Abstract
The morbidity and mortality associated with Vibrio-mediated waterborne diseases necessitates the development of sensitive detection technologies that are able to elucidate the identity, potential pathogenicity, susceptibility, and viability of contaminating bacteria in a timely manner. For this purpose, we have designed a single multiplex PCR assay to simultaneously amplify 95 diagnostic regions (encompassing species/serogroup-specific, antimicrobial resistance, and known toxin markers) and combined it with a long oligonucleotide microarray to create a platform capable of rapidly detecting and discriminating the major human pathogenic species from the genus Vibrio: V. cholerae, V. parahaemolyticus, V. vulnificus, and V. mimicus. We were able to validate this strategy by testing 100 geographically and temporally distributed isolates and observed an excellent concordance between species- and serotype-level microarray-based identification and traditional typing methods. In addition to accurate identification, the microarray simultaneously provided evidence of antibiotic resistance genes and mobile genetic elements, such as sulfamethoxazole-trimethoprim constins and class I integrons, and common toxin (ctxAB, rtxA, hap, hlyA, tl, tdh, trh, vvhA, vlly, and vmhA) and pathogenicity (tcpA, type III secretion system) genes that are associated with pathogenic Vibrio. The versatility of this method was further underscored by its ability to detect the expression of known toxin and virulence genes from potentially harmful viable but nonculturable organisms. The results suggest that this molecular identification method provides rapid and definitive information that would be of value in epidemiological, environmental, and health risk assessment surveillance.
Keywords: pathogen detection, molecular diagnostics, cholera
Members of the genus Vibrio are Gram-negative motile bacteria that are naturally occurring, free-living inhabitants of marine and estuarine environments throughout the world. Whereas the vast majority of Vibrio are nonpathogenic to humans, select strains from four species, V. cholerae, V. parahaemolyticus, V. vulnificus, and V. mimicus, are known to be important human pathogens that are predominantly associated with food and waterborne illness. V. cholerae serogroup O1 and V. cholerae serogroup O139, the most notable of the pathogenic Vibrio spp., are the etiologic agents of the severe diarrheal disease known as cholera (1, 2). The strains responsible for endemic, epidemic, and pandemic cholera are known to harbor two critical virulence factors, cholera toxin (CT) and the toxin coregulated pilus (2); however, the overall pathology of V. cholerae infections appears to be determined by the coordinated action of several virulence factors (3). Although closely related to V. cholerae (and each other), the diseases caused by V. parahaemolyticus (gastroenteritis, wound infections, septicemia), V. vulnificus (severe necrotizing wound infections, invasive fulminating septicemia), and V. mimicus (gastroenteritis) are associated with different sets of virulence factors (4, 5). Despite these differences in the mechanisms of disease, members from each of these species have become and continue to be formidable pathogens, especially V. cholerae O1 and V. parahaemolyticus serotype O3:K6, which are responsible for the only two existing bacterial pandemics (1, 2, 6, 7). Thus, the potential morbidity and mortality associated with Vibrio infections, combined with the fact that these organisms cannot be eradicated from the aquatic environment (1), has placed a premium on the development of technologies that can facilitate the rapid detection and identification of virulent strains from environmental and clinical sources.
The laboratory methods most often used to detect and identify the pathogenic Vibrio spp. rely on culture followed by conventional biochemical, serological, and susceptibility testing. However, these methods are time consuming, labor intensive, and reagent intensive and usually do not directly characterize the virulence factors associated with human illness. Although easier to use, commercially available products for the identification of Vibrio spp. are limited to genus- and species-level designations, often require supplemental tests for confirmation, and are known to present difficulties in obtaining accurate identifications (8). Thus, current pathogen identification methods are vulnerable to incomplete or misidentification of potentially harmful Vibrio. The shortcomings of current detection methods are further exacerbated by the ability of these microorganisms to enter a viable but nonculturable (VBNC) state (9) and their proclivity to acquire mobile genetic elements (specifically those that harbor virulence and antimicrobial resistance genes) (10–18).
Molecular genetic identification methods have the ability to not only accurately identify these bacterial pathogens, but to also provide information pertaining to the pathogenic and biological potential of the organism and in doing so can directly address a number of the problems encountered by currently used methods (19). As such, we have designed a 90-plex PCR assay and combined it with a long oligonucleotide DNA microarray that can simultaneously identify diagnostic regions specific for species, serogroup, biotype, antimicrobial resistance, and pathogenicity markers for V. cholerae, V. parahaemolyticus, V. vulnificus, and V. mimicus. Herein we describe the validation of this method by using 100 temporally and geographically distributed isolates and demonstrate its utility in accurately identifying human pathogenic strains, as a surveillance tool for monitoring genetic heterogeneity, and as a method capable of detecting VBNC cells.
Materials and Methods
Vibrio Isolates. Of the 100 Vibrio isolates chosen for this study, >90% were clinical in origin, collected between 1921 and 2004, and archived at the Centers for Disease Control and Prevention (Atlanta) (Table 3, which is published as supporting information on the PNAS web site). The isolates were originally identified and characterized by standard methods. PCR was used to test V. cholerae and V. parahaemolyticus isolates for the presence of the ctxA and tdh/trh genes, respectively.
DNA Amplification. Each multiplex PCR contained 90 primer pairs (Table 4, which is published as supporting information on the PNAS web site). The 90-plex PCRs were performed in 50-μl volumes containing 1× PCR buffer (Qiagen, Valencia, CA), 2.5 mM MgCl2, 400 μM dATP, dGTP, and dTTP, 40 μM dCTP, 40 μM Cy5-dCTP (Amersham Pharmacia Biosciences), 200 nM of each primer [except groEL-F and groEL-R (800 nM), pVC-F (400 nM), trhF3 and trhR3 (400 nM)], 15 units of TaqDNA polymerase (Qiagen), and 50 ng of genomic DNA. The amplification reactions were performed with an initial denaturation at 94°C for 5 min followed by 35 cycles of: 94°C for 30 s, 59°C for 60 s, 72°C for 90 s, and a final extension at 72°C for 7 min.
Microarray Hybridization and Processing. Oligonucleotide probes (34–70 mers) were designed, synthesized, and covalently immobilized as described (20) (Table 5, which is published as supporting information on the PNAS web site). Once constructed, the spotted microarrays were blocked with a 3% BSA-casein solution (BSA-C) for 15 min at room temperature, and the slides were outfitted with MAUI Mixer DC hybridization chambers (BioMicro Systems, Salt Lake City). Hybridization reactions (10 μl of multiplex PCR amplicons, 4 μl of 20 × SSC, 4 μl of formamide, 1 μl of 3% BSA-C, 1 μl of hybridization-positive control, and 0.4 μl of 10% SDS) were denatured for 3 min at 95°C, applied to the microarray, and allowed to incubate for 2 h at 63°C on a MAUI Hybridization System (BioMicro Systems). Posthybridization, the slides were washed twice with 4 × SSC/0.2% SDS buffer for 3 min at 63°C and twice with 2 × SSC buffer for 1 min at room temperature, rinsed once with distilled water, and dried with compressed air. Images were captured with a ScanArray Lite confocal laser scanning system (PerkinElmer) at laser power 80 and Photo Multiplier Tube gain 80. Of the 100 DNA extracts sent to the Naval Research Laboratory from the Centers for Disease Control and Prevention for microarray-based identification, 70 were tested in a masked fashion.
VBNC Vibrio. V. cholerae O1 (strain N16961), V. parahaemolyticus O3:K6 (F5828), and V. vulnificus (F7472) were grown in alkaline peptone water to midlogarithmic phase and harvested by centrifugation. Cell pellets were rinsed twice, resuspended in 5 ml of artificial seawater (ASW; 1% salinity, Instant Ocean Aquarium Systems, Mentor, OH), and used to inoculate 995 ml of sterile ASW at a final concentration of ≈106 cells per ml. The 1-liter cell suspensions were maintained at 4°C on a rotary shaker, and plate counts on tryptic soy agar with ASW dilutions were performed in triplicate at 7-day intervals. When the bacteria could no longer be cultured from ASW dilutions, confirmatory plating experiments with cells that had been collected by centrifugation from 1-ml aliquots of the inoculated ASW were performed. Each ASW suspension was considered to be in the nonculturable state when <0.3 colony-forming units/ml could be detected. Total bacterial RNA was then extracted from 100 ml of each original ASW suspension and 10 ml of matched newly inoculated overnight alkaline peptone water cultures by using the RiboPure Bacteria Kit (Ambion, Austin, TX) with the optional DNase digestion step to ensure the complete removal of genomic DNA. To obtain expression profiles for both sets of samples, 50 ng of RNA from each sample was amplified by using the SuperScript III Reverse Transcriptase Kit (Invitrogen) and subsequent 90-plex PCR. Multiplex amplification conditions and hybridization reactions were performed as described above with the exception that the entire multiplex amplification reaction was applied to the microarray.
Antibiograms. The disk diffusion technique was used to determine susceptibilities to a standard panel of 11 antimicrobial agents [ampicillin, chloramphenicol, cephalothin, ciprofloxacin, furazolidone, sulfisoxazole, kanamycin, nalidixic acid, streptomycin, sulfamethoxazole-trimethoprim (SXT), and tetracycline]. The interpretive criteria for assessing V. cholerae susceptibility profiles have been described (21).
Results
Microarray Design and Testing. A total of 95 oligonucleotide probes were designed to enable the detection of species-specific, serogroup-specific, toxin, and antimicrobial susceptibility genes from V. cholerae, V. parahaemolyticus, V. vulnificus, and V. mimicus (Table 5 and Table 6, which is published as supporting information on the PNAS web site). A special emphasis was placed on the design of probes that would permit the accurate identification of genetic markers associated with the two current pandemic (V. cholerae O1 El Tor and V. parahaemolyticus O3:K6) and potentially pandemic (V. cholerae O139) strains (Table 7, which is published as supporting information on the PNAS web site) (1, 7). Thirty-two probes on the microarray were dedicated to the detection of putative antimicrobial resistance markers that confer resistance to the antibiotics most often used for the treatment of cholera or were often associated with epidemic V. cholerae strains such as the genes that reside on the O139 SXT or O1 El Tor SXTET constin (18, 22) and class I integrons (10, 23, 24).
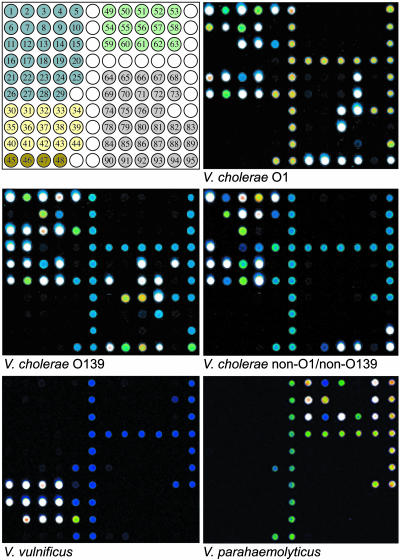
To determine the validity of this approach, we tested 100 temporally and geographically distributed isolates (65 V. cholerae, 13 V. parahaemolyticus, 11 V. vulnificus, 7 V. mimicus, 3 V. hollisae, and 1 V. fluvialis) that had previously been identified by using standard biochemical and typing methods. The hybridization results from five representative isolates are shown in Fig. 1. The microarray-based approach was 100% concordant with the standard methods of identification for V. cholerae O1 El Tor (n = 35), V. cholerae O1 atypical (n = 2), V. cholerae O139 (n = 10), V. cholerae non-O1/non-O139 (n = 9), V. vulnificus and V. parahaemolyticus isolates, 89% concordant for V. cholerae O1 classical (n = 9) and 71% concordant in its identification of V. mimicus isolates (Table 3). The accuracy and reproducibility of the amplification and microarray hybridization results were confirmed by multiple reamplification and hybridization experiments, PCR-based genotyping using single primer pairs, PCR amplicon sequencing, and previously described random amplification methods (25) (data not shown).
Fig. 1.
Microarray hybridization profiles of human pathogenic Vibrio. (Top Left) Microarray template. V. cholerae O1 (C6607T1, Top Right), V. cholerae O139 (K0020, Middle Left), V. cholerae non-O1/non-O139 (serogroup O141) (3522-00, Middle Right), V. vulnificus (F7472, Bottom Left), and V. parahaemolyticus O3:K6 (F5828, Bottom Right). V. cholerae (blue): 1, groEL; 2, 16S-23S IGS; 3, ompU;4, ompW;5, toxR;6, rfbN;7, nanH;8, luxO;9, tcpI; 10, rstRCalc; 11, wbfR; 12, wbfO; 13, hlyAET; 14, tcpAET; 15, rstRET; 16, otnA; 17, otnB; 18, hlyACL; 19, tcpACL; 20, rstRCL; 21, epsM; 22, rtxA; 23, rtxAET; 24, hap; 25, stn/sto; 26, ctxA; 27, ctxB; 28, ace; 29, zot; V. vulnificus (yellow): 30, groEL; 31, wcvH; 32, wza; 33, vvhA; 34, vlly; 35, hml; 36, vvp; 37, rtxA; 38, vpl; 39, hlyIII; 40, d-vph; 41, VV0601; 42, VV1546; 43, VV0795; 44, VV0914; V. mimicus (brown): 45, groEL; 46, vmhA; 47, vmc; 48, phlA; V. parahaemolyticus (green): 49, groEL; 50, 16S-23S IGS; 51, gyrB; 52, pss; 53, VP1696 (TTSS); 54, tl; 55, tdh; 56, trh 1; 57, trh 2; 58, VPA1339 (TTSS); 59, orf8; 60, VPA0896-VPA0898 and VP1569-VP1571; 61, toxRS/new; 62, HU-α ORF; 63, VPA1346 (TTSS); antimicrobial resistance (gray): 64, dfrA1; 65, dfrA5; 66, dfrA12; 67, dfrA15; 68, dfrA18; 69, floR; 70, aadA1; 71, aadA2; 72, sulI; 73, aac(6′)-Ib; 74, ereA2; 75, aphAI; 76, sulII; 77, qacEΔ1; 78, tetA; 79, tetB; 80, tetC; 81, tetD; 82, tetE; 83, tetG; 84, blaAmpC; 85, blaPER-2; 86, blaCTX-M; 87, blaTEM; 88, blaSHV; 89, vcmA; 90, strA; 91, strB; 92, vceA; 93, vceB; 94, vcaM; 95, vcrM. Unrelated hybridization positive controls (white).
Microarray-Based Identification of Genetic Heterogeneity. One of the principal benefits of using microarray-based genotyping methods (in which the presence of several genes are interrogated simultaneously) was the ability to identify uncommon or unexpected genetic assemblages (Table 1). The microarray reproducibly detected isolates with previously recognized genotypes such as V. cholerae O1 with incomplete CTXΦ genomes, a toxigenic (i.e., CT positive) sixth pandemic V. cholerae O1 classical isolate that may have predated the acquisition of Vibrio pathogenicity island-2 (VPI-2) (13), V. cholerae O1 strains that were neither the classical nor El Tor biotype (atypical), and toxigenic V. cholerae non-O1/non-O139 with multiple CTXΦ. The microarray also revealed uncommon genotypes such as V. cholerae O139 isolates with multiple trimethoprim resistance genes and completely unique genotypes such as V. mimicus strains that harbored type III secretion system (TTSS) genes. Unique observations were verified by gene-specific PCR, sequence verification, and similarity comparisons (data not shown).
Table 1. Microarray-based identification of genetic heterogeneity.
| Organism (strain) | Profile* | Implication |
|---|---|---|
| V. cholerae O1 (E8261) | ctxA–, ctxB–, ace+, zot+, rstRCL+ | Incomplete CTXΦ genome (nontoxigenic V. cholerae O1, classical) |
| V. cholerae O1 (75) | rfbN+, nanH–, ctxA+, ctxB+, ace+, zot+, rstRCL+ | Sixth pandemic (1921) toxigenic V. cholerae O1, classical isolate that may predate the acquisition of VPI-2 |
| V. cholerae O1 (3535–02) | rfbN+, nanH–, hlyAET+, rtxA+, hap+, tcpA–, tcpI–, ctxA–, ctxB–, ace–, zot–, rstR– | Nontoxigenic V. cholerae O1, atypical isolate lacking VPI-2 |
| V. cholerae non-O1/non-O139 (3522–00)† | rfbN–, nanH+, wbfR–, wbfO–, otnA+, otnB–, ctxA+, ctxB+, ace+, zot+, rstRCL+, rstRCalc+ | Toxigenic non-O1/non-O139 isolate (serogroup O141) with VPI-2 neuraminidase and multiple (CTXCL and CTXCalc) prophages |
| V. cholerae O139 (K0020)† | dfrA12+, dfrA18+ | Multiple trimethoprim resistance determinants |
| V. parahaemolyticus (F5052)‡ | groEL+, 16S-23S IGS+, gyrB+, pss+, VP1696+, tl+, tdh+, trh+, toxRS/new+ | Nonpandemic group member with the thermolabile hemolysin, thermostable direct hemolysin, thermostable direct hemolysin-related hemolysin, and a chromosome I TTSS marker only§ |
| V. vulnificus (F9546)‡ | wcvH+, vvhA+, vllY+, vvp+, rtxA+, hlyIII+, VV0601+, VV1546+, VV0914+ | Cytolysin-hemolysin, hemolysin, vibriolysin, repeats-in-toxin cytotoxin, and putative hemolysins |
| V. mimicus (2419–94) | tdh+, nanH+, hap+, strA+, strB+, VPA1339+, VPA1346+, VP1696+, vmhA+, vmc+ | Thermostable direct hemolysin, VPI-2 neuraminidase, streptomycin resistance, and putative TTSS genes homologous to escC, yopP, and yscC§ |
Partial profile from microarray-based genotyping
The entire hybridization positive profile has been presented. Profile differences can be seen in comparison to the species-matched profiles in Fig. 1
V. parahaemolyticus open reading frames VPA1339 (escC) and VPA1346 (yopP) are found on chromosome 2 and VP1696 (yscC) is found on chromosome 1 (5)
Antimicrobial Resistance Profiles. Based on the hybridization profiles of the 65 V. cholerae isolates tested, 31 appeared to harbor multiple genes (not including efflux pump genes) that are associated with resistance to commonly used antimicrobial compounds. To assess whether the detected genotype correlated with a resistance phenotype, we compared the genotypic antimicrobial resistance profiles generated by the microarray with phenotypic disk diffusion data (Table 2). As expected, V. cholerae O139 strains F851 and K0020 (collected post-1992) harbored the SXT constin as indicated by the presence of the sulII, strA, strB, dfrA18, and floR genes (22). Similarly, the presence of the sulII, strA, strB, dfrA1, and floR genes suggested that V. cholerae O1 El Tor isolates 3517-98 and K0007 harbored the SXTET constin (22). Each of these four isolates demonstrated the appropriate corresponding phenotypes for streptomycin (strA, strB), trimethoprim (dfrA), and sulfamethoxazole (sulII) resistance but not chloramphenicol (floR) resistance (18, 22). Four of the 10 isolates tested appeared to harbor class I integrons based on the codetection of the sulI and qacEΔ1 genes that characterize the 3′ terminus of these mobile genetic elements (10, 23, 24). In each case, sulI was a reliable indicator of the sulfonamide resistance phenotype. Taken together, the data suggested that the identification of proposed resistance genes that encoded enzymes involved in the modification or inactivation of antibiotics (i.e., reductases, nucleotidyltransferases, phosphotransferases) were reliable indicators of their corresponding phenotype, whereas drug efflux/export protein genes, although present, tended to be variable or unreliable markers for phenotype prediction. Substrates for the vcrM and qacEΔ1 gene products were not tested, and nontargeted genetic mechanisms appeared to be responsible for the observed ampicillin and cephalothin resistance phenotypes.
Table 2. Vibrio spp. genetic and phenotypic antimicrobial resistance profiles.
| Isolate (strain) | Origin (year) | Antibiogram phenotype (corresponding genotype)* | Additional markers† |
|---|---|---|---|
| V. cholerae O1, classical (9060–79) | ATCC 14035 (1949) | Pan susceptible | tetC, vceA, vceB, vcaM, vcmA |
| V. cholerae O1, El Tor (1029–84) | Florida (1984) | AM‡ | vceA, vceB, vcaM, vcrM |
| V. cholerae O1, El Tor (C6607T1)§ | Malawi (1990) | C(vceA, vceB), SXT(dfrA15, suIII), G(suII, suIII), S(strA, strB, vcmA), AM | qacEΔ1, tetC, vcrM |
| V. cholerae O139 (F851) | Bangladesh (1993) | SXT(dfrA18, suIII), G(suIII), S(strA, strB, vcmA), C†(floR, vceA, vceB), FX, AM‡ | vcrM |
| V. cholerae O1, El Tor (3500–96) | Guatemala (1996) | G(suII), S(aadA1, aadA2, vcmA), FX, AM‡ | qacEΔ1, vceA, vceB, vcrM |
| V. cholerae O1, El Tor (3517–97) | China (1997) | TE(tetA), G(suIII), S(strA, strB, vcmA), FX | floR, vceA, vceB, vcrM |
| V. cholerae O1, El Tor (3517–98) | India (1998) | SXT(dfrA1, suIII), NA(vceA, vceB), G(suIII), S(strA, strB, vcmA), FX | floR, vcrM |
| V. cholerae O139 (K0020)§ | Hong Kong (2000) | SXT(drfA12, dfrA18, suIII), TE(tetD), G(suII, suIII), S(aadA2, strA, strB, vcmA), K(aphA1, vcmA) | floR, qacEΔ1, vceA, vceB, vcrM |
| V. cholerae O1, El Tor (K0007) | Bangladesh (2003) | SXT(dfrA1, suIII), NA(vceA, vceB), G(suIII), S(strA, strB, vcmA), FX | floR, vcrM |
Antibiotics: ampicillin (AM), chloramphenicol (C), cephalothin (CF), furazolidone (FX), sulfisoxazole (G), kanamycin (K), nalidixic acid (NA), streptomycin (S), SXT, tetracycline (TE).
Combined, the entire antimicrobial resistance profile for each strain has been presented
Genes that were present (hybridization positive) in the microarray analyses but whose corresponding phenotype was not found or was not tested for in the antibiogram analyses
Intermediate resistance
Microarray hybridization profiles presented in Fig. 1
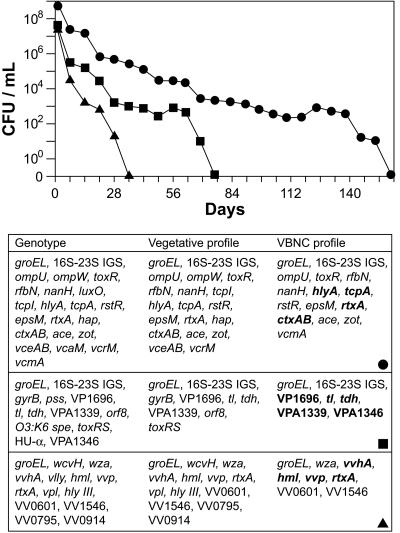
Detection of VBNC Vibrio. Members of the family Vibrionaceae enter a VBNC state in response to low temperatures (26, 27). Once in the VBNC state, these bacteria can no longer be detected by conventional culturing methods. As the presence of bacterial mRNA is a good indicator of cellular function and viability, we attempted to detect mRNA species from ASW Vibrio cultures that were no longer culturable by combining RT-PCR/90-plex PCR amplification with microarray hybridization. V. cholerae O1, V. parahaemolyticus O3:K6, and V. vulnificus were incubated in ASW at 4°C and driven into a nonculturable state in 157, 76, and 35 days, respectively (Fig. 2 Upper). To assess the viability and potential molecular virulence characteristics of these nonculturable Vibrio, we used the microarray to compare mRNA expression profiles generated from matched vegetative (viable and culturable) and ASW cultures with their corresponding genotypes (Fig. 2 Lower). As expected, expression profiles from the vegetative cultures were more robust than their nonculturable counterparts for all three isolates in that most of the genes present (as determined by the genotype) were expressed. However, the unambiguous detection of mRNA species in each of the three nonculturable ASW cultures confirmed that (i) these bacteria were indeed VBNC, (ii) bacteria in the VBNC state could be detected in this manner, and (iii) although VBNC, these strains continued to express known toxin (ctxAB, rtxA, hlyA, tl, tdh, vvhA) and virulence (tcpA, TTSS) genes, thus retaining their pathogenic potential. DNA polymerase control experiments using the total bacterial RNA extracts as template did not generate discernable hybridization signals, thus confirming the absence of genomic DNA contamination (data not shown).
Fig. 2.
Induction and expression profiles of VBNC Vibrio. (Upper) Induction of VBNC V. cholerae O1 (•), V. parahaemolyticus O3:K6 (▪), and V. vulnificus (▴) in ASW. CFU, colony-forming units. (Lower) Comparison of microarray-based genotypes, vegetative RNA expression profiles, and VBNC RNA expression profiles. The RNA used to establish the VBNC expression profiles was obtained from the V. cholerae, V. parahaemolyticus, and V. vulnificus ASW cultures a minimum of 7 weeks after they were deemed nonculturable. Boldface type indicates known toxin and virulence factor genes.
Discussion
Although effective at preliminary identification, conventional methods are generally unable to accurately assess the potential pathogenicity of bacteria that readily acquire and use mobile genetic elements containing toxin, virulence factor, and multi-drug resistance genes (5, 10–18, 22–24, 28, 29). Furthermore, they are completely ineffective at detecting bacteria in the VBNC state. As a result, these methods are prone to misidentify or underestimate the importance of circulating emerging toxigenic V. cholerae non-O1/non-O139 serogroups (12), nontoxigenic V. cholerae O139 (30), naturally occurring genetic hybrids (31, 32), rapidly evolving and reassorting strains (28, 33) and VBNC Vibrio. In this study, we have developed a molecular genetic identification method that circumvents the constraints of conventional microbiological identification by directly addressing the two issues, biological potential and organism viability, that are the most relevant for human health risk assessment.
The issue of biological potential, as it relates to the ability of the Vibrio spp. to infect and cause disease in humans, is of critical importance as most of the major known virulence-associated factors attributed to Vibrio are believed to have been acquired by horizontal transfer (5, 13). We have demonstrated that the use of microarray-based methods, which offer the ability to simultaneously interrogate for the presence of several genes, are ideally suited for the determination of genotype, and hence, biological potential. A salient example from this study was provided by the discovery that two of the V. mimicus isolates (548-77 and 2419-94) tested harbored TTSS genes. Although this result is not entirely surprising based on recent findings (5, 34), to our knowledge the presence of TTSS genes in V. mimicus isolates has not been previously demonstrated. In addition to providing basic microbiological or molecular epidemiological information, this ability to simultaneously assess genetic heterogeneity may have immediate practical implications pertinent for V. cholerae infections when applied to the detection of antimicrobial resistance genes and tracking of SXT constins (35, 36), class I integrons (10, 23, 24), resistance genes for commonly used antibiotics, and widespread multidrug-resistant strains (10, 23, 24, 29).
The second issue of organism viability, especially in the context of the VBNC state, has received increased attention as ≈60 species of bacteria are now known to enter this state (9), and recent evidence indicates that VBNC human pathogenic bacteria can act as reservoirs for antimicrobial resistance genes (37), retain their virulence (26), reside in the environment during interepidemic periods (38), and be fully resuscitated to cause disease (39). Like other bacteria, the Vibrionaceae enter a VBNC state as a survival strategy in response to environmental stresses such as low temperatures (26, 27). When in this state, these bacteria cannot be detected by conventional culture methods as they are incapable of the sustained cellular division required for colony formation. Yet Vibrio in the VBNC state remain metabolically active, retain their virulence properties, and under the appropriate conditions (such as ingestion) recover to become fully vegetative, culturable (40), and pathogenic (39). Thus, whereas conventional detection methods are prone to underestimating the potential risk associated with VBNC Vibrio-contaminated sources, molecular methods that exclusively interrogate DNA may overestimate the potential risk based on the detection of extracellular DNA or DNA from nonviable organisms. In light of these difficulties, RT-PCR, which targets short-lived mRNA molecules, has become an increasingly popular method for assessing the viability of bacterial cells and a more accurate method of assessing the potential risk of VBNC bacteria-contaminated samples.
In this study, we detected VBNC V. cholerae, V. parahaemolyticus, and V. vulnificus by combining the microarray with a modified amplification protocol designed to target short-lived mRNA species and demonstrated that each VBNC species continued to express a number of genes, some of which were known virulence determinant and toxin genes (Fig. 2). Although somewhat surprising, these findings were not unprecedented as the detected expression of the cytolysin-hemolysin gene (vvhA) in VBNC V. vulnificus was in agreement with published findings (41). Moreover, the transcription of toxin-encoding genes while in this physiological condition does not appear to be restricted to the genus Vibrio as Escherichia coli O157:H7 continue to express the shiga-like toxin I gene (stxI) when in the VBNC state (42) and have been implicated in VBNC-mediated foodborne illness (43). A more recent investigation of VBNC V. parahaemolyticus found that two selected housekeeping genes (16S-23S rDNA and rpoS) could be detected by RT-PCR but two targeted toxin genes (tdh1 and tdh2) could not (44). Although our results confirm the expression of the 16S-23S rDNA housekeeping gene in VBNC V. parahaemolyticus, we also detected the expression of the tdh gene, a result that contradicts the previous finding (44). As there are a number of experimental (assay sensitivity, VBNC induction period) and biological (strain, gene copy number, regulation of expression) variations, it is difficult to meaningfully reconcile this discrepancy.
Although limited, this study demonstrates a successful attempt at microarray-based expression profiling of bacteria in the VBNC state. In contrast to single-target RT-PCR assays, the combination of an appropriate amplification strategy and microarray permits the simultaneous interrogation of multiple targets. Regardless of whether the mRNA transcripts detected are from housekeeping (42, 44), pathogenicity (41, 42), or antimicrobial resistance (37) genes, a positive identification still provides evidence for the presence of VBNC bacteria. Furthermore, screening in this manner alleviates the necessity to determine the behavior and genetic regulation of appropriate genetic targets before experimentation. Although the results of this study outline an approach for the detection of bacteria in the VBNC state and provide data that highlight the potential risk and expressed virulence characteristics of VBNC Vibrio, they do not help to explain the details of the physiological condition itself. Future investigations using whole-genome expression profiling time course studies will be required to understand the purpose and genetic program responsible for entry into, maintenance during, and resuscitation from the VBNC state.
We have demonstrated that the microarray provides a means to simultaneously estimate the viability and pathogenic potential of VBNC Vibrio in a timely manner. Although this study has served as a proof-of-concept demonstration, this capability may have utility in a number of practical applications. It is known that the Vibrio spp. can initiate infections from the VBNC state (26, 27, 39, 40), and our results and those of others (41, 42) suggest that certain bacteria in the VBNC state continue to express known toxin and virulence-associated genes. Although energetically expensive, this strategy may represent a survival and preparedness mechanism for bacterial pathogens awaiting a more suitable environment for propagation. Therefore, methods such as the one described may be useful in monitoring natural environments where the pathogen is known to exist but cannot be detected with conventional methods because of unfavorable climate conditions (38, 45) or in conjunction with conventional assays for the testing of suspect food matrices in an attempt to reduce VBNC bacteria-related foodborne illness (43, 46, 47). This method may also be suitable for evaluating the reliability of cold pasteurization processes, such as high hydrostatic pressure (48) or irradiation (49), for microbial inactivation.
This study has outlined the development of a molecular detection technology that addresses the constraints characteristic of conventional microbiological detection and identification methods. In contrast to other molecular typing methods often used to characterize pathogenic Vibrio spp., this method directly and simultaneously characterizes genetic markers valuable for the accurate detection of pandemic isolates, disease causation, and potential treatment-based resolution. These findings suggest that this microarray-based method (and further elaborations) can provide a valuable tool for the identification of genetic assemblages associated with particular types of infection, VBNC bacteria-contaminated samples, epidemiological tracking, and environmental surveillance efforts.
Supplementary Material
Acknowledgments
We thank Bala Swaminathan for critical evaluation of this manuscript and Jessie Hagger, Katherine Greene, and Nancy Puhr for excellent technical assistance. This work was supported by the Office of Naval Research.
Author contributions: G.J.V. and C.E.M. designed research; G.J.V., C.E.M., and M.M.B. performed research; G.J.V., M.M.B., and C.A.B. contributed new reagents/analytic tools; G.J.V., C.E.M., M.M.B., C.A.B., and J.D.A. analyzed data; and G.J.V., J.D.A., and D.A.S. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: VBNC, viable but nonculturable; CT, cholera toxin; SXT, sulfamethoxazoletrimethoprim; ASW, artificial seawater; TTSS, type III secretion system; VPI-2, Vibrio pathogenicity island-2.
References
- 1.Sack, D. A., Sack, R. B., Nair, G. B. & Siddique, A. K. (2004) Lancet 363, 223-233. [DOI] [PubMed] [Google Scholar]
- 2.Faruque, S. M., Albert, M. J. & Mekalanos, J. J. (1998) Microbiol. Mol. Biol. Rev. 62, 1301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Faruque, S. M., Chowdhury, N., Kamruzzaman, M., Dziejman, M., Rahman, M. H., Sack, D. A., Nair, G. B. & Mekalanos, J. J. (2004) Proc. Natl. Acad. Sci. USA 101, 2123-2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, C. Y., Wu, K. M., Chang, Y. C., Chang, C. H., Tsai, H. C., Liao, T. L., Liu, Y. M., Chen, H. J., Shen, A. B., Li, J. C., et al. (2003) Genome Res. 13, 2577-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Makino, K., Oshima, K., Kurokawa, K., Yokoyama, K., Uda, T., Tagomori, K., Iijima, Y., Najima, M., Nakano, M., Yamashita, A., et al. (2003) Lancet 361, 743-749. [DOI] [PubMed] [Google Scholar]
- 6.Chowdhury, N. R., Chakraborty, S., Ramamurthy, T., Nishibuchi, M., Yamasaki, S., Takeda, Y. & Nair, G. B. (2000) Emerg. Infect. Dis. 6, 631-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsumoto, C., Okuda, J., Ishibashi, M., Iwanaga, M., Garg, P., Rammamurthy, T., Wong, H. C., Depaola, A., Kim, Y. B., Albert, M. J. & Nishibuchi, M. (2000) J. Clin. Microbiol. 38, 578-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Hara, C. M., Sowers, E. G., Bopp, C. A., Duda, S. B. & Strockbine, N. A. (2003) J. Clin. Microbiol. 41, 5654-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oliver, J. D. (2005) J. Microbiol. 43, 93-100. [PubMed] [Google Scholar]
- 10.Amita, Chowdhury, S. R., Thungapathra, M., Ramamurthy, T., Nair, G. B. & Ghosh, A. (2003) Emerg. Infect. Dis. 9, 500-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyd, E. F., Moyer, K. E., Shi, L. & Waldor, M. K. (2000) Infect. Immun. 68, 1507-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalsgaard, A., Serichantalergs, O., Forslund, A., Lin, W., Mekalanos, J., Mintz, E., Shimada, T. & Wells, J. G. (2001) J. Clin. Microbiol. 39, 4086-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faruque, S. M. & Mekalanos, J. J. (2003) Trends Microbiol. 11, 505-510. [DOI] [PubMed] [Google Scholar]
- 14.Jermyn, W. S. & Boyd, E. F. (2002) Microbiology 148, 3681-3693. [DOI] [PubMed] [Google Scholar]
- 15.Karaolis, D. K., Johnson, J. A., Bailey, C. C., Boedeker, E. C., Kaper, J. B. & Reeves, P. R. (1998) Proc. Natl. Acad. Sci. USA 95, 3134-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Terai, A., Baba, K., Shirai, H., Yoshida, O., Takeda, Y. & Nishibuchi, M. (1991) J. Bacteriol. 173, 5036-5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waldor, M. K. & Mekalanos, J. J. (1996) Science 272, 1910-1914. [DOI] [PubMed] [Google Scholar]
- 18.Waldor, M. K., Tschape, H. & Mekalanos, J. J. (1996) J. Bacteriol. 178, 4157-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Panicker, G., Call, D. R., Krug, M. J. & Bej, A. K. (2004) Appl. Environ. Microbiol. 70, 7436-7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charles, P. T., Vora, G. J., Andreadis, J. D., Fortney, A. J., Meador, C. E., Dulcey, C. S. & Stenger, D. A. (2003) Langmuir 19, 1586-1591. [Google Scholar]
- 21.Steinberg, E. B., Greene, K. D., Bopp, C. A., Cameron, D. N., Wells, J. G. & Mintz, E. D. (2001) J. Infect. Dis. 184, 799-802. [DOI] [PubMed] [Google Scholar]
- 22.Hochhut, B., Lotfi, Y., Mazel, D., Faruque, S. M., Woodgate, R. & Waldor, M. K. (2001) Antimicrob. Agents Chemother. 45, 2991-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dalsgaard, A., Forslund, A., Serichantalergs, O. & Sandvang, D. (2000) Antimicrob. Agents Chemother. 44, 1315-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thungapathra, M., Amita, Sinha, K. K., Chaudhuri, S. R., Garg, P., Ramamurthy, T., Nair, G. B. & Ghosh, A. (2002) Antimicrob. Agents Chemother. 46, 2948-2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vora, G. J., Meador, C. E., Stenger, D. A. & Andreadis, J. D. (2004) Appl. Environ. Microbiol. 70, 3047-3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baffone, W., Citterio, B., Vittoria, E., Casaroli, A., Campana, R., Falzano, L. & Donelli, G. (2003) Int. J. Food Microbiol. 89, 31-39. [DOI] [PubMed] [Google Scholar]
- 27.Wong, H. C., Shen, C. T., Chang, C. N., Lee, Y. S. & Oliver, J. D. (2004) J. Food Protein 67, 2430-2435. [DOI] [PubMed] [Google Scholar]
- 28.Kimsey, H. H., Nair, G. B., Ghosh, A. & Waldor, M. K. (1998) Lancet 352, 457-458. [DOI] [PubMed] [Google Scholar]
- 29.Petroni, A., Corso, A., Melano, R., Cacace, M. L., Bru, A. M., Rossi, A. & Galas, M. (2002) Antimicrob. Agents Chemother. 46, 1462-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh, D. V., Bhanumathi, R. & Colwell, R. R. (2004) J. Clin. Microbiol. 42, 1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nair, G. B., Faruque, S. M., Bhuiyan, N. A., Kamruzzaman, M., Siddique, A. K. & Sack, D. A. (2002) J. Clin. Microbiol. 40, 3296-3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ansaruzzaman, M., Bhuiyan, N. A., Nair, G. B., Sack, D. A., Lucas, M., Deen, J. L., Ampuero, J. & Chaignat, C.-L. (2004) Emerg. Infect. Dis. 10, 2057-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Faruque, S. M., Chowdhury, N., Kamruzzaman, M., Ahmad, Q. S., Faruque, A. S., Salam, M. A., Ramamurthy, T., Nair, G. B., Weintraub, A. & Sack, D. A. (2003) Emerg. Infect. Dis. 9, 1116-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dziejman, M., Serruto, D., Tam, V. C., Sturtevant, D., Diraphat, P., Faruque, S. M., Rahman, M. H., Heidelberg, J. F., Decker, J., Li, L., et al. (2005) Proc. Natl. Acad. Sci. USA 102, 3465-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahmed, A. M., Shinoda, S. & Shimamoto, T. (2005) FEMS Microbiol. Lett. 242, 241-247. [DOI] [PubMed] [Google Scholar]
- 36.Toma, C., Nakasone, N., Song, T. & Iwanaga, M. (2005) Emerg. Infect. Dis. 11, 346-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lleo, M. M., Bonato, B., Signoretto, C. & Canepari, P. (2003) Antimicrob. Agents Chemother. 47, 1154-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Binsztein, N., Costagliola, M. C., Pichel, M., Jurquiza, V., Ramirez, F. C., Akselman, R., Vacchino, M., Huq, A. & Colwell, R. (2004) Appl. Environ. Microbiol. 70, 7481-7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oliver, J. D. & Bockian, R. (1995) Appl. Environ. Microbiol. 61, 2620-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Colwell, R. R., Brayton, P., Herrington, D., Tall, B., Huq, A. & Levine, M. M. (1996) World J. Microbiol. Biotechnol. 12, 28-31. [DOI] [PubMed] [Google Scholar]
- 41.Fischer-Le Saux, M., Hervio-Heath, D., Loaec, S., Colwell, R. R. & Pommepuy, M. (2002) Appl. Environ. Microbiol. 68, 5641-5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yaron, S. & Matthews, K. R. (2002) J. Appl. Microbiol. 92, 633-640. [DOI] [PubMed] [Google Scholar]
- 43.Makino, S. I., Kii, T., Asakura, H., Shirahata, T., Ikeda, T., Takeshi, K. & Itoh, K. (2000) Appl. Environ. Microbiol. 66, 5536-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coutard, F., Pommepuy, M., Loaec, S. & Hervio-Heath, D. (2005) J. Appl. Microbiol. 98, 951-961. [DOI] [PubMed] [Google Scholar]
- 45.Hlady, W. G. & Klontz, K. C. (1996) J. Infect. Dis. 173, 1176-1183. [DOI] [PubMed] [Google Scholar]
- 46.Oliver, J. D. (2005) in Foodborne Pathogens: Microbiology and Molecular Biology, eds. Fratamico, P. M., Bhunia, A. K. & Smith, J. L. (Horizon Scientific, Norfolk, U.K.), pp. 99-112.
- 47.Oliver, J. D. (2000) in Nonculturable Microorganisms in the Environment, eds. Colwell, R. R. & Grimes, D. J. (Am. Soc. Microbiol., Washington, DC), pp. 277-299.
- 48.Berlin, D. L., Herson, D. S., Hicks, D. T. & Hoover, D. G. (1999) Appl. Environ. Microbiol. 65, 2776-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lowder, M., Unge, A., Maraha, N., Jansson, J. K., Swiggett, J. & Oliver, J. D. (2000) Appl. Environ. Microbiol. 66, 3160-3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.