Abstract
Alternative splicing is a sophisticated nuclear process that regulates gene expression. It represents an important mechanism for enhancing the functional diversity of proteins. Our current knowledge of alternatively spliced variants is derived mainly from mRNA transcripts, and very little is known about their protein tertiary structures. We carried out a large-scale analysis of known alternatively spliced variants at both protein sequence and structure levels and have shown that threading is, in general, a viable approach for modeling structures of alternatively spliced variants. An examination of alternative splicing at the protein sequence level revealed that the size of splicing events follows the power law distribution and the majority of splicing isoforms harbor only one or two alternations. We examined alternative splicing in the context of protein 3D structures and found that the boundaries of alternative splicing events generally happen in coil regions of secondary structures and exposed residues and the majority of the sequences involved in splicing are located on the surface of proteins. In light of these findings, we then proceeded to demonstrate that threading represents a useful tool for structure prediction of alternative splicing isoforms and addressed the fold stability issue of threading-based structure prediction by molecular dynamics simulation. Our analysis and the insights gained have helped to establish a viable method for structure prediction of alternatively spliced isoforms at the genome scale.
Keywords: threading, structure prediction, protein variants
Alternative splicing is a eukaryotic cellular process where multiple mature mRNAs can be produced from the same pre-mRNA by inclusion of different portions of coding sequences (1). With the progress of genome sequencing and accumulation of EST data, it has been estimated that 45–60% of human genes undergo alternative splicing (2, 3). The large number of splicing variants produced raises intriguing questions about the regulation of splicing process and the functional roles of splicing products. Numerous studies have demonstrated that alternative splicing plays key roles in vital biological processes such as apoptosis, heart development, and neuron differentiation. Splicing isoforms involved in those processes differ in their peptide sequences and hence can provide different and sometimes even antagonist functions (4–7).
Alternative splicing has significant impacts on protein functions such as ligand binding, enzyme activity, and protein–protein interaction (8–10), and defects in mRNA splicing are natural causes of human diseases. An examination of the Human Gene Mutation Database (January 2004), which documented 39,415 mutations in regulatory, splicing, and coding regions of human nuclear genes, revealed that ≈10% (3,783) annotated mutations affect canonical splicing sites (11), and numerous cases have been documented in terms of having different isoforms in a variety of diseases such as myotonic dystrophy (12), Azoospermia (13, 14), and Alzheimer's disease (15–17). The most striking examples of aberrant splicing-related diseases are cancers. Studies in the past 20 years have documented numerous cancer-specific alternatively spliced isoforms that affect the functions of proteins, including transcription factors, cellular signaling components, apoptosis regulators, and components of extracellular matrix (18). A prominent example is tumor suppressor P53. A recent survey found 29 different P53 splicing site mutations in >12 types of cancers that result in numerous cancer-specific splicing isoforms (19). Cancer-specific P53 splicing isoforms generally contain the intact DNA binding domain and harbor different sizes of deletions in the C- or N-terminal domain (20). Some of the isoforms have been shown to counteract the tumor suppressor activity of P53 and may be critical to our understanding of tumorgenesis given the central role of P53 as a tumor suppressor (20).
Alternative splicing was realized by exclusion of introns and inclusion of exons in a combinatorial manner. Diversity of splicing was further enhanced by cryptic splicing sites whose sequences resemble authentic sites and could be activated because of mutations and splicing regulators that could include introns in final splicing products, change the sizes of exons, and introduce frameshifts (21, 22). When alternatively spliced isoforms were examined at the protein level, three types of alternative splicing events could be established with the full-length protein as a reference. Splicing isoforms can harbor deletions where part of the sequence of the full-length protein is removed, which can be accomplished either through exon exclusion or use of a noncanonical splicing site that uses a shortened exon. An insertion in an isoform sequence is usually caused by the use of cryptic splicing sites that allow intron retention. Substitutions are most commonly caused by the use of mutually exclusive exons, cryptic splicing sites that create frameshifts, or a combination of exon exclusion and intron retention.
Despite significant efforts that have been put into the study of the mechanisms of alternative splicing and functions of different isoforms, our knowledge of the structures of splicing isoforms is very limited. There are currently <10 spliced isoforms with documented structures in the Protein Data Bank (PDB) (January 2005). This clearly represents a major knowledge gap as structures hold key information regarding how different isoforms with largely identical sequences perform different functions. Modeling structures for splicing isoforms provide a unique opportunity for threading-based computational prediction methods because isoforms share significant portions of their sequences. In this work, we have carried out a large-scale analysis of alternative splicing at the protein sequence level and in the context of protein 3D structures. This analysis has shown that splicing isoforms are dominated by small-sized alternations and there are structural constraints that limit the locations within a protein fold that splicing events could occur. We established threading as a viable method for structure prediction of splicing isoforms.
Materials and Methods
Alternative Splicing Data Set. The alternatively spliced protein data set used throughout this work was obtained from SWISS-PROT protein database release 42. The three types of splicing events (deletion, insertion, and substitution) are annotated in SWISS-PROT under keyword VARSPLIC. Information about splicing events was extracted from SWISS-PROT by using the Sequence Retrieval System, which is a tool in the SWISS-PROT package for querying databases. The sequences of full-length and alternatively spliced proteins were extracted from SWISS-PROT by using perl scripts developed in our laboratory.
Mapping Human Isoforms to the Human Genome. The human May 2004 (hg17) assembly was downloaded from the University of California at Santa Cruz genome bioinformatics web site (http://hgdownload.cse.ucsc.edu/downloads.html). The human splicing isoform sequences were mapped to the human genome by using a copy of locally installed blat program.
Structures of Full-Length Proteins and the Method for Structure Prediction. The 3D structures of full-length proteins were obtained from two sources. We first obtained a nonredundant set of full-length proteins that have documented splicing isoforms in SWISS-PROT. We then examined the intersection of the full-length protein data set and the PDB. This query returned PDB structures for 351 full-length proteins. We then carried out structure prediction for the rest of the full-length proteins in our data set by using the threading program prospect (23). The threading results were first filtered to only include structures with a threading z score ≥ 20, which indicates the probabilities of having correct fold prediction and correct secondary structure prediction are extremely high (P << 10–8) based on the threading alignments by prospect. The atomic-level model structures were then generated by using modeller (version 6.0) (24). The modeled structures were further examined by using procheck (25), and only structures with at least 80% residues in core regions were retained, which resulted in 858 structural models. The combined PDB hits and threading structures yielded a data set of 1,209 unique proteins that were used in structural genomics analysis. The data set of protein structures used in our analysis is available from the authors.
Secondary Structure, Solvent Accessibility, and Mapping Splicing Events to Structures. The secondary structures and solvent accessibilities of residues were assigned by using the program dssp (26). Residues with <6.9% water-accessible surface area are considered to be buried, residues with >36% water-accessible surface area are considered to be exposed, and the rest residues are considered to be half-buried. perl scripts developed in our laboratory were used to map the boundaries of splicing events to the 3D structure of full-length protein, and the secondary structure and solvent accessibility information of the two residues at the end of splicing events was extracted for statistical analysis. The solvent accessibility information was also used to determine whether a fragment of protein was located on the surface or in the interior of a protein structure. If the fragment has four or more consecutive residues that are buried, those buried residues are regarded as in the interior of proteins.
Modeling Structures for Splicing Isoforms. The structures of splicing isoforms were modeled with the same procedure as full-length proteins. We chose to use the default prospect template library (nonredundant PDB structures), because there are examples of splicing isoforms in our analysis that could have significantly different structures from their full-length counterparts caused by large splicing alternations. High-quality isoform structure models generated in this work are available from the authors.
Core Secondary Structure Elements. A helix or sheet is considered to be in the core secondary structure elements if it satisfies the following criteria: (i) an α-helix has a minimum length of four or a β-strand has a minimum length of three; and (ii) a core secondary element should have residual contacts with other structural cores. Cores were identified by using a perl script developed in-house.
Molecular Dynamics Simulation. NPT (constant mole number, pressure, and temperature) molecular dynamics simulation was performed with namd (27) by using the charmm22 force field (28) with an explicit water model. The time step was 1 fs, and trajectory was saved every 1 ps. The particle mesh Ewald method (29) was used to treat long-range electrostatic interactions. Constant temperature was controlled by Langevin dynamics, and pressure was maintained by using Nosé-Hoover Langevin piston pressure control.
Results
The Length of Splicing Events Follows Power Law. An analysis of the SWISS-PROT database revealed that there are 4,399 proteins with documented splicing isoforms. The majority of those proteins are from well studied organisms such as human (2,412 proteins), mouse (1,101 proteins), and rat (358 proteins). There are total of 8,220 isoforms averaging about two isoforms per gene. An examination of the length of the alternative splicing events revealed an interesting feature. The size (length) of alternative splicing events versus the number of events with such size is plotted in Fig. 1a. When the size of alternative splicing events is small (<20 aa), the number of such splicing events seems to follow a uniform distribution. For splicing events >20 aa, it appears to follow the power law distribution in that splicing events become rare as their sizes increase. This observation is in concert with the report that the size of constitutive exons follows a normal distribution, whereas the distribution of alternatively spliced exons is skewed toward smaller ones (30).
Fig. 1.
Distribution of splicing events. (a) The size of splicing events follows the power law distribution. The size of alternative splicing events and the number of corresponding events are plotted on a log vs. log graph. The majority of alternative splicing events are of small size, and events become rare as size increases. For events with size >20, they appear to follow the power law distribution (power law factor –1.07, linear regression adjusted R2 0.75). (b) The number of isoforms decreases exponentially as the number of splicing events per isoform increases. The log of the number of splicing isoforms has a strong linear relationship with the number of splicing events per isoform with adjusted R2 0.93.
The Majority of Splicing Isoforms Have Only a Single Splicing Alternation. We next examined the distribution of splicing events on splicing isoforms and the patterns by which splicing events were organized in isoforms. The three types of splicing events are not equally abundant. Deletion is the most abundant one that accounts for 57% of all splicing events. Substitution comes in second at 38%, whereas insertion only accounts for 5% of all splicing events. The number of splicing isoforms was plotted against the number of splicing events they harbor in Fig. 1b. The log of the number of splicing isoforms has a strong linear relationship with the number of splicing events per isoform with a linear regression adjusted R2 of 0.93. This linear relationship indicates that the number of splicing isoforms decreases exponentially as the number of splicing events per isoform increases and the isoforms that harbor one or two splicing events make up 88% of all splicing isoforms. We then examined how the three splicing events were organized in each splicing isoform. The ways by which splicing events were organized were classified into different patterns (see Table 3, which is published as supporting information on the PNAS web site). All patterns that are adopted by >100 splicing isoforms harbor only one or two splicing events, and patterns with four or more splicing events are very rare (all adopted by 10 or fewer isoforms). The alternations are strongly biased toward terminal regions as 57% of all isoforms have splicing alternations at N or C terminals. An interesting combination of splicing events observed is “SDC” where a substitution was followed by a deletion to the C-terminal of proteins. This combination appears in 22 patterns and was adopted by 1,659 splicing isoforms. We mapped 815 human isoforms with the SDC pattern to human genome and found that ≈12% of them introduce frameshifts in exons and the remaining isoforms involve intron retention.
Our analysis of the length, distribution, and pattern of splicing events indicates that the majority of splicing isoforms harbor only one or two splicing events whose length is small (60% splicing events are <50 aa). Thus it appears the effects of splicing on isoform structures are most likely to be local: splicing may only influence the structural domain that harbors that event, which may be well accommodated without drastic change to its original structural fold.
Boundaries of Splicing Events Strongly Prefer Coil Regions and Exposed Residues. Current protein evolution theory suggests that introns can separate complete functional domains (31), and previous studies have suggested that exon-intron boundaries tend to be located in coil regions (32). The boundaries of splicing events will only include exon-intron boundaries adjacent to alternative exons and with additional boundaries created by using noncanonical splicing sites. It is of great interest to examine structural locations of the boundaries of splicing events, because those sites might hold useful information about locations chosen by nature to alter existing proteins to create new proteins with stable structures and different functions. We first collected high-quality structures for 1,209 proteins that have documented splicing isoforms in the SWISS-PROT database (see Materials and Methods for details). There are 286,624 aa in the data set of full-length protein structure with 42% in coil regions, 41% in helices, and 17% in extended strands. We then mapped boundaries of alternative splicing events onto the available structures and examined their distributions in terms of secondary structure types and regions with different levels of solvent accessibilities. In terms of secondary structure types (Table 1), it immediately becomes evident that the ends of deletions have a strong tendency to avoid helices and prefer loop regions. Deletions located in helices are found significantly less often than expected, whereas deletions in loop regions are found significantly more often than expected. The distribution of the ends of deletions suggests that deletions in alternatively spliced isoforms tend to remove entire secondary structure units, and a quick examination shows that among all deletions whose two ends are mapped to known structures ≈40% have entire secondary structure units removed. Substitutions, on the other hand, do not display such preferences. Although the numbers of insertions in helices and sheets are lower than expected, the overall number of insertions that can be mapped to structures is too small to draw any reliable conclusion. In terms of preference to the level of solvent accessibilities by splicing events (Table 2), we have observed that ends of deletions have a strong preference for exposed residues (i.e., tend to avoid residues that are buried) and substitutions also display a significant preference toward exposed residues but it is not as strong as deletions. As for insertions, our sample size is too small to draw any reliable conclusion in regard to preferences to any particular solvent accessibility. The overall distribution of splicing events in the context of protein 3D structure suggests that deletion has a rather drastic destabilizing effect on protein fold and there is significant evolutionary pressure to restrict the structural environment under which a deletion could happen. Substitution, on the other hand, seems to be a milder alternation and could be well accommodated. Although no statistical significant conclusion can be drawn, our data suggest that insertion could be a factor of destabilization and its location in 3D structure is preferably in loop regions and exposed areas.
Table 1. Observed and expected frequencies of secondary structure elements to which the boundaries of alternative splicing events are mapped.
| Splicing event | dssp secondary structure | Observed (expected) | χ2 |
|---|---|---|---|
| Insertion | C | 36 (29) | 1.68 |
| H | 17 (28) | 4.57 | |
| E | 16 (12) | 1.68 | |
| Deletion | C | 537 (372) | 73.43 |
| H | 235 (364) | 45.55 | |
| E | 112 (148) | 8.98 | |
| Substitution | C | 323 (314) | 0.22 |
| H | 322 (308) | 0.66 | |
| E | 103 (126) | 4.09 |
Coil, extended strand, and helix are identified by the letters C, E, and H, respectively. Statistical significances of observed differences were calculated by using χ2 statistics with four degrees of freedom. Statistically significant differences are highlighted in bold (P < 0.001).
Table 2. Observed and expected frequencies of residues of different solvent accessibilities (exposed, half-buried, and buried) that are mapped to the boundaries of alternative splicing events.
| Splicing event | Solvent accessibility | Observed (expected) | χ2 |
|---|---|---|---|
| Insertion | Exposed | 38 (28) | 3.79 |
| Half-buried | 23 (23) | 0.00 | |
| Buried | 8 (18) | 5.69 | |
| Deletion | Exposed | 471 (355) | 37.53 |
| Half-buried | 258 (296) | 4.82 | |
| Buried | 155 (233) | 25.97 | |
| Substitution | Exposed | 355 (301) | 9.76 |
| Half-buried | 253 (250) | 0.03 | |
| Buried | 140 (197) | 16.46 |
Statistical significances of observed differences were calculated with χ2 distribution with four degrees of freedom. Statistically significant differences are highlighted in bold (P < 0.001).
The Majority of Alternative Splicing Alternations Happen on Protein Surface. We have examined the structural locations of the entire sequences involved in splicing (Fig. 2). A fragment of protein was considered to be in the interior of a protein if it has four or more consecutive amino acids that are buried. For the insertion events that can be mapped to protein structures, all of them happen in regions located on protein surface. For substitutions and deletions, small-sized events are predominantly located in surface regions. As the size increases, splicing events start to involve regions that are in the interior of proteins. However, events covering regions both on the surface and in the interior of protein are only a small portion of all events (25%), whereas the majority of splicing events (75%) involve only regions on protein surface and there is no event that involves only regions in the interior of protein. Because residues buried inside a protein are responsible mainly for folding, whereas surface residues contribute to functionalities of proteins, this observation may suggest that splicing largely may not alter the structural fold of the full-length proteins and splicing isoforms achieve diversity by altering surface residues involved in protein functions.
Fig. 2.
Distribution of protein fragments involved in splicing on the surface and in the interior of protein structures. The fragments were partitioned according to their sizes. The filled bars show the frequencies of fragments mapped to protein surfaces only, and the empty bars show the frequencies of fragments mapped to both surface and interior. (a) Substitution events. (b) Deletions.
High-Quality Structures Can Be Modeled for the Majority of Alternative Splicing Isoforms. The results presented so far suggest that structures of the majority of isoforms may be readily obtainable because their splicing alternations are of small size, are located on protein surface, and prefer to end in coil regions. Encouraged by the above analysis results, we carried out a genome-scale structure prediction for alternatively spliced isoforms by using a threading approach. We chose to model the structures for 708 splicing isoforms whose alternations could be mapped to known structures. Modeling structures for other isoforms was not done because their regions with splicing alternations could not be modeled because of the lack of templates as indicated by the results of our efforts to model structures for full-length proteins. Because the structures of splicing isoforms would have various degrees of alternations, we chose to carry out threading against all nonredundant PDB structures to capture potential changes of isoform structures. The majority of the isoform structures (65.7%; 464 of 708) are of superior quality as they have threading z score ≥ 20 by prospect and have at least 80% residues in the core region as determined by procheck (see Fig. 5, which is published as supporting information on the PNAS web site). The number of isoforms with reliably modeled structures is actually >464 because our past threading experiences indicate that z score 8 is a good cutoff. The stricter cutoff was used in this study because it can retain a sufficiently large set of structures and introduces no or little errors associated with low-quality structures. Using a lower z-score cutoff (such as 8) and the same procheck cutoff leads to a higher number of predicted structures. However, we found that using a lower cutoff makes our discussion in the following more complex because of the possible inclusion of false predictions. For practical applications, we believe that using a lower cutoff might be desirable.
Splicing Isoforms Share the Same Structural Folds as Their Full-Length Counterparts. With the large data set of high-quality isoform structures at hand, we set out to determine the general structural principle that underlies the functional variations of splicing isoforms. The key question to ask is whether alternations of splicing isoforms change the fold adopted by isoforms. To determine whether an isoform shares the same structural fold as its full-length counterpart, we compared the scop classification of the structure template used to generate the isoform structure with the scop classification of the template used to generate the structure of the corresponding full-length protein (or the scop classification of a full-length protein's PDB structure if there is any). For the 464 isoforms with high-quality structures, 344 (74%) use the same template as their full-length counterparts, 116 (25%) use a template in the same scop family, 2 (0.4%) use a template in the same SCOP superfamily, and 2 pick the template in the same fold (see Fig. 6, which is published as supporting information on the PNAS web site). All of the checked isoforms maintain the same fold, and the majority of them have very similar structures to those of the full-length proteins because their threading templates are the same. In our data set, there are 244 isoforms whose modeled structures are not of superior quality and are not included in our scop analysis. Thus there may be some splicing isoforms that adopt different structural folds.
The Quality of Isoform Structures Is Inversely Correlated with the Percentage of Core Secondary Structure Elements Altered. Our analysis of splicing events indicates that there could be some splicing isoforms that have potential fold destabilizing alternations such as removal of long sequences containing multiple helices and sheets or removal of fully buried fragments. Thus it is important to determine whether such isoforms exist and if so what the qualities and fold stabilities of their modeled structures are. We started by adopting the notion of core secondary structure elements (CSSEs) that denotes the regions of a protein structure important for folding (33). We calculated the CSSEs of the structures of full-length proteins. Then for each splicing isoform with a modeled structure, we mapped the splicing events harbored by the isoform to the CSSEs of corresponding full-length protein and calculated the percentage of CSSEs altered by splicing. We then investigated the relationship of structure quality and percentage of CSSEs altered. It appears that isoform structure quality is inversely correlated with the percentage of CSSEs altered by it. For splicing isoforms with single substitution, CSSE and z score have a correlation coefficient of –0.39 (Fig. 3a). Isoforms with a single deletion have a stronger correlation coefficient of –0.55 (Fig. 3b), and isoforms with multiple alternations have a correlation coefficient –0.61 (Fig. 3c). Overall the quality of modeled structures deteriorates as the percentage of CSSEs altered increases. Lower-quality isoform structures (with a threading z score <20 and residues in core region <80%) are dominated by isoforms with a large alternation of CSSEs as 60% of these isoforms alter >20% of CSSEs. Most of the high-quality isoform models have only small CSSE alternation. For the 464 high-quality isoform structures, 278 of 464 (60%) isoforms alter <10% of CSSEs and only 120 of 464 (25%) isoforms alter >20% of CSSEs.
Fig. 3.
The threading z score of splicing isoforms was inversely correlated with the percentage of the core secondary structure elements altered. Splicing isoforms were partitioned into three types: isoforms with single substitution (a), isoforms with single deletion (b), and isoforms with multiple alternations (c). The threading z score of splicing isoforms was then plotted against the percentage of CSSEs altered by the isoforms.
Molecular Dynamics Simulations Demonstrate the Fold Stability of Modeled Isoform Structures. For the majority of the isoform models, threading z score and procheck results are good indications of their structure qualities because they have small or no CSSE alternation. However, for isoforms with substantial alternation of CSSEs, such static measurements may not fully reflect the quality of modeled structures because the alternation they harbored could potentially destabilize the fold. We set out to determine whether modeled structures of isoforms with substantial CSSE alternation are stable by using molecular dynamics simulation. Because molecular dynamics is a rather time-consuming approach, we chose to illustrate the fold stability of isoform structures by using two examples with increased level of CSSE alternations and different splicing alternations.
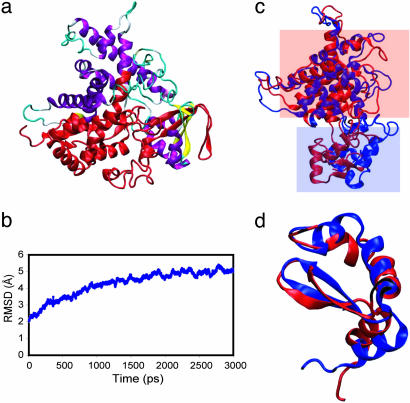
The first example is O64636-2, which is the isoform 2 of the Arabidopsis protein cytochrome P450 76C1. O64636-2 is a protein with 322 aa, which harbors a 16-aa substitution and a 190-aa deletion that alters 43% of the O64636-1 CSSE (Fig. 4a). Despite the large splicing alternation, the modeled O64636-2 structure is of high quality with a threading z score 52.23 and has 84.2% residues in core regions according to procheck. The molecular dynamics simulation indicated that the structure of O64636-2 stabilized 2 ns into simulation, and the rms deviation in the last 1 ns simulation has a mean of 4.96 Å and a SD of 0.157 Å (Fig. 4b). The structure of O64636-2 at 0 ns of simulation was aligned with that of 3 ns of simulation (Fig. 4c). The major dynamics of O64636-2 structure is the movement of the structural unit consisting of three helices and two strands (enclosed in the blue box in Fig. 4c) toward the structure unit enclosed in the red box in Fig. 4c. At the end of 3-ns simulation, the backbone of the structure unit enclosed in the blue box has moved 11.3 Å on average. Such movement closed the gaps in modeled structures created by splicing alternation without significant change of secondary structures and their packing because the structural alignment showed both the structure unit in the red box (Fig. 4c) and the structure unit in the blue box (Fig. 4d) are largely unchanged. This simulation has clearly demonstrated the fold stability of the modeled isoform structure even in the presence of large splicing alternations. It also shows that for some modeled structures it is necessary to perform further energy minimization to refine the structures. However, even with large movements introduced by energy minimization, the structure remains stable and fold is still the same. An additional simulation was performed with splicing isoform Q9JLT4-4 and it also clearly demonstrated the fold stability of modeled isoform structures (see Supporting Text and Fig. 7, which are published as supporting information on the PNAS web site).
Fig. 4.
Molecular dynamics simulation of O64636-2. (a) The structure of O64636-1 is shown in with the region substituted or deleted colored in red. (b) The backbone rms deviation (RMSD) of O64636-2 from initial structure as a function of time. (c) The alignment of O64636-2 structures at 0 ns (blue) and 3 ns (red) of simulation. The transformation matrix used in alignment was calculated by using only helices highlighted in the red box in the O64636-2 structure to demonstrate the movement of the O64636-2 structure highlighted in the blue box. (d) The three helices and two strands highlighted in blue box are aligned. The structure at 0-ns simulation is colored blue, and the structure at 3-ns simulation is colored red.
Discussion
Alternative splicing has emerged as the central paradigm for enhancing proteome diversity. As >50% of human genes undergo alternative splicing and are often with multiple isoforms, the majority of human proteomes are actually alternatively spliced protein products. Given the extremely limited availability of splicing isoform structures in the PDB, we urgently need effective methods to model the structures of splicing isoforms. Here, we have provided a useful solution by demonstrating the viability of a threading method to model the structures for splicing isoforms. For the 708 isoforms included in our modeling efforts, high-quality structures for 464 isoforms are obtained as they are of high threading z scores, and excellent procheck results (structures for some of remaining isoforms can be obtained if we use a lower threading cutoff). The fold stability of models was further confirmed with molecular dynamics simulations. Establishment of threading as a useful method that can generate high-quality structure models for large numbers of splicing isoforms is of immense value. This result will significantly expand our knowledge of the structures and functions of alternative splicing isoforms and help to answer many important questions in the development and pathogenesis of many diseases where splicing was involved.
An important observation of modeled isoform structures is that they share the same structural folds as their full-length counterparts. Thus it appears that splicing isoforms have adopted the “Occam's razor” principle, also known as the principle of parsimony in that the simplest model from a set of otherwise equivalent models of a given phenomenon should be adopted, in achieving different functions: instead of adopting a completely different fold, the same fold was used with alternations mostly of small size and on protein surface. This observation implies that the functions of alternatively spliced isoforms can be effectively accessed by an homology-based method like multiple sequence alignment without worrying about isoforms adopting a completely different structural fold. The tendency of isoforms to adopt the same folds of their full-length counterparts can also be used to greatly improve the speed of isoform structure modeling. In practice we can simply carry out threading only with templates from the same family or same superfamily of the full-length protein. This simplification could potentially reduce the time needed to run threading by hundreds of fold.
The fact that high-quality structures couldn't be generated for some of the isoforms raises two interesting questions. First, it points out the fundamental limitation of threading-based structure modeling methods, which is that low-quality models are expected if no suitable templates are available. For some splicing isoforms with large alternations, it is necessary to use experimental methods such as x-ray crystallography or NMR or wait for the accumulation of new fold templates to predict their structures. Another interesting possibility is that structures of those isoforms may not need to be modeled at all. About one-third of alternative spliced mRNAs are subjected to nonsense-mediated decay (NMD) and no protein products exist (34). It is possible that isoforms subjected to NMD are associated with bad structures because they have no pressure to harbor splicing events resulting in stable folds.
From a protein engineering point of view, each alternative splicing isoform can be viewed as an engineered protein product by altering an existing protein with insertion, deletion, or substitution. Thus the entire set of alternative splicing events can be viewed as nature's toolbox for protein engineering, and valuable insights can be learned by examining nature's choices. Here, we have performed a preliminary analysis of available splicing events. From our analysis, it is apparent that the boundaries of splicing events have significant differences in their preferences for structural regions. As we have seen, boundaries of deletions have strong preferences for loop regions in secondary structure and exposed regions, whereas boundaries of substitutions prefer exposed regions but do not display preference for any particular secondary structure elements. These observations suggest that deletions are potential fold-destabilizing events, and there are evolutionary pressures to select deletions in splicing isoforms that remove entire domains or entire secondary structural units. Substitutions, on the other hand, are not restricted by secondary structures. These observations could serve as valuable guides in protein design where proteins with altered functions could possibly be generated through careful selections of substitutions and deletions.
Supplementary Material
Acknowledgments
This research was supported in part by a Distinguished Cancer Scholar grant from the Georgia Cancer Coalition, National Science Foundation Grants NSF/DBI-0354771 and NSF/ITR-IIS-0407204, and the U.S. Department of Energy's Genomes to Life program under project “Carbon Sequestration in Synechococcus sp.: From Molecular Machines to Hierarchical Modeling.”
Author contributions: P.W. and Y.X. designed research; P.W., B.Y., and J.-t.G. performed research; P.W., B.Y., J.-t.G., and C.H. analyzed data; and P.W. and Y.X. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PDB, Protein Data Bank; CSSE, core secondary structure element.
References
- 1.Black, D. L. (2003) Annu. Rev. Biochem. 72, 291–336. [DOI] [PubMed] [Google Scholar]
- 2.Brett, D., Hanke, J., Lehmann, G., Haase, S., Delbruck, S., Krueger, S., Reich, J. & Bork, P. (2000) FEBS Lett. 474, 83–86. [DOI] [PubMed] [Google Scholar]
- 3.Johnson, J. M., Castle, J., Garrett-Engele, P., Kan, Z., Loerch, P. M., Armour, C. D., Santos, R., Schadt, E. E., Stoughton, R. & Shoemaker, D. D. (2003) Science 302, 2141–2144. [DOI] [PubMed] [Google Scholar]
- 4.Xu, X., Yang, D., Ding, J. H., Wang, W., Chu, P. H., Dalton, N. D., Wang, H. Y., Bermingham, J. R., Jr., Ye, Z., Liu, F., et al. (2005) Cell 120, 59–72. [DOI] [PubMed] [Google Scholar]
- 5.Johnson, C. R. & Jarvis, W. D. (2004) Apoptosis 9, 423–427. [DOI] [PubMed] [Google Scholar]
- 6.Xerri, L., Hassoun, J., Devilard, E., Birnbaum, D. & Birg, F. (1998) Leuk. Lymphoma 28, 451–458. [DOI] [PubMed] [Google Scholar]
- 7.Bartel, F., Taubert, H. & Harris, L. C. (2002) Cancer Cell 2, 9–15. [DOI] [PubMed] [Google Scholar]
- 8.Minneman, K. P. (2001) Mol. Interv. 1, 108–116. [PubMed] [Google Scholar]
- 9.Thai, T. H. & Kearney, J. F. (2004) J. Immunol. 173, 4009–4019. [DOI] [PubMed] [Google Scholar]
- 10.Scheper, W., Zwart, R. & Baas, F. (2004) Neurogenetics 5, 223–227. [DOI] [PubMed] [Google Scholar]
- 11.Stenson, P. D., Ball, E. V., Mort, M., Phillips, A. D., Shiel, J. A., Thomas, N. S., Abeysinghe, S., Krawczak, M. & Cooper, D. N. (2003) Hum. Mutat. 21, 577–581. [DOI] [PubMed] [Google Scholar]
- 12.Roberts, R., Timchenko, N. A., Miller, J. W., Reddy, S., Caskey, C. T., Swanson, M. S. & Timchenko, L. T. (1997) Proc. Natl. Acad. Sci. USA 94, 13221–13226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma, K., Inglis, J. D., Sharkey, A., Bickmore, W. A., Hill, R. E., Prosser, E. J., Speed, R. M., Thomson, E. J., Jobling, M., Taylor, K., et al. (1993) Cell 75, 1287–1295. [DOI] [PubMed] [Google Scholar]
- 14.Venables, J. P., Elliott, D. J., Makarova, O. V., Makarov, E. M., Cooke, H. J. & Eperon, I. C. (2000) Hum. Mol. Genet. 9, 685–694. [DOI] [PubMed] [Google Scholar]
- 15.Lovestone, S., Reynolds, C. H., Latimer, D., Davis, D. R., Anderton, B. H., Gallo, J. M., Hanger, D., Mulot, S., Marquardt, B., Stabel, S., et al. (1994) Curr. Biol. 4, 1077–1086. [DOI] [PubMed] [Google Scholar]
- 16.Hernandez, F., Perez, M., Lucas, J. J., Mata, A. M., Bhat, R. & Avila, J. (2004) J. Biol. Chem. 279, 3801–3806. [DOI] [PubMed] [Google Scholar]
- 17.Manabe, T., Katayama, T., Sato, N., Gomi, F., Hitomi, J., Yanagita, T., Kudo, T., Honda, A., Mori, Y., Matsuzaki, S., et al. (2003) Cell Death Differ. 10, 698–708. [DOI] [PubMed] [Google Scholar]
- 18.Venables, J. P. (2004) Cancer Res. 64, 7647–7654. [DOI] [PubMed] [Google Scholar]
- 19.Holmila, R., Fouquet, C., Cadranel, J., Zalcman, G. & Soussi, T. (2003) Hum. Mutat. 21, 101–102. [DOI] [PubMed] [Google Scholar]
- 20.Courtois, S., de Fromentel, C. C. & Hainaut, P. (2004) Oncogene 23, 631–638. [DOI] [PubMed] [Google Scholar]
- 21.Thanaraj, T. A., Stamm, S., Clark, F., Riethoven, J. J., Le Texier, V. & Muilu, J. (2004) Nucleic Acids Res. 32, D64–D69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clark, F. & Thanaraj, T. A. (2002) Hum. Mol. Genet. 11, 451–464. [DOI] [PubMed] [Google Scholar]
- 23.Xu, Y. & Xu, D. (2000) Proteins 40, 343–354. [PubMed] [Google Scholar]
- 24.Sali, A. & Blundell, T. L. (1993) J. Mol. Biol. 234, 779–815. [DOI] [PubMed] [Google Scholar]
- 25.Laskowski, R. A., Rullmannn, J. A., MacArthur, M. W., Kaptein, R. & Thornton, J. M. (1996) J. Biomol. NMR 8, 477–486. [DOI] [PubMed] [Google Scholar]
- 26.Kabsch, W. & Sander, C. (1983) Biopolymers 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- 27.Kalé, L., Skeel, R., Bhandarkar, M., Brunner, R., Gursoy, A., Krawetz, N., Phillips, J., Shinozaki, A., Varadarajan, K. & Schulten, K. (1999) J. Comput. Phys. 151, 283–312. [Google Scholar]
- 28.Brooks, B. R., Bruccoleri, R. E., Olafson, B. D., States, D. J., Swaminathan, S. & Karplus, M. (1983) J. Comp. Chem. 4, 187–217. [Google Scholar]
- 29.Essmann, U., Perera, L., Berkowitz, M. L., Darden, T., Lee, H. & Pedersen, L. G. (1995) J. Chem. Phys. B. 101, 8577–8593. [Google Scholar]
- 30.Stamm, S., Zhu, J., Nakai, K., Stoilov, P., Stoss, O. & Zhang, M. Q. (2000) DNA Cell Biol. 19, 739–756. [DOI] [PubMed] [Google Scholar]
- 31.Chothia, C., Gough, J., Vogel, C. & Teichmann, S. A. (2003) Science 300, 1701–1703. [DOI] [PubMed] [Google Scholar]
- 32.Contreras-Moreira, B., Jonsson, P. F. & Bates, P. A. (2003) J. Mol. Biol. 333, 1045–1059. [DOI] [PubMed] [Google Scholar]
- 33.Xu, Y., Xu, D. & Uberbacher, E. C. (1998) J. Comput. Biol. 5, 597–614. [DOI] [PubMed] [Google Scholar]
- 34.Lewis, B. P., Green, R. E. & Brenner, S. E. (2003) Proc. Natl. Acad. Sci. USA 100, 189–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.