540. [Google Scholar]
- 27.Thirumalai D., Woodson S.A. Kinetics of folding of proteins and RNA. Accounts Chem. Res. 1996;29:433–439. [Google Scholar]
- 28.Lozupone C., Changayil S., Majerfeld I., Yarus M. Selection of the simplest RNA that binds isoleucine. RNA. 2003;9:1315–1322. doi: 10.1261/rna.5114503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan T., Uhlenbeck O.C. In vitro selection of RNAs that undergo autolytic cleavage with Pb2+ Biochemistry. 1992;31:3887–3895. doi: 10.1021/bi00131a001. [DOI] [PubMed] [Google Scholar]
- 30.Bittker J.A., Le B.V., Liu D.R. Nucleic acid evolution and minimization by nonhomologous random recombination. Nat. Biotechnol. 2002;20:1024–1029. doi: 10.1038/nbt736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartel D.P., Zapp M.L., Green M.R., Szostak J.W. HIV-1 Rev regulation involves recognition of non-Watson–Crick base pairs in viral RNA. Cell. 1991;67:529–536. doi: 10.1016/0092-8674(91)90527-6. [DOI] [PubMed] [Google Scholar]
- 32.Anderson P.C., Mecozzi S. Unusually short RNA sequences: design of a 13-mer RNA that selectively binds and recognizes theophylline. J. Am. Chem. Soc. 2005;127:5290–5291. doi: 10.1021/ja0432463. [DOI] [PubMed] [Google Scholar]
- 33.Zimmermann G.R., Jenison R.D., Wick C.L., Simorre J.P., Pardi A. Interlocking structural motifs mediate molecular discrimination by a theophylline-binding RNA. Nature Struct. Biol. 1997;4:644–649. doi: 10.1038/nsb0897-644. [DOI] [PubMed] [Google Scholar]
- 34.Mohamadi F., et al. J. Comput. Chem. 1990;11:440–467. [Google Scholar]
- 35.Weiner S.J., Kollman P.A., Case D.A., Singh U.C., Ghio C., Alagona G., Profeta S., Weiner P. A new force field for molecular mechanical simulation of nucleic acids and proteins. J. Am. Chem. Soc. 1984;106:765–784. [Google Scholar]
- 36.Still W.C., Tempczyk A., Hawley R.C., Hendrickson T. Semianalytical treatment of solvation for molecular mechanics and dynamics. J. Am. Chem. Soc. 1990;112:6127–6129. [Google Scholar]
- 37.Case D.A., et al. 2002.
- 38.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 39.Wang J., Cieplak P., Kollman P.A. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 2000;21:1049–1074. [Google Scholar]
- 40.Wang J., Wolf R.M., Caldwell J.W., Kollman P.A., Case D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 41.Darden T.A., York D.M., Pedersen L. Particle mesh Ewald: an N-log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 42.Ryckaert J.-P., Ciccotti G., Berendsen H.J.C. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Computat. Phys. 1977;23:327–341. [Google Scholar]
- 43.Berendsen H.J.C., Postma J.P.M., van Gunsteren W.F., DiNola A., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 44.Lauhon C.T., Szostak J.W. RNA aptamers that bind flavin and nicotinamide redox cofactors. J. Am. Chem. Soc. 1995;117:1246–1257. doi: 10.1021/ja00109a008. [DOI] [PubMed] [Google Scholar]
- 45.Frisch M.J., et al. Pittsburgh PA: Gaussian, Inc.; 1998. Gaussian 98 (Revision A.11.3) [Google Scholar]