Abstract
Human T cell leukemia virus type-1 (HTLV-1) is an oncogenic retrovirus etiologically causal of adult T cell leukemia. The virus encodes a Tax oncoprotein that functions in transcriptional regulation, cell cycle control, and transformation. Because adult T cell leukemia like many other human cancers is a disease of genomic instability with frequent gains and losses of chromosomes, to understand this disease it is important to comprehend how HTLV-1 engenders aneuploidy in host cells. In this regard, loss of cell cycle checkpoints permits tolerance of aneuploidy but does not explain how aneuploidy is created. We show here that HTLV-1 Tax causes abnormal centrosome fragmentation in the mitotic phase of the cell cycle. We report that Tax directly binds Ran and Ran-binding protein-1, locates to centrosomes/spindle poles, and causes supernumerary centrosomes.
Keywords: oncogene, transformation, aneuploidy
Human T cell leukemia virus type-1 (HTLV-1) is the etiological agent for adult T cell leukemia (ATL) (1–5). This human retrovirus encodes a 40-kDa nuclear oncoprotein named Tax (6–8). How T cells are transformed by HTLV-1 is currently incompletely understood. However, it is thought that cellular transformation by the virus is linked to Tax's capacity to activate pro-proliferation genes (9–13), to interact with cell-cycle factors (8, 14–21), and to dysregulate checkpoints and DNA damage repair pathways (22–24).
ATL cells, like many other cancer cells, possess a chromosomal instability phenotype (24, 25). The nuclei of ATL cells have a typical “flower” configuration consistent with their high degree of aneuploidy. Recent findings suggest that the emergence of aneuploidy in cells precedes malignant transformation (25, 26), although how aneuploidy causes transformation continues to be debated. Aneuploidy may arise in diploid cells in several ways. Two routes to aneuploidy are the improper segregation of chromosomes during mitosis and the occurrence of multinucleated polyploid cells from failed cytokinesis. In both settings, supernumerary centrosomes are common.
The centrosome is the major microtubule-organizing center in animal cells. During the normal cell cycle, the single centrosome, composed of two centrioles (27–30), is duplicated once and only once. Normal centrosome replication is coupled to DNA synthesis, and the newly replicated centrosome is completed during the G2/M phase of the cell cycle to give rise to two spindle poles in mitosis. Mistakes in centrosome duplication, accumulation, and fragmentation have been linked to supernumerary centrosomes, multipolar mitosis, aneuploidy, and cancer development (31–35). Indeed, supernumerary centrosomes are frequent in many human cancers, including breast, lung, and colon (36–38).
To investigate whether centrosome abnormalities contribute to ATL development, we asked whether the HTLV-1 Tax oncoprotein might affect this organelle's function in cells. Here we show that Tax locates to centrosomes and interacts with the Ran–GTP network. Our results suggest that during mitosis Tax targets Ran-binding protein-1 (RanBP1) to cause aberrant centrosome fragmentation.
Materials and Methods
Antibodies and Plasmids. Rabbit anti-Tax or mouse monoclonal anti-Tax were from National Institutes of Health AIDS Reagent Program. α-Tubulin was from Sigma (clone B-5-1-2). Goat anti-Ran (clone 20), anti-RanBP1 (clone M-19 for murine cells and clone C19 for human cells), and control short interfering RNA (siRNA) were from Santa Cruz Biotechnology. Anti-hemagglutinin (HA) was from Sigma. Centrosomes were visualized with polyclonal pericentrin antibody (catalog no. PRB-432C, Covance, Richmond, CA). siRNA against RanBP1 was from Invitrogen. Wild-type Ran N-HA and nuclear mutant Ran D-HA plasmids were from P. Wong (Temple University School of Medicine, Philadelphia) (39).
Cells, Transfection, and Synchronization. Mouse embryonic fibroblast (MEF E6i), human fibroblast (BJ), and HeLa cells were propagated in Dulbecco's modified Eagle's medium with 10% FCS and transfected according to the manufacturer's protocol using Lipofectamine and Plus reagent (Invitrogen). Where indicated (see Fig. 1), transfected cells were synchronized starting 3 h after transfection. For synchronizations, cells were maintained in 0.5% FCS for 48 h before transfection and stimulated to reenter the cell cycle by raising the FCS concentration to 15%. Cells were treated with 2 mM hydroxyurea (Sigma) for 24 h to analyze G1–S arrest or with 0.2 mg/ml of nocodazole (Sigma) to analyze G2–M arrest. Cells were analyzed by FACS (BD Biosciences, Franklin Lakes, NJ) with modfit software (Verity Software House, Topsham, ME).
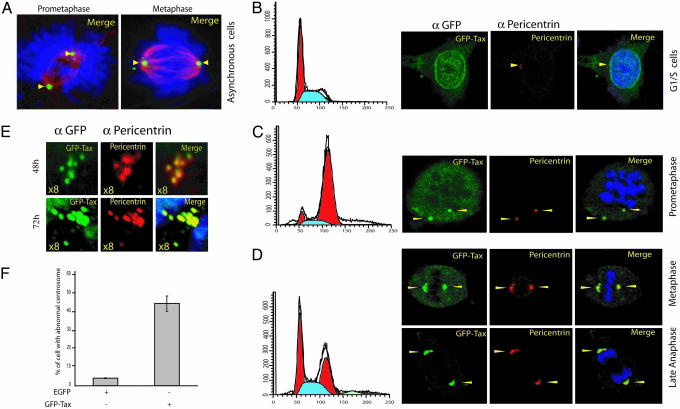
Fig. 1.
Tax is associated with centrosomes and induces abnormal centrosome morphology and number. Tax associates with centrosome during mitosis. (A) Asynchronous MEFs were transiently transfected with GFP–Tax and fixed with methanol. Signals from GFP–Tax (Alexa Fluor 488), γ-tubulin (Alexa Fluor 568), and DNA (Hoechst 33342) were merged. (B and C) Three hours after transfection, cells were synchronized for 24 h with 2 mM hydroxyurea (B) or 0.2 mg/ml nocodazole (C) and fixed with methanol. GFP–Tax and pericentrin were revealed with Alexa Fluor 488- and 568-conjugated secondary antibodies, respectively. Signals were then merged and are shown. The histograms show a parallel set of cells that were fixed in 70% ethanol and stained with propidium iodide for FACS. (D and E) Tax induces abnormal centrosome amplification. MEFs were transiently transfected with HTLV-1 Tax C-terminally fused to GFP (GFP–Tax). (E) Several enlarged examples of fragmentation of Tax-associated centrosomes. GFP–Tax and pericentrin were revealed with Alexa Fluor 488- and 568-conjugated secondary antibodies, respectively. DNA was counterstained with Hoechst 33342. (F) Centrosomal aberration was many fold higher in Tax-expressing cells than in control cells (300 cells were counted). Arrows point to centrosomes.
Immunofluorescence. Cells were cultured on glass coverslips, washed in PBS, and fixed in methanol for 10 min at –20°C. To prevent nonspecific binding, cells were first incubated with 1% BSA in PBS for 30 min followed by primary antibodies for 1 h at room temperature. Alexa Fluor 488- or 568-conjugated secondary antibodies were then added for 45 min at room temperature. DNA was counterstained with 0.1 μg/ml Hoechst 33342. Coverslips were mounted in Prolong Antifade (Molecular Probes) on glass slides and visualized with a Leica TCS-NP/SP confocal microscope.
Nickel Agarose Chromatography. MEFs were transfected with pCDNA3TaxHis, pRan–HA, or pRanBP1–HA vectors and lysed in nickel agarose lysis buffer (50 mM NaH2PO4/300 mM NaCl/5mM imidazole/0.05% Tween 20/Complete protease inhibitor). His-conjugated proteins were purified by nickel chromatography (Ni-NTA agarose, Qiagen, Valencia, CA). Beads were washed with nickel agarose wash buffer (50 mM NaH2PO4/300 mM NaCl/10 mM imidazole/0.05% Tween 20). Nickel-binding proteins were resuspended in 2× sample buffer supplemented with 200 mM imidazole and heated for 10 min at 95°C before Western blotting.
In Vitro Pull Down and Immunoprecipitation. GST and GST–Tax were expressed in BL21 cells. After isopropyl-β-d-thioglactoside induction for 5 h at 37°C (1 mM final concentration), cells were lysed with NENT buffer (20 mM Tris·HCl, pH 8.0/100 mM NaCl/1 mM EDTA/0.5% Nonidet P-40/1 mM PMSF/protease inhibitors). GST proteins were recovered by binding to gluthatione Sepharose resin and were incubated with Jurkat extracts. Proteins bound to beads were detected by using anti-Ran. For coimmunoprecipitations, HeLa and MEFs were transfected with Ran–HA or RanBP1–HA plus GFP-tagged Tax wild type or mutants using Lipofectamine and Plus reagent (Invitrogen). Cell pellets were resuspended in RIPA buffer (50 mM Hepes, pH 7.3/150 mM NaCl/2 mM EDTA/20 mM β-glycerophosphate/0.1 mM Na3VO4/1mMNaF/0.5mMDTT/protease inhibitors). Total cell lysates were immunoprecipitated with monoclonal anti-HA agarose or anti-GFP/protein A/G beads (Sigma–Aldrich) overnight at 4°C. The coimmunoprecipitates were analyzed by SDS/PAGE and immunoblotted.
Results
Tax Locates to Centrosomes During Mitosis. Tax is the HTLV-1 oncoprotein that dysregulates cellular checkpoints and is linked to genomic instability (8, 13). Of note, Tax expression induces multinucleated cells, which, at a low stochastic frequency, leads to the accumulation and emergence of aneuploid cells (18, 40). How a normal T cell infected by HTLV-1 transits from euploidy to aneuploidy is poorly understood. Recently, a role for the centrosome in chromosomal instability and carcinogenesis has been proposed for many solid tumors (41), and we wondered whether centrosome abnormalities may also explain aneuploidy in Tax-expressing cells.
To ask whether Tax causes centrosome pathology, we transfected MEFs with GFP–control or GFP–Tax plasmid. Centrosomes compose the poles of the cell's mitotic spindles. During interphase, the centrosome is a cytoplasmic organelle and the Tax protein is nuclear. When we transfected asynchronous cells in culture, we observed that mitotic cells were seen at a high frequency with Tax at the centrosome of the spindle poles (Fig. 1A). To understand when Tax first appears at centrosomes, we synchronized newly transfected MEFs either at the G1/S junction by using hydroxyurea (Fig. 1B) or at M by using nocodazole (Fig. 1C). In G1/S cells, no GFP–Tax was with centrosomes (stained with anti-pericentrin) (Fig. 1B); by contrast, nocodazole-treated cells in early M showed clear colocalization of GFP–Tax with pericentrin (an integral centrosome protein) (Fig. 1C).
If cells are released from nocodazole arrest (Fig. 1C) and allowed to progress to metaphase and anaphase, we frequently observed distinct changes in the morphology and intactness of Tax-associated centrosomes. For example, many anti-pericentrin-stained spindle pole bodies became misshaped and multiply fragmented (Fig. 1 D and E). Centrosome fragmentation has been reported to be a consequence of genotoxicity and DNA damage (31, 42). In our experiments, we noted that Tax expression increased the prevalence of centrosome abnormalities in cells by 8-fold over control cells (Fig. 1F).
Tax Interacts with Ran-GTPase. The centrosome is a complex organelle composed of two barrel-shaped centrioles surrounded by a dense pericentriolar matrix (30, 43) with many associated proteins. Ran-GTPase, a well known regulator of nucleocytoplasmic transport (44–46), and several Ran binding proteins are associated with the centrosome (46). Ran has been suggested to drive mitotic spindle assembly by controlling the availability of aster-promoting activities (47).
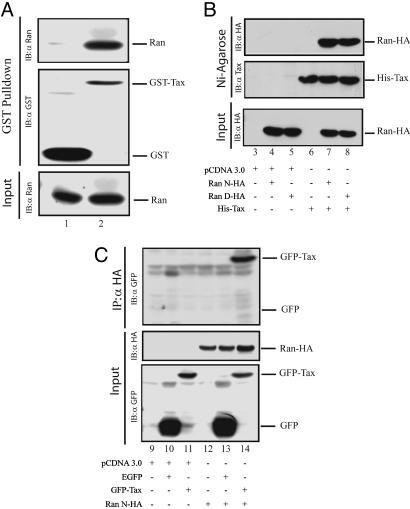
Because Ran and Tax (48) shuttle between the nucleus and the cytoplasm and both associate with centrosomes, we wondered whether Tax might target Ran to create centrosome pathology. To test this hypothesis, we asked whether Tax binds Ran. We incubated GST or GST–Tax separately with Jurkat cell extract and queried whether the latter but not the former might capture Ran. Using anti-Ran in Western blotting, we found that Ran was bound by GST–Tax (Fig. 2A, lane 2) but not by GST (Fig. 2A, lane 1).
Fig. 2.
HTLV-1 Tax binds Ran-GTPase. (A) In vitro interaction between Tax and Ran–GTPase (Ran). (Top and Middle) MEF extracts were incubated with GST or GST–Tax, and pull-downs were probed and detected with anti-Ran (Top) and anti-GST (Middle) by Western blot. (Bottom) Amount of Ran in cell extract was verified. Lane 1, incubation of extract with GST; lane 2, incubation of extract with GST–Tax. (B and C) Coimmunoprecipitation of Tax and Ran. (B) MEFs were transfected with empty vector (lane 3), Ran wild type (Ran N-HA) (lanes 4 and 7) or nuclear mutant (Ran D-HA) (lanes 5 and 8), and a His-tagged Tax vector (His-Tax) (lanes 6–8). His-tagged Tax protein was pulled-down by Ni-agarose beads. Pull downs were probed with anti-HA or anti-Tax. Amount of Ran in the extract was monitored by using anti-HA. (C) MEFs were transfected with empty vector (lane 9) or a Ran vector (Ran N-HA) (lanes 12–14) with either GFP (EGFP) (lanes 10 and 13) or GFP–Tax (lanes 11 and 14). Ran–HA was immunoprecipitated by anti-HA. GFP–Tax was detected by anti-GFP. Amounts of Ran–HA and GFP–Tax in cell extract were monitored by using anti-HA and anti-GFP. IB, immunoblot.
We next tested Tax–Ran interaction in freshly cultured primary MEFs. We transfected MEFs with a HA-tagged Ran plasmid or a His-tagged Tax plasmid, with or without a separate GFP–Tax plasmid. Cells lysates were prepared and equilibrated individually with anti-Ran-agarose beads or Ni-agarose beads. Beads were pelleted and washed, and bead-captured proteins were probed for Tax or Ran (Fig. 2 B and C). Consistent with the Jurkat results, a specific association between Ran and Tax was reproduced in primary MEFs (Fig. 2 B, lane 7 and 8, and C, lane 14).
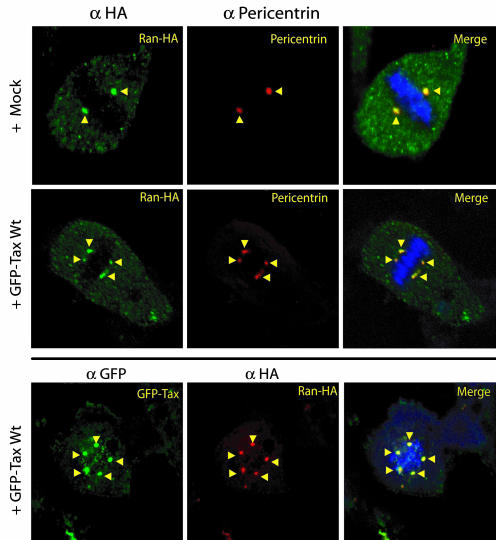
We next performed confocal immunofluorescent studies in MEFs transfected with Ran and/or Tax plasmids. MEFs transfected with Ran–HA and/or GFP–Tax were fixed and stained with anti-HA, anti-pericentrin, or anti-GFP (Fig. 3). In mitotic cells, Ran was at centrosomes; however, overexpression of Ran neither perturbed centrosome morphology nor number (Fig. 3 Top). When Ran was coexpressed with Tax, both were together at centrosomes; notably, in this setting, aberrantly numbered and morphologically fragmented centrosomes were seen (Fig. 3 Middle and Bottom). The observed association between Ran and Tax was not an artifact of plasmid overexpression given that we found the same interaction between cell endogenous Ran and physiologically expressed Tax in HTLV-1-transformed T cells (e.g., C81–6645 and MT4) (data not shown).
Fig. 3.
Tax interacts with Ran in cells. Tax and Ran colocalize at centrosomes, and Tax association with Ran disrupts centrosome integrity. MEFs were cotransfected with GFP–Tax and Ran–HA. Anti-HA, anti-pericentrin, and anti-GFP were used to reveal the subcellular localization of Ran and Tax. DNA was stained with Hoechst 33342. Signals were then merged. (Top and Middle) With or without Tax, Ran localized with centrosomes in mitotic cells (arrows). (Middle and Bottom) In the presence of Tax, abnormally amplified centrosomes were observed.
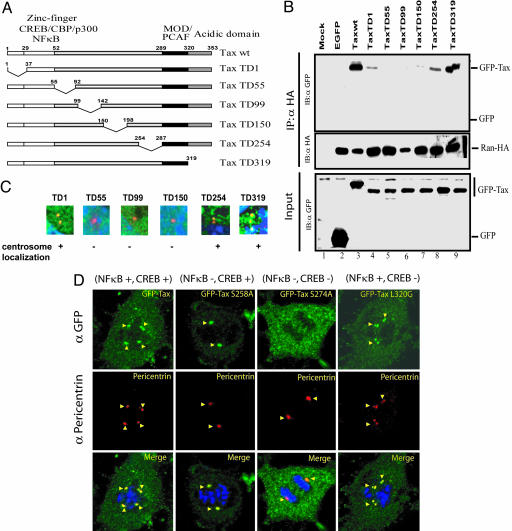
A Domain for Ran Interaction Falls Within Tax Amino Acids 55–198. To map Tax–Ran interaction, we created six GFP-tagged Tax-deletion mutants (Fig. 4A). We transfected MEFs with Ran–HA and individual GFP–Tax wild types or mutants. Transfected cell lysates were immunoprecipitated with anti-Ran, and we queried for co-immunoprecipitated Tax (Fig. 4B). Ran coimmunoprecipitated wild-type GFP–Tax (Fig. 4B, lane 3), but not GFP alone (Fig. 4B, lane 2). Two Tax deletion mutants, TD1 and TD254, bound Ran weakly (Fig. 4B, lanes 4 and 8), whereas a third Tax mutant, TD319, associated with Ran strongly (Fig. 4B, lane 9). By contrast, three other Tax mutants, TD55, TD99 and TD150, showed no detectable binding to Ran. These results map the Tax–Ran interaction domain to fall between amino acids 55 and 198 of Tax.
Fig. 4.
Characterization of Tax amino acids important for Ran interaction. (A) A schematic overview of the different Tax mutants fused to GFP. CBP, CREB-binding protein; MOD, modulator domain; PCAF, p300–CREB-binding protein-associated factor. (B) Delineation of HTLV-1 Tax residues important for Tax–Ran interaction and centrosome localization. MEFs were cotransfected with Ran–HA and Tax deletion mutants (lanes 4–9). (Top) Ran–HA was immunoprecipitated with anti-HA. GFP fusion protein was detected by anti-GFP. (Middle and Bottom) Amount of Ran–HA in the immunoprecipitate (IP) and GFP–Tax in cell extracts were checked with anti-HA and anti-GFP. (C) Anti-pericentrin and anti-GFP were used to reveal the centrosomal localization of Tax in MEFs transfected with GFP–Tax deletion mutants. In these enlarged images, TaxTD1, TD254, and TD319 but not TD55, TD99, or TD150 localized to centrosomes. (D) Tax localization to the centrosome is insufficient to induce aberrant amplification. MEFs were cotransfected with GFP–Tax and Ran–HA. Anti-HA, anti-pericentrin, and anti-GFP were used to reveal the subcellular localizations. DNA was stained with Hoechst 33342. Whereas wild-type Tax (NF-κB+/CREB+) induced centrosome amplification, Tax S258A (NF-κB–/CREB+), a mutant unable to activate NF-κB, localized to the centrosome (arrowheads) but did not induce centrosome amplification. In contrast, Tax S 274A (NF-κB–/CREB–), a CREB-defective mutant, can bind Ran but did not localize to the centrosome (arrowheads). Tax L320G (NF-κB+/CREB–) behaved like GFP–Tax.
We next transfected individual GFP–Tax mutants into MEFs and stained for Tax (GFP), centrosome (pericentrin), and DNA (Fig. 4C). Three Tax mutants (TD1, TD254, and TD319) distributed to centrosomes like wild-type Tax. By contrast, three other mutants deleted in the central region of Tax (TD99, TD55, and TD150) did not appear at centrosomes. Hence, the central region of Tax, in addition to determining Ran binding, specifies localization to centrosomes.
Tax activates two major signal transduction pathways, cAMP-response element-binding protein (CREB) and NF-κB, in cells (49). With the exception of TD319, all Tax deletion mutants used above are inactive in both CREB and NF-κB pathways (17). To clarify whether its signal transduction role might contribute to Tax's centrosome function, we tested the Tax point mutations Tax S258A, Tax S274A, and Tax L320G, which have NF-κB–/CREB+, NF-κB–/CREB–, and NF-κB+/CREB– functional phenotypes, respectively. Interestingly, we observed that only the GFP–Tax L320G mutant induced supernumerary centrosomes, whereas neither GFP–Tax S258A (capable of centrosome localization) nor GFP–Tax274A (incapable of centrosome localization) did so (Fig. 4D). These results argued that Tax localization to the centrosome may be necessary but is not in itself sufficient to provoke supernumerary aberrancies. The results also suggested that the ability of Tax to signal through NF-κB but not CREB may be needed to disrupt normal centrosome physiology.
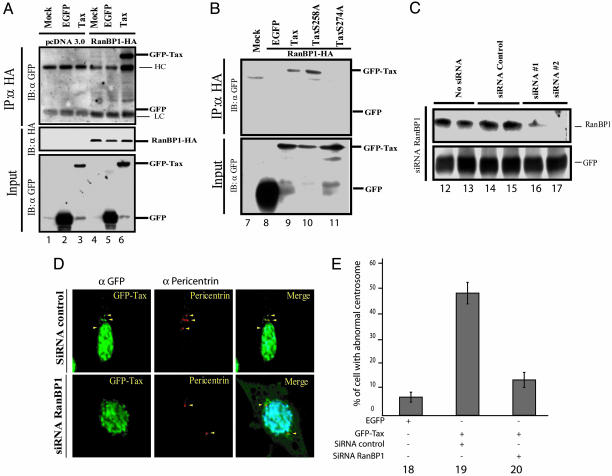
RanBP1 Is Targeted by Tax. Separately, we found that the TaxS274A mutant can bind Ran (data not shown) but that such binding was insufficient to dictate either centrosome localization or numerical aberrancies (Fig. 4D). Moreover, GFP–Tax S258A mutant, which does localize to centrosomes (Fig. 4D), was found not to bind Ran (data not shown). These unexpected conundrums suggested that another centrosome protein other than Ran may be targeted by Tax. Recently, RanBP1, an important partner of Ran–GTPase, has been reported to play a role in regulating centrosome stability during mitosis (47). We next hypothesized that perhaps RanBP1 is the functional centrosome target of Tax.
To ask whether Tax and RanBP1 interact, we transfected MEFs with HA-tagged RanBP1 and GFP–Tax. When we immunoprecipated cell lysates with anti-HA and probed for a Tax–RanBP1 complex by Western blotting with anti-GFP (Fig. 5A), we observed specific coimmunoprecipitation of Tax with RanBP1 (Fig. 5A, lane 6). As controls, we used two Tax point mutants, GFP–Tax S258A and GFP–Tax S274A. Interestingly GFP–Tax S274A, which does not localize to centrosomes (Fig. 4D), did not bind RanBP1 (Fig. 5B, lane 11), whereas GFP–Tax S258A, which does localize to centrosomes (Fig. 4D) but does not to bind Ran (data not shown), did bind RanBP1 (Fig. 5B, lane 10). These results correlated Tax binding of RanBP1 with its ability to locate to centrosomes.
Fig. 5.
RanBP1 is important for Tax association with centrosome and induction of pathology. (A and B) MEFs were cotransfected with RanBP1–HA (lanes 4–6 and 9–11), and wild-type Tax (lanes 3, 6, and 9) or different Tax point mutants (lanes 10 and 11). RanBP1–HA was immunoprecipitated by using anti-HA. GFP-tagged protein was detected with anti-GFP. Amounts of RanBP1–HA in the immunoprecipitate and GFP–Tax in cell extract were verified by using anti-HA or anti-GFP. (C) MEFs were cotransfected with 0.5 μg of EGFP vector and 1.5 μg of control siRNA (lanes 14 and 15) or siRNA targeted against RanBP1 (lanes 16 and 17). (D) MEFs were cotransfected with 0.5 μg of GFP–Tax and 1.5 μg of control siRNA (Upper) or siRNA against RanBP1 (Lower). Anti-pericentrin and anti-GFP were used to reveal the subcellular localization of centrosomes and Tax. DNA was stained with Hoechst 33342. (E) Whereas wild-type Tax induced centrosome destabilization and amplification (lane 19), knockdown of RanBP1 by siRNA abrogated centrosomal amplification by Tax (lane 20); 500 cells with supernumerary centrosomes were counted.
The above transfection/overexpression experiments do not comment on the role of cell-endogenous RanBP1 in Tax-engendered centrosomal pathology. To address the latter issue, we used two siRNAs to knock down RanBP1 (Fig. 5C, lanes 16 and 17). When MEFs were transfected with siRNA against RanBP1, we found that Tax no longer localized to centrosomes (Fig. 5D Lower). Correspondingly, siRNA knockdown of RanBP1 also abrogated Tax's induction of supernumerary centrosomes in transfected cells (Fig. 5E, lanes 19 and 20). Taken together, these findings support a role for RanBP1 in Tax's localization to centrosomes and in its induction of centrosome pathology. Similar siRNA experiments targeting Ran were difficult to interpret because knockdown of Ran appeared to provoke general cytotoxicity (data not shown).
Discussion
ATL develops in ≈2–5% of HTLV-1 infected individuals after a prolonged latency period (50). This long latency for ATL development suggests that multiple discrete events in a normal cell need to be subverted by HTLV-1 in its transformation of cells. It has been suggested that impairment of cell cycle checkpoints by Tax contributes to ATL transformation (13). However, although abrogation of checkpoints allows for tolerance of chromosomal imbalances, such inactivation cannot mechanistically create aneuploidy in ATL cells (8, 15–17, 22–24).
Alterations in centrosome number or function are frequent in transformed cells (25, 26, 51). Because centrosomes dictate the formation of a bipolar mitotic spindle, which is needed for the proper segregation of duplicated chromosomes, aberrancies in centrosome number/function would unbalance chromosome partitioning. A recent study suggested that Tax can trigger chromosomal missegregation in mitosis through its interaction with the anaphase-promoting complex incurring premature degradation of securin (18); in our assays, we have not observed a significant securin–APC effect (S. V. Sheleg and J.-M.P., unpublished data). Here we show Tax's induction of supernumerary centrosomes as a mechanism for HTLV-1 to create aneuploidy in cells.
Supernumerary centrosomes can occur in at least three ways. First, centrosome replication in late G1 can become dysregulated, leading to amplification in number. Second, various genotoxic insults can result in fragmentation of mitotic centrosomes, leading to the production of multipolar spindles (32, 35, 52). Third, failed cytokinesis with the formation of multinucleated cells can accumulate abnormal numbers of centrosomes. In other viral oncoprotein systems, such as human papillomavirus E7, supernumerary moieties are shown to occur via dysregulated interphase replication of the centrosome (53–56). In contrast, Tax does not affect the interphase duplication of centrosomes; instead, the HTLV-1 oncoprotein causes centrosomes to fragment in mitosis, creating numerical aberrancies. Our data indicate that Tax first interacts with centrosomes after nuclear envelope breakdown has occurred in mitosis (Fig. 1).
How does Tax cause centrosome fragmentation? The Ran–GTPase network is involved in the control of nucleocytoplasmic transport of macromolecules (46). Recent evidence offers another function for the Ran network in mitosis. Ran also has been shown to regulate mitotic microtubule nucleation and spindle formation (46, 47, 57–60). Interestingly, during mitosis, Tax, Ran, and RanBP1 are found together on centrosomes/spindle poles. Tax can bind Ran independent of RanBP1, although our evidence indicates that Tax binding to RanBP1, not to Ran, is the critical event that targets Tax to centrosomes (Figs. 4 and 5). Our siRNA knockdown experiments show that RanBP1 is required for Tax to a create centrosome fragmentation (Fig. 5); however, because of the complications of generalized cytotoxicity when Ran knockdowns were attempted, we cannot formally exclude a role for Ran in Tax's activity on centrosomes. It is conceivable that RanBP1 is responsible for targeting Tax to centrosomes, but, once Tax locates to the centrosome, a ternary complex of Ran/RanBP1/Tax creates centrosomal aberrations.
That RanBP1 may be a cellular factor required for Tax's induction of supernumerary centrosomes (Fig. 5) is not necessarily surprising. Elsewhere, RanBP1 is overexpressed in many tumors that have multipolar spindles, and RanBP1 has a demonstrated role in regulating the cohesion of duplicated centrioles (57). Abnormal RanBP1 function is known to alter microtubule dynamic at the spindle pole and to induce the aberrant separation of mother and daughter centrioles (47, 61). Our current data are compatible with a scenario in which, once the nuclear envelope dissolves and the cell enters mitosis, nuclear Tax protein goes to the spindle pole, where it subverts normal RanBP1 function. We hypothesize that a Tax/Ran/RanBP1 complex then alters the otherwise normal tethering of duplicated centrioles leading to their premature splitting and causing the formation of multipolar spindles. Multipolar mitosis then leads to chromosomal missegregation and aneuploid daughter cells.
In summary, here we show that Tax induces abnormal centrosome numbers in cells. HTLV-1 and human papillomavirus (4, 54), two very different human-transforming viruses, appear to share a common objective to create numerical chromosomal mistakes in cells, albeit through different mechanisms. Elsewhere, Tax has also been shown to cause multinucleated cells (8). Currently, multinucleated cells cannot be excluded as a separate means through which HTLV-1 can further create numerical centrosome abnormalities. Multipolar mitosis and the missegregation of chromosomes would normally be expected to be arrested by the mitotic spindle assembly checkpoint. Intriguingly, the mitotic spindle assembly sentinel was previously shown to be disabled by Tax (23). Hence, Tax can altogether create numerical chromosomal mistakes (i.e., multipolar mitosis) and inactivate the mitotic checkpoint (i.e., inactivation of mitotic spindle checkpoint), which would otherwise censor those errors. Thus, HTLV-1 Tax appears to be a singularly sufficient oncoprotein that creates errors while compelling the progression of error-ridden mitosis to evolve aneuploid progeny cells.
Acknowledgments
We thank Dr. P. Wong for generously providing plasmids, members of the K.-T.J. laboratory and A. Dayton for critically reading the manuscript, and R. Plishka and A. Buckler-White for assistance with DNA sequencing and oligonucleotide synthesis. This research was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases.
Author contributions: J.-M.P., K.H., and K.-T.J. designed research; J.-M.P., K.H., and A.M. performed research; J.-M.P., K.H., and K.-T.J. analyzed data; and J.-M.P. and K.-T.J. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ATL, adult T cell leukemia; CREB, cAMP-response element-binding protein; HA, hemagglutinin; HTLV-1, human T cell leukemia virus type-1; MEF, mouse embryonic fibroblast; RanBP1, Ran-binding protein-1; siRNA, short interfering RNA.
References
- 1.Poiesz, B. J., Ruscetti, F. W., Gazdar, A. F., Bunn, P. A., Minna, J. D. & Gallo, R. C. (1980) Proc. Natl. Acad. Sci. USA 12, 7415–7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hinuma, Y., Nagata, K., Hanaoka, M., Nakai, M., Matsumoto, T., Kinoshita, K. I., Shirakawa, S. & Miyoshi, I. (1981) Proc. Natl. Acad. Sci. USA 10, 6476–6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoshida, M., Miyoshi, I. & Hinuma, Y. (1982) Proc. Natl. Acad. Sci. USA 79, 2031–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gallo, R. C. (2005) Retrovirology 2, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takatsuki, K. (2005) Retrovirology 2, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Green, P. L. & Chen, I. S. (1990) FASEB J. 4, 169–175. [DOI] [PubMed] [Google Scholar]
- 7.Jeang, K.-T. (2001) Cytokine Growth Factor Rev. 12, 207–217. [DOI] [PubMed] [Google Scholar]
- 8.Marriott, S. J., Lemoine, F. J. & Jeang, K. T. (2002) J. Biomed. Sci. 9, 292–298. [DOI] [PubMed] [Google Scholar]
- 9.Fujii, M., Niki, T., Mori, T., Matsuda, T., Matsui, M., Nomura, N. & Seiki, M. (1991) Oncogene 6, 1023–1029. [PubMed] [Google Scholar]
- 10.Ng, P. W., Iha, H., Iwanaga, Y., Bittner, M., Chen, Y., Jiang, Y., Gooden, G., Trent, J., Meltzer, P., Jeang, K. T. & Zeichner, S. L. (2000) Oncogene 20, 4484–4496. [DOI] [PubMed] [Google Scholar]
- 11.Ross, T. M., Narayan, M., Fang, Z. Y., Minella, A. C. & Green, P. L. (2000) J. Virol. 74, 2655–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Twizere, J. C., Kruys, V., Lefebvre, L., Vanderplasschen, A., Collete, D., Debacq, C., Lai, W. S., Jauniaux, J. C., Bernstein, L. R., Semmes, O. J., et al. (2003) J. Natl. Cancer Inst. 95, 1846–1859. [DOI] [PubMed] [Google Scholar]
- 13.Jeang, K. T., Giam, C. Z., Majone, F. & Aboud, M. (2004) J. Biol. Chem. 279, 31991–31994. [DOI] [PubMed] [Google Scholar]
- 14.Schmitt, I., Rosin, O., Rohwer, P., Gossen, M. & Grassmann, R. (1998) J. Virol. 72, 633–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kibler, K. V. & Jeang, K.-T. (2001) J. Virol. 5, 2161–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemoine, F. J., Wycuff, D. R. & Marriott, S. J. (2001) Dis. Marker 17, 129–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haller, K., Wu, Y., Derow, E., Schmitt, I., Jeang, K. T. & Grassmann, R. (2002) Mol. Cell. Biol. 10, 3327–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, B., Liang, M. H., Kuo, Y. L., Liao, W., Boros, I., Kleinberger, T., Blancato, J. & Giam, C. Z. (2003) Mol. Cell. Biol. 15, 5269–5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haoudi, A., Daniels, R. C., Wong, E., Kupfer, G. & Semmes, O. J. (2003) J. Biol. Chem. 278, 37736–37744. [DOI] [PubMed] [Google Scholar]
- 20.Kehn, K., Deng, L., de la Fuente, C., Strouss, K., Wu, K., Maddukuri, A., Baylor, S., Rufner, R., Pumfery, A., Bottazzi, M. E. & Kashanchi, F. (2004) Retrovirology 1, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu, K., Bottazzi, M. E., de la Fuente, C., Deng, L., Gitlin, S. D., Maddukuri, A., Dadgar, S., Li, H., Vertes, A., Pumfery, A. & Kashanchi, F. (2004) J. Biol. Chem. 279, 495–508. [DOI] [PubMed] [Google Scholar]
- 22.Suzuki, T., Kitao, S., Matsushime, H. & Yoshida, M. (1996) EMBO J. 15, 1607–1614. [PMC free article] [PubMed] [Google Scholar]
- 23.Jin, D. Y., Spencer, F. & Jeang, K. T. (1998) Cell 93, 81–91. [DOI] [PubMed] [Google Scholar]
- 24.Kasai, T., Iwanaga, Y., Iha, H. & Jeang, K. T. (2002) J. Biol. Chem. 277, 5187–5193. [DOI] [PubMed] [Google Scholar]
- 25.Rajagopalan, H. & Lengauer, C. (2004) Nature 432, 338–341. [DOI] [PubMed] [Google Scholar]
- 26.Lengauer, C., Kinzler, K. W. & Vogelstein, B. (1998) Nature 396, 643–649. [DOI] [PubMed] [Google Scholar]
- 27.Glover, D. M., Gonzalez, C. & Raff, J. W. (1993) Sci. Am. 268, 62–68. [DOI] [PubMed] [Google Scholar]
- 28.Winey, M. (1999) Curr. Biol. 9, R449–R452. [DOI] [PubMed] [Google Scholar]
- 29.Young, A., Tuft, R., Carrington, W. & Doxsey, S. J. (1999) Methods Cell Biol. 58, 223–238. [DOI] [PubMed] [Google Scholar]
- 30.Meraldi, P. & Nigg, E. A. (2002) FEBS Lett. 521, 9–13. [DOI] [PubMed] [Google Scholar]
- 31.Pihan, G. A., Purohit, A., Wallace, J., Knecht, H., Woda, B., Quesenberry, P. & Doxsey, S. J. (1998) Cancer Res. 58, 3974–3985. [PubMed] [Google Scholar]
- 32.D'Assoro, A. B., Lingle, W. L. & Salisbury, J. L. (2002) Oncogene 21, 6146–6153. [DOI] [PubMed] [Google Scholar]
- 33.Kramer, A., Neben, K. & Ho, A. (2002) Leukemia 16, 767–775. [DOI] [PubMed] [Google Scholar]
- 34.Nigg, E. A. (2002) Nat. Rev. Cancer 2, 815–825. [DOI] [PubMed] [Google Scholar]
- 35.Wang, Q., Hirohashi, Y., Furuuchi, K., Zhao, H., Liu, Q., Zhang, H., Murali, R., Berezov, A., Du, X., Li, B. & Greene, M. I. (2004) DNA Cell Biol. 23, 475–489. [DOI] [PubMed] [Google Scholar]
- 36.Pihan, G. A., Purohit, A., Wallace, J., Malhotra, R., Liotta, L. & Doxsey, S. J. (2001) Cancer Res. 61, 2212–2219. [PubMed] [Google Scholar]
- 37.Schneeweiss, A., Sinn, H. P., Ehemann, V., Khbeis, T., Neben, K., Krause, U., Ho, A. D., Bastert, G. & Kramer, A. (2003) Int. J. Cancer 107, 346–352. [DOI] [PubMed] [Google Scholar]
- 38.Salisbury, J. L., D'Assoro, A. B. & Lingle, W. L. (2004) J. Mammary Gland Biol. Neoplasia 9, 275–283. [DOI] [PubMed] [Google Scholar]
- 39.Chung, S. W., Huang, X. Y., Song, J., Thomas, R. & Wong, P. M. (2004) Front Biosci. 9, 3374–3383. [DOI] [PubMed] [Google Scholar]
- 40.Liang, M. H., Geisbert, T., Yao, Y., Hinrichs, S. H. & Giam, C. Z. (2002) J. Virol. 76, 4022–4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emdad, L., Sarkar, D., Su, Z. Z. & Fisher, P. B. (2005) Front Biosci. 10, 728–742. [DOI] [PubMed] [Google Scholar]
- 42.Kramer, A. & Ho, A. D. (2001) Onkologie 24, 538–544. [DOI] [PubMed] [Google Scholar]
- 43.Lange, B. M. (2004) Zellbiol. Aktuell 30, 18–20. [Google Scholar]
- 44.Moore, M. S. & Blobel, G. (1993) Nature 365, 661–663. [DOI] [PubMed] [Google Scholar]
- 45.Kunzler, M. & Hurt, E. (2001) J. Cell Sci. 114, 3233–3241. [DOI] [PubMed] [Google Scholar]
- 46.Dasso, M. (2002) Curr. Biol. 12, R502–8. [DOI] [PubMed] [Google Scholar]
- 47.Di Fiore, B., Ciciarello, M., Mangiacasale, R., Palena, A., Tassin, A. M., Cundari, E. & Lavia, P. (2003) J. Cell Sci. 116, 3399–3411. [DOI] [PubMed] [Google Scholar]
- 48.Burton, M., Upadhyaya, C. D., Maier, B., Hope, T. J. & Semmes, O. J. (2000) J. Virol. 74, 2351–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neuveut, C. & Jeang, K. T. (2000) Prog. Cell Cycle Res. 4, 157–162. [DOI] [PubMed] [Google Scholar]
- 50.Matsuoka, M. (2005) Retrovirology 2, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lengauer, C. (2005) Semin. Cancer Biol. 15, 1. [DOI] [PubMed] [Google Scholar]
- 52.Kuntziger, T. & Bornens, M. (2000) Curr. Top Dev. Biol. 49, 1–25. [DOI] [PubMed] [Google Scholar]
- 53.Minn, A. J., Boise, L. H. & Thompson, C. B. (1996) Genes Dev. 10, 2621–2631. [DOI] [PubMed] [Google Scholar]
- 54.Duensing, S. & Munger, K. (2002) Oncogene 21, 6241–6248. [DOI] [PubMed] [Google Scholar]
- 55.Tarapore, P. & Fukasawa, K. (2002) Oncogene 21, 6234–6240. [DOI] [PubMed] [Google Scholar]
- 56.Haoudi, A. & Semmes, O. J. (2003) Virology 305, 229–239. [DOI] [PubMed] [Google Scholar]
- 57.Di Matteo, G., Fuschi, P., Zerfass, K., Moretti, S., Ricordy, R., Cenciarelli, C., Tripodi, M., Jansen-Durr, P. & Lavia, P. (1995) Cell Growth Differ. 6, 1213–1224. [PubMed] [Google Scholar]
- 58.Kahana, J. A. & Cleveland, D. W. (1999) J. Cell Biol. 146, 1205–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nishimoto, T. (1999) Biochem. Biophys. Res. Commun. 262, 571–574. [DOI] [PubMed] [Google Scholar]
- 60.Sazer, S. & Dasso, M. (2000) J. Cell Sci. 113, 1111–1118. [DOI] [PubMed] [Google Scholar]
- 61.Guarguaglini, G., Renzi, L., D'Ottavio, F., Di Fiore, B., Casenghi, M., Cundari, E. & Lavia, P. (2000) Cell Growth Differ. 11, 455–465. [PubMed] [Google Scholar]