Abstract
The T cell receptor (TCR) αβ heterodimer determines the peptide and MHC specificity of a T cell. It has been proposed that in vivo selection processes maintain low TCR affinities because T cells with higher-affinity TCRs would (i) have reduced functional capacity or (ii) cross-react with self-peptides resulting in clonal deletion. We used the class II-restricted T cell clone 3.L2, specific for murine hemoglobin (Hb/I-Ek), to explore these possibilities by engineering higher-affinity TCR mutants. A 3.L2 single-chain TCR (Vβ-linker-Vα) was mutagenized and selected for thermal stability and surface expression in a yeast display system. Stabilized mutants were used to generate a library with CDR3 mutations that were selected with Hb/I-Ek to isolate a panel of affinity mutants with KD values as low as 25 nM. Kinetic analysis of soluble single-chain TCRs showed that increased affinities were the result of both faster on-rates and slower off-rates. T cells transfected with the mutant TCRs and wild-type TCR responded to similar concentrations of peptide, indicating that the increased affinity was not detrimental to T cell activation. T cell transfectants maintained exquisite hemoglobin peptide specificity, but an altered peptide ligand that acted as an antagonist for the wild-type TCR was converted to a strong agonist with higher-affinity TCRs. These results show that T cells with high-affinity class II reactive TCRs are functional, but there is an affinity threshold above which an increase in affinity does not result in significant enhancement of T cell activation.
Keywords: antigen specificity, major histocompatibility complex, T cell activity
T cell activation depends upon interactions between the T cell receptor (TCR) and an antigenic peptide bound to a product of the major histocompatibility complex (pMHC). Developing T cells undergo selection in the thymus in which survival depends on a minimal affinity of TCR for self-pMHC (positive selection) (1). If the affinity of the TCR is too high for the self-pMHC, the T cell will be deleted (negative selection). These selection processes yield a population of T cells that are not self-reactive but that have the potential to recognize foreign peptides bound to an MHC product. The affinities of TCRs from peripheral T cells for foreign pMHC have been shown to be restricted to a relatively narrow range of KD values of ≈5-90 μM or half-lives of ≈3-30 s (2-4). The affinities of TCRs associated with alloreactions, such as the 2C/Ld system, may be higher than those with self-MHC (5), but a comprehensive comparative study has not been performed.
Although a controversy over whether affinity or half-life of the TCR:pMHC interaction correlates with T cell activity remains unresolved (2), it is clear that one of the initial consequences of pMHC binding by a TCR is the ordered phosphorylation of immunoreceptor tyrosine-based activation motifs on the ζ chain (6). Several models have been proposed to explain how T cells initiate these signals. The kinetic proofreading model proposes that a TCR must bind to pMHC sufficiently long for subsequent signaling steps to occur. This model implies that a TCR:pMHC interaction with a short half-life would not function as efficiently as one with a longer half-life (7). An extension of this model, the serial triggering hypothesis, holds that the half-life of the TCR bound to pMHC must be sufficiently short to allow a single pMHC complex to engage multiple TCRs in succession (8). This hypothesis led to the prediction that there would be an optimal dwell time of a TCR:pMHC interaction and longer half-lives would be detrimental to T cell activation and function (9). Studies with mutants of the class I-restricted TCR from T cell 2C did not provide evidence of an optimal dwell time (10, 11). More recently, a T cell activation model has been proposed where the basic unit of signaling is a TCR pseudodimer comprised of one TCR bound to an agonist pMHC and a second TCR bound to a self-pMHC after recruitment by CD4 (12).
Because higher-affinity TCRs are not present in the mature T cell repertoire, the functional activity and peptide specificity of T cells with high-affinity TCRs remain largely unexplored. To examine these issues, we have engineered here the mouse T helper clone 3.L2. Clone 3.L2 recognizes a peptide (GKKVITAFNEGLK) from the minor d allele of the β chain of mouse hemoglobin bound to I-Ek (Hb/I-Ek) (13). The wild-type (WT) 3.L2 TCR binds to Hb/I-Ek with a KD value of 12 μM (14). Single amino acid variants of Hb (altered peptide ligands; APLs) have been shown to elicit dramatically different responses in 3.L2 T cell function (14, 15). For example, the conserved peptide variant D73 (Glu-73 → Asp) acted as an antagonist (13, 16). The structure of Hb/I-Ek showed that Glu-73 is a MHC anchor residue and that the absence of the methyl group in D73/I-Ek resulted in 0.5- to 2.0-Å changes in several TCR-accessible peptide residues (17).
In this study we have generated and characterized the binding and functional properties of high-affinity class II-restricted TCRs engineered in vitro, thereby providing a unique perspective on antigen specificity and the fundamental properties of T cell recognition and function. We used yeast display (18) to engineer a panel of 3.L2 class II restricted TCRs with higher affinities. Engineering of these mutants enabled expression of the stabilized proteins as single-chain TCRs (scTCRs) (Vβ-linker-Vα) in Escherichia coli. Surface plasmon resonance (SPR) studies of the refolded scTCRs showed that the mutations in three complementarity-determining regions (CDRs) contributed additively to an 800-fold increase in affinity compared with the WT 3.L2 TCR. The majority of the increased binding free energy was due to a faster on-rate (≈50-fold), although the highest-affinity mutant also exhibited a 15-fold slower off-rate (t1/2 = 104 s).
Functional studies of T cells transfected with the WT 3.L2 and high-affinity TCRs were performed to examine the role of affinity or kinetics in T cell activity. We found that T cells transfected with each of the higher-affinity TCRs were stimulated efficiently by the agonist peptide Hb, a finding that is contradictory to a continuing view that higher-affinity and/or longer TCR:pMHC half-lives result in impaired T cell activity (19). The Hb peptide dose-response profiles of T cells expressing the higher-affinity TCRs were similar to the WT 3.L2 TCR. This result indicates that T cells have a switch-like response with exquisite antigen sensitivity and a binding threshold, above which there is not a significant enhancement in activity. This finding thus establishes the binding and kinetic parameters that correspond to the threshold that was predicted to exist (20) and that has more recently been shown to operate in vivo for class II-restricted T cells (21). Despite the 800-fold increase in affinity and 15-fold increase in half-life, T cell transfectants retained remarkable antigen peptide specificity. Accordingly, T cells were not stimulated by the spectrum of self-peptides expressed by H-2k antigen-presenting cells (APCs) or by exogenous null peptide (from moth cytochrome c; MCC). However, the D73/I-Ek ligand was converted into a strong agonist for T cells that expressed each of the higher-affinity TCRs. The inability to detect T cells with high affinities in vivo may in part be a consequence of our finding that T cells exhibited minimal functional improvement and thus may not be preferentially expanded in vivo. This property may distinguish TCRs from antibodies, which have well-known advantages associated with high-affinity interactions (22). Alternatively, the absence of high-affinity T cells could be the result of a lower threshold associated with TCR binding affinities during negative selection in the thymus, compared with the stimulation of mature T cells.
Materials and Methods
Library Construction. A 3.L2 scTCR gene was constructed by using primers specific for the 3.L2 Vβ and Vα regions (National Center for Biotechnology Information accession nos. U46841 and U46581) and a 17-aa linker. The 3.L2-scTCR (Vβ-linker-Vα) was created by overlap extension PCR (SOE-PCR). The 783-bp construct consisted of residues 1-117 of the mature 3.L2β chain, the 17-aa linker (GSADDAKKDAAKKDGKS), residues 1-118 of the mature 3.L2α chain, and a 10-aa c-myc tag. The PCR product was ligated into the yeast display vector pCT302 as a NheI-BglII fragment. Selected plasmids were used as templates for error-prone PCR or site-directed mutagenesis of the CDR3α or CDR3β regions as described in ref. 23. Mutagenized PCR products were electroporated along with NheI-BglII digested vector into EBY100 yeast to generate libraries by homologous recombination. Stability and affinity libraries of ≈1.5 × 106 clones were generated.
Flow Cytometry and Library Sorting. Yeast libraries were cultured, and surface scTCR expression was induced as described in ref. 24. For selections, cells were incubated with 3.L2 clonotypic antibody (CAb) or Hb/I-Ek-Ig dimer, followed by staining with biotinylated goat anti-mouse IgG and streptavidin phycoerythrin. Fluorescent cells were selected through four rounds of growth and sorting by using a Cytomation MoFlo sorter. Hb/I-Ek-Ig dimers were produced and purified as described in ref. 25. I-Ek genes were cloned into the plasmids encoding genes for I-Adα-Fc and I-Ag7 β chains-Fc, a kind gift of J. Bluestone (University of California, San Francisco). Empty dimers were loaded with Hb peptide for 3 h at 37°C at pH 5.0, neutralized, and purified (25).
scTCR and Hb/I-Ek Protein Production. 3.L2 scTCR mutants were cloned into pET28a, and protein was produced in BL21 cells. Cells were passed through a microfluidizer and proteins were refolded from E. coli inclusion bodies as described in ref. 26. Proteins were purified with Ni agarose resin (Qiagen, Valencia, CA) followed by gel filtration (Superdex 200). Hb/I-Ek was refolded from E. coli inclusion bodies with Hb peptide (GKKVITAFNEGLK) following a published protocol (27).
SPR. 3.L2 scTCR binding to WT Hb/I-Ek was analyzed by SPR on a Biacore 3000. Activated CM5 chips were coupled with Neutravidin, and purified pMHC with in vitro-labeled biotin was immobilized to a total resonance [response units (RU)] value of 100-250. All measurements were baseline corrected by subtracting TCR injected over a blank surface, and were performed two to four times. Each scTCR (40 μl) was injected in PBS containing 0.005% surfactant P20 at a flow rate of 30 μl/min at 25°C, over various concentration ranges. Typically, six concentrations of protein for each scTCR were measured and data were fitted to a 1:1 Langmuir binding model (biaeval, Biacore). Equilibrium values were calculated by determining RU at equilibrium for each scTCR at various ligand concentrations, and those values were plotted vs. the RU divided by concentration (RU/C) (28).
T Cell Hybridoma Transfections and Assays. T cell hybridomas were generated by cotransfecting a 58α-β-CD4+ T cell hybridoma with plasmids pcDNAzeo and pcDNAneo containing the full-length 3.L2 TCRα and TCRβ genes, with CDR mutations (29). Cells were selected in zeocin and G418 and subcloned, and individual clones were tested for responses to Hb. Various peptide-reactive clones were further selected for TCR levels (Vβ8.3) similar to that of the previously generated 3.L2 hybridoma that expresses full-length 3.L2 TCRα and TCRβ chains (29). Several clones were tested, and identical results were observed. To assess peptide stimulation, 5 × 105 T cell hybridoma cells were cocultured with 3 × 105 CH27 and various doses of Hb peptide for 24 h. Supernatants were removed, and the level of IL-2 produced was quantified by a bioassay using the IL-2 reactive cell, CTLL. EC50 values were calculated from the concentration of Hb that stimulated 50% of the maximal response.
Results and Discussion
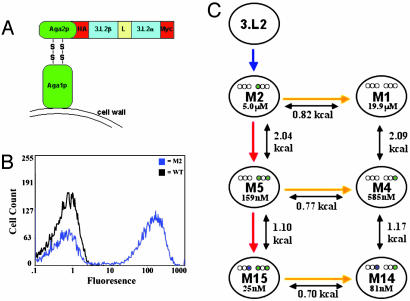
Yeast Surface Display of a Stabilized 3.L2 scTCR. To explore the issues relating to TCR affinity, function, and specificity, we used yeast display and in vitro evolution to generate a stabilized form of the 3.L2 scTCR and higher-affinity scTCR mutants. The Hb/I-Ek-specific 3.L2 scTCR (Vβ8.3-linker-Vα18) was cloned into the yeast display vector as a C-terminal fusion to the yeast surface protein Aga2 (Fig. 1A). To enhance surface display levels, four mutations (G17βE, H47βY, L81βS, I118αN) that were shown to stabilize the 2C scTCR on the surface of yeast were included in the 3.L2 scTCR gene (30-32). However, the resulting 3.L2 scTCR was not detected on the surface of yeast by flow cytometry with the 3.L2 clonotypic antibody (CAb) (Fig. 1B). To generate a stabilized form of the 3.L2 scTCR, the protein was evolved for yeast surface display by using random PCR mutagenesis followed by selection with CAb (Fig. 1C). This approach yielded two unique mutants (Clone 1 and Clone 5) that stained moderately positive for CAb (see Fig. 5A, which is published as supporting information on the PNAS web site). These clones contained two or four mutations in the variable (V) coding regions (T68βA, L78βV or E17βG, T87βS, K25αE, and T28αS) (see Fig. 6, which is published as supporting information on the PNAS web site).
Fig. 1.
Yeast display of 3.L2 scTCR and overview of engineering process. (A) Schematic of the 3.L2 scTCR (Vβ-linker-Vα), hemagluttinin (HA) tag, and c-myc tag expressed as an Aga-2 fusion protein on the surface of yeast. (B) Histogram overlay of yeast displayed scTCRs (WT and M2) stained with CAb. (C) Overview of 3.L2 engineering showing the selection process for each clone, SPR measured KD, free energy of binding differences (kcal/mol), and the location of CDR affinity mutations (purple or green circles represent mutations in respective CDR regions: Vβ1-2-3 and Vα1-2-3).
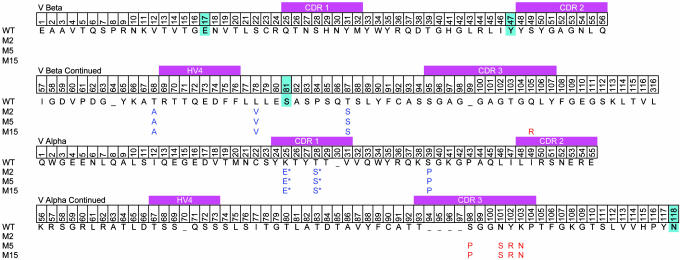
Previous studies with the 2C scTCR showed that selection of scTCR with increased resistance to thermal denaturation yielded mutants that had significantly higher levels of display on yeast and that were produced at higher levels in a secretion system (32). To generate such mutants for 3.L2, the two mutants from the first round of selection were used as templates for a second round of random mutagenesis, followed by thermal denaturation of the library and selection with CAb. After sorting the thermally denatured library, a number of clones with increased thermal stability and surface levels of CAb staining were isolated (Figs. 5B and 6; see also Fig. 7, which is published as supporting information on the PNAS web site). Clone M2 had the highest levels of CAb staining and was used as the template for affinity maturation (Fig. 1 B and C). Clone M2 contained a total of six mutations, four in framework regions (T68βA, L78βV, T87βS, and S39αP) and two in CDR1α (K25αE and T28αS) (Fig. 2).
Fig. 2.
Vα and Vβ sequences of 3.L2 TCR and mutants. TCR alignment shows 3.L2 WT amino acids and the mutations isolated during the selection for yeast surface display (blue) and affinity selection (red). The four 2C stabilizing mutations cloned into WT are highlighted (cyan).
Engineering a Panel of Higher-Affinity 3.L2 scTCR Mutants. Clone M2 served as the template for the construction of a library of mutants at five amino acid positions in the CDR3α region (99-103). The Hb/I-Ek-Ig dimer was used for sorting the CDR3α library, and after four rounds of selection an affinity mutant called M5 was isolated (Fig. 1C; three rounds were selected with 2 μM ligand and the fourth round with 400 nM ligand). Flow cytometry of mutant M5 with 2 μM Hb/I-Ek-Ig dimer showed a distinct shift in comparison with M2 (Fig. 3A). Titrations with the Hb/I-Ek-Ig dimer indicated that the avidity of M5 was at least 10-fold improved compared with M2 (see Fig. 8, which is published as supporting information on the PNAS web site). M5 contained four CDR3α mutations: S98αP, N101αS, Y102αR, and K103αN (Fig. 2). One of these mutations (S98αP) was in a codon that flanked the site-directed primer and presumably arose from a PCR error. Two of the WT residues within the mutated region, Gly-99 and Gly-100, were retained. We have observed a preference for retaining the WT Gly residues within the CDR3β region of the 2C TCR in all affinity-matured TCRs (23). Glycines appear to be important residues within CDR3 regions, and retention of the two glycines in M5 may indicate that CDR3α maintains a degree of flexibility required for Hb/I-Ek binding and specificity.
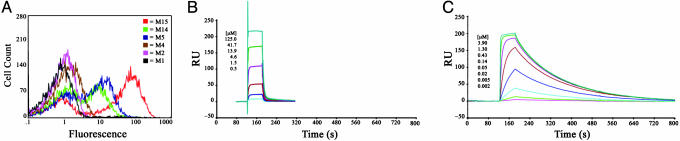
Fig. 3.
Hb/I-Ek binding by 3.L2 scTCR mutants. (A) Histograms of yeast displayed 3.L2 scTCR clones stained with 2μM Hb/I-Ek-Ig dimer. (B and C) Sample SPR traces of immobilized Hb/I-Ek binding by soluble scTCR 3.L2 mutants, M1 (B) and M15 (C), at the indicated concentrations.
To determine whether a TCR with higher affinity than M5 could be generated, two separate CDR3β libraries were constructed using M5 as a template (Fig. 1C). These libraries each spanned five residues in the CDR3β (96-101 and 102-106). The libraries were pooled and selected with lower concentrations of Hb/I-Ek-Ig dimer than was used for the first round of affinity selection (three rounds were selected with 4 nM ligand and the fourth round with 2 nM ligand). After the fourth round of sorting, clone M15 with enhanced staining compared with M5 was identified (Fig. 3A). Titrations with the Hb/I-Ek-Ig dimer indicated that the avidity of M15 was at least 25-fold improved compared with M2 (Fig. 8). M15 contained the same four mutations in the CDR3α as M5 and had only a single mutation in the CDR3β, Gln-105β-Arg (Fig. 2). Thus, the CDR3β retained each of the other WT residues, including five Gly residues.
Because both M5 and M15 were derived from the stabilized mutant M2, each of these mutants also contained the two stabilizing mutations that were present in CDR1α. To examine the possible effect of these mutations on binding, three additional mutants in which the CDR1α was reverted to WT were made (Fig. 1C). These mutants were called M1 (no CDR mutations), M4 (CDR3α mutations only), and M14 (CDR3α and CDR3β mutations). Flow cytometry with the Hb/I-Ek-Ig dimer showed that the CDR1α mutations contributed to the binding affinity and that the scTCR with CDR3α mutations alone or in combination with CDR3β were higher affinity than clone M1 with WT CDRs (Fig. 3A).
Binding Affinities and Kinetics of 3.L2 TCR Mutants. To determine the binding affinities and kinetics of 3.L2 mutants, scTCR genes were cloned and expressed in E. coli. Previous efforts to express and refold the 3.L2 WT scTCR in E. coli had been unsuccessful (D.L.D., unpublished data). Based on the observation that stabilizing mutations in the 2C scTCR resulted in higher levels of scTCR secretion from yeast and in greater solubility (31), we reasoned that the stabilized 3.L2 mutants (M1, M2, M4, M5, M14, and M15) might yield higher levels of refolded proteins compared with the 3.L2 WT scTCR. Consistent with this prediction, each of the scTCR proteins could be refolded from E. coli inclusion bodies and concentrated to >10 mg/ml.
Binding and kinetic measurements with each of the scTCR proteins were performed by using SPR (28). Biotinylated Hb/I-Ek was immobilized on a Neutravidin-coated chip, and the various scTCR mutants were analyzed at 25°C (Fig. 3 B and C; see also Fig. 9, which is published as supporting information on the PNAS web site; and data not shown). The equilibrium-binding and kinetic constants of the scTCR-Hb/I-Ek interactions are summarized in Table 1. KD values determined by equilibrium-binding titrations vs. kinetic analysis were very similar. Mutant M15 had a binding affinity of 25 nM, an 800-fold increase in affinity compared with the KD value (19.9 μM) of M1 that contains WT CDRs. This increase in affinity was achieved through a 50-fold faster on-rate and a 15-fold slower off-rate. The relative contributions of on-rate and off-rate to changes in binding affinity can be quantitatively assessed through the parameter φ, where an affinity change totally due to off-rate (no increase in on-rate) would yield a value of 0 and an affinity change solely due to on-rate would yield a value of 1 (28). For the M15 TCR, the φ value is 0.597, indicating that on-rate predominates in the M15 affinity increase. Furthermore, the similar affinities and kinetics of the bacterial M1 scTCR (KD = 19.9 μM; kon = 5,225 M-1·s-1) and the full-length, baculovirus 3.L2 TCR (KD = 12.0 μM; kon = 5,557 M-1·s-1) (14) indicates that the single-chain construct and mutations outside of the CDRs do not significantly affect pMHC binding.
Table 1. Kinetic and equilibrium binding properties of panel of 3.L2 mutants.
| Mutant | CDR mutations | kon, M–1·S–1 | koff, S–1 | Half-life, * S | KD (Koff/kon), nM | KD, equilibrium, nM | ΔΔGeq,† kcal/mol | Φ‡ |
|---|---|---|---|---|---|---|---|---|
| M1 | None | 5,220 ± 410 | 0.1023 ± 0.0107 | 6.8 ± 0.7 | 19,860 ± 3,630 | 16,490 ± 3,420 | – | – |
| M2 | CDR1α | 19,450 ± 1,650 | 0.0966 ± 0.0065 | 7.2 ± 0.5 | 4,970 ± 75 | 6,165 ± 735 | –0.82 | 0.95 |
| M4 | CDR3α | 42,670 ± 17,430 | 0.0243 ± 0.0007 | 28.5 ± 0.8 | 580 ± 185 | 805 ± 305 | –2.09 | 0.60 |
| M5 | CDR1α/CDR3α | 81,270 ± 29,600 | 0.0096 ± 0.0001 | 71.6 ± 0.9 | 159 ± 41 | 447 ± 235 | –2.86 | 0.57 |
| M14 | CDR3α/CDR3β | 173,500 ± 3,500 | 0.0141 ± 0.0007 | 48.9 ± 2.4 | 81 ± 2 | 83 ± 1 | –3.26 | 0.64 |
| M15 | CDR1α/CDR3α/CDR3β | 281,500 ± 34,800 | 0.0068 ± 0.0006 | 104.4 ± 9.1 | 25 ± 5 | 51 ± 7 | –3.96 | 0.60 |
| M15/D73 | CDR1α/CDR3α/CDR3β | 164,500 ± 5,500 | 0.0596 ± 0.0009 | 11.6 ± 0.2 | 363 ± 6 | 399 ± 14 | –2.37 | 0.20§ |
The location of the affinity mutations for each 3.L2 mutant is listed as well as well kinetic measurements (kon, koff, half-life, KD) and equilibrium values (KD) measured by SPR (see ref. 28). ΔG and Φ values are relative to M1.
t1/2 = 0.69/koff
ΔΔG = RTIn(KD,mu/KD,WT), where R is the gas constant (R = 0.001987 kcal/mol) and T is temperature in Kelvin. WT = M1
Phi values (Φ) are calculated as activation energy (ΔEa) normalized by ΔG. Φ = RTIn(kon,WT/kon,mut)/RTIn(KD,mut/KD,WT)
The M15/D73 Φ value is relative to M15/Hb
To examine the relative contributions of mutations in each of the three CDRs, the affinities of individual mutants were examined. Based on this analysis, CDR1α, CDR3α, and CDR3β mutations increased the affinity by ≈4-, 30-, and 7-fold, respectively (Table 1). Furthermore, the CDR variants were associated with both increased kinetic on-rates and half-lives. The increase in on-rates observed for each of the mutants suggests that there is a significant effect of the mutations on the conformational stability of the TCR. This result is not unexpected for the CDR1α mutations (φ = 0.95, M2 vs. M1) because these mutants were selected based on increased expression levels (i.e., stability) and not on higher binding affinity for Hb/I-Ek. However, CDR3 mutations that increase pMHC affinity now have also been shown to be dominated by faster on-rates in the 3.L2 and 2C T cell systems (this work and ref. 11).
Comparison of the changes in binding free energy among the different CDR mutants (Fig. 1C and Table 1) indicated that the energetic contributions of individual mutated CDRs to increased affinity were largely additive. This finding suggests that the grouped CDR mutations acted in an independent manner, without significant negative or positive cooperativity. This finding contrasts with other affinity-matured proteins in which some coupling of mutations, or cooperativity, has been observed (e.g., refs. 33 and 34). However, it has been shown that coselected mutations generated in a single library show more cooperativity than mutations generated in successive evolution of independent libraries (35). Thus, the CDR1α, CDR3α, and CDR3β mutants act independently, whereas it is possible the mutations identified in a single library (e.g., the four CDR3α mutations) may indeed act cooperatively.
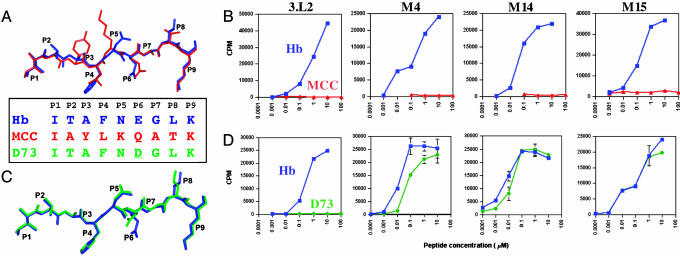
Functional Consequences of Higher-Affinity 3.L2 TCRs. To test the function of the higher-affinity mutant receptors, we transfected full-length 3.L2 TCR α and β chains, containing the corresponding CDR mutations (without framework mutations) for M4, M14, and M15, into the 58α-β-CD4+ T cell hybridoma line. Transfectants expressed approximately the same level of TCRs as measured by flow cytometry with Vβ-specific antibodies (data not shown). Their responses to Hb and MCC peptides were compared with a 3.L2 CD4+ hybridoma that expressed WT 3.L2 TCR chains (29). MCC is a peptide similar to Hb and is also presented by I-Ek and recognized by the T cell 2B4 (28, 36). Hb and MCC share the same anchor residues at P1 and P9, but they differ at the four surface-exposed residues P2, P3, P5, and P8 (Fig. 4A). M4, M14, and M15 T cells all responded strongly to Hb and not at all to MCC (Fig. 4B). There was also no response to APCs alone (Fig. 4B) or to H-2k spleen cells (data not shown), indicating that the M4, M14, and M15 T cells retained a high degree of specificity and were not autoreactive. Thus, the 800-fold increase in affinity of the M15 TCR did not result in a corresponding loss of peptide specificity, as judged by the lack of stimulation by endogenous peptides presented by I-Ek. This finding differs in this respect from our previous study with a 2C TCR (m33α) engineered to have higher affinity for a foreign peptide/MHC called SIYR/Kb (37). T cells transfected with the m33α TCR were reactive with self-peptide/Kb complexes including the peptide dEV8 that is structurally related to SIYR. This reactivity would explain why T cells with the m33α TCR would not be found in vivo: they would most likely have been negatively selected in the thymus of Kb-bearing mice. It is possible that T cells bearing the M15 TCR (or more appropriately, the M4 and M14 TCRs that have only CDR3 mutations) undergo negative selection in the thymus because the threshold for deletion is lower than the threshold for peripheral T cell activation (38), but this possibility remains to be determined.
Fig. 4.
Hb/MCC and Hb/D73 peptide overlays and peptide-mediated IL-2 release from T cell transfected with WT 3.L2 TCR or high-affinity mutant TCRs. (A) Hb/MCC peptide overlay with Hb shown in red and MCC in blue and sequence alignment of Hb, MCC, and D73. Coordinates of Hb/I-Ek [Protein Data Bank (PDB) ID code 1FNG] and MCC/I-Ek (PDB ID code 1KT2) were used to compare the peptides by aligning the I-Eα chains using the insight ii suite of programs (Molecular Simulations, Waltham, MA). (B) IL-2 release measured in a biological indicator assay (cpm) at various concentrations of WT Hb (blue curves) or MCC (red curves) presented by APCs. (C)Hb/D73 peptide overlay with Hb shown in blue and D73 in green, using coordinates of Hb/I-Ek and D73/I-Ek (PDB ID code 1FNE) as described above. (D) IL-2 release measured at various concentrations of the WT Hb (blue curves) or D73 peptide (green curves) presented by APCs.
With higher-affinity mutants, especially M15, we anticipated an increased sensitivity in the response to Hb. Surprisingly, T cells expressing the affinity-matured TCRs responded to Hb with similar sensitivity compared with WT 3.L2 (Fig. 4B). A compilation of multiple experiments revealed no statistical difference in the EC50 values among 3.L2, M4, M14, and M15 (EC50 = 510 ± 130, 321 ± 41, 369 ± 140, and 393 ± 61 nM, respectively). This result indicated that although the longer half-lives of the TCRs were not detrimental to T cell activation (8), the large increases in affinity (35-, 240-, and 800-fold, respectively, for the M4, M14, and M15 TCRs) did not result in a corresponding increase in peptide sensitivity. This result suggests that there is a cellular activation threshold for T cells that provides T cells with maximal sensitivity at low antigen levels. WT 3.L2, with a KD of ≈12 μM and a half-life of ≈10 s, appears to be near this affinity threshold so that dramatic increases in T cell sensitivity are not observed. Our previous studies with the class I-restricted TCR 2C also showed an activation threshold, but for CD8+ cells this threshold appeared to be a KD value of ≈3 μM (11). As discussed (11), this affinity threshold may vary to some degree depending on the stability/cell surface lifetime of the pMHC ligand and/or on the contribution from accessory molecules CD4 or CD8. Furthermore, it remains to be seen whether other functions of T cells, such as proliferation, differ in their affinity threshold.
The Hb variant at position P6, called D73 (Glu-73 → Asp), acts as an antagonist for 3.L2 T cells (13, 16). The structures of Hb/I-Ek and D73/I-Ek are very similar, with both Glu-73 and Asp-73 serving as anchor residues that point toward the I-Ek floor (17) (Fig. 4 A and C). However, the removal of a single MHC buried methylene group resulted in small but significant changes (≈0.5-2.0 Å) in three residues, Asn-72, Gly-74, and Leu-75, that are surface exposed and accessible to the TCR (Fig. 4C). Although T cells with the high-affinity TCRs were not reactive with unrelated peptides, it was possible that they could now respond to the structurally similar D73 ligand. Transfected T cells were assayed for their response to D73 loaded APCs, but unlike WT 3.L2 T cells, all of the higher-affinity TCR transfectants recognized D73 as a strong agonist (Fig. 4D). The agonist activity was observed for all of the affinity-matured clones including M4, which has only the CDR3α mutations. Although the affinity of the WT TCR for D73/I-Ek is apparently below the threshold for T cell activation, the affinity of the mutant TCRs are presumably above the threshold.
To determine the affinity of M15 TCR for D73/I-Ek, SPR was performed with immobilized D73/I-Ek. M15 had a KD value of 363 nM and a half-life of 11.6 s (Table 1). These binding and kinetic constants are within the range of values that were observed for the binding of the WT Hb/I-Ek complex to the 3.L2 WT and mutant TCRs. Interestingly, these binding constants represent a 15-fold lower KD value, dominated by a 9-fold faster off-rate and only a 1.7-fold slower on-rate than the M15:Hb/I-Ek interaction (φ = 0.2). The more predominant effect on the half-life of the interaction is consistent with the fact that the ligand has changed in this case, whereas the TCR is the same and thus shows the same degree of flexibility (compared, for example, with the different TCRs, where the ligand is fixed). Thus, the half-life of the interaction appears to be influenced by the subtle but significant changes that exist at the position of peptide residues N72, G74, and L75 (17).
In summary, we show here that it is possible to engineer functional high-affinity TCRs that retain exquisite peptide specificity. The demonstration of comparable antigen sensitivity of the various T cell transfectants, despite increases in affinity up to 800-fold, has significant implications for T cell biology. Other studies have concluded that there is a T cell signaling threshold dependent on the number of pMHC ligands. Thus, both CD4 and CD8 T cells show similar high levels of sensitivity (for <10 pMHC), despite different binding affinities of the TCRs (39-41). McHeyser-Williams and colleagues (21) examined the maturation of a T cell response and found that T cells from an early response were replaced by T cells with higher-affinity TCRs, up to a particular “affinity” threshold. Although their study did not reveal the explicit KD values (or half-lives) that correlate with the affinity threshold, our findings suggest that a KD value of ≈12 μM and a half-life of ≈10 s (measured at 25°C) are likely to be near this threshold. From these and other studies, it has been concluded that T cells have a relatively low signaling threshold. Thus, T cells have evolved to be maximally sensitive to low amounts of antigen, and once this threshold is reached, they ignore any further receptor engagement. It is perhaps not surprising that sensitivity cannot be improved below this number, especially because it is becoming clearer that endogenous peptides enhance this sensitivity (36, 42, 43). It is possible that the complex nature of the TCR proximal signaling apparatus is directly involved in setting this threshold. In contrast, the B cell receptor proximal signaling machinery may be less complicated, and B cells have been shown to sense a range of different affinities. B cells, unlike T cells, undergo affinity maturation at a single-cell level, and thus enhanced sensitivity to antigen with higher-affinity surface Ig provides an important selective advantage for B cells (22).
If T cells with such high-affinity TCRs are not functionally impaired, nor are they reactive with self-peptide/MHC as peripheral T cells, then why have they not been found in vivo? One likely possibility stems from the lower threshold for negative selection in the thymus than for peripheral T cell activation. Thus, T cells with M15-like properties would cross-react with self-peptides and be deleted during thymic development. Another possibility arises from the threshold of activity that was observed among T cells that expressed these high-affinity TCRs. Without selective advantages in vivo, as with high-affinity antibodies, T cells with high-affinity TCRs may not expand at the expense of T cells with low-affinity TCRs, as recently observed in studies examining clonal selection of helper T cells (21). Accordingly, they may exist, but they may be too rare to detect. Finally, there could be other functions of T cells, such as proliferation or trafficking, that are affected negatively by higher affinity. Each of these possibilities remains to be tested.
Supplementary Material
Acknowledgments
We thank Daved Fremont and Tom Brett for numerous useful discussions and overall contributions to the project, Barbara Pilas and Ben Montez for assistance with cell sorting, Liping Wang for assistance with SPR, Steve Horvath for peptide synthesis and protein production, and Jennifer Alexander for initial help with SPR. This work was supported by National Institutes of Health Grants AI24157 and AI61173 (to P.M.A.) and GM55767 (to D.M.K.). K.S.W. was supported in part by a National Institutes of Health training grant.
Author contributions: K.S.W., D.L.D., P.M.A., and D.M.K. designed research; K.S.W. and D.L.D. performed research; K.S.W., D.L.D., P.M.A., and D.M.K. analyzed data; and K.S.W., D.L.D., P.M.A., and D.M.K. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TCR, T cell receptor; scTCR, single-chain TCR; CDR, complementarity determining region; V, variable region; pMHC, product of the MHC; MCC, moth cytochrome c; CAb, clonotypic Ab; SPR, surface plasmon resonance; APC, antigen-presenting cell.
References
- 1.Starr, T. K., Jameson, S. C. & Hogquist, K. A. (2003) Annu. Rev. Immunol. 21, 139-176. [DOI] [PubMed] [Google Scholar]
- 2.Davis, M. M., Boniface, J. J., Reich, Z., Lyons, D., Hampl, J., Arden, B. & Chien, Y. (1998) Annu. Rev. Immunol. 16, 523-544. [DOI] [PubMed] [Google Scholar]
- 3.Williams, C. B., Engle, D. L., Kersh, G. J., Michael White, J. & Allen, P. M. (1999) J. Exp. Med. 189, 1531-1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gascoigne, N. R. J., Zai, T. & Alam, S. M. (2001) Exp. Rev. Mol. Med., http://www-ermm.cbcu.cam.ac.uk/01002502h.htm. [DOI] [PubMed]
- 5.Sykulev, Y., Brunmark, A., Tsomides, T. J., Kageyama, S., Jackson, M., Peterson, P. A. & Eisen, H. N. (1994) Proc. Natl. Acad. Sci. USA 91, 11487-11491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kersh, E. N., Shaw, A. S. & Allen, P. M. (1998) Science 281, 572-575. [DOI] [PubMed] [Google Scholar]
- 7.McKeithan, T. W. (1995) Proc. Natl. Acad. Sci. USA 92, 5042-5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valitutti, S., Muller, S., Cella, M., Padovan, E. & Lanzavecchia, A. (1995) Nature 375, 148-151. [DOI] [PubMed] [Google Scholar]
- 9.Kalergis, A. M., Boucheron, N., Doucey, M. A., Palmieri, E., Goyarts, E. C., Vegh, Z., Luescher, I. F. & Nathenson, S. G. (2001) Nat. Immunol. 2, 229-234. [DOI] [PubMed] [Google Scholar]
- 10.Holler, P. D., Lim, A. R., Cho, B. K., Rund, L. A. & Kranz, D. M. (2001) J. Exp. Med. 194, 1043-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holler, P. D. & Kranz, D. M. (2003) Immunity 18, 255-264. [DOI] [PubMed] [Google Scholar]
- 12.Li, Q. J., Dinner, A. R., Qi, S., Irvine, D. J., Huppa, J. B., Davis, M. M. & Chakraborty, A. K. (2004) Nat. Immunol. 5, 791-799. [DOI] [PubMed] [Google Scholar]
- 13.Evavold, B. D. & Allen, P. M. (1991) Science 252, 1308-1310. [DOI] [PubMed] [Google Scholar]
- 14.Kersh, G. J., Kersh, E. N., Fremont, D. H. & Allen, P. M. (1998) Immunity 9, 817-826. [DOI] [PubMed] [Google Scholar]
- 15.Sloan-Lancaster, J. & Allen, P. M. (1996) Annu. Rev. Immunol. 14, 1-27. [DOI] [PubMed] [Google Scholar]
- 16.Evavold, B. D., Williams, S. G., Hsu, B. L., Buus, S. & Allen, P. M. (1992) J. Immunol. 148, 347-353. [PubMed] [Google Scholar]
- 17.Kersh, G. J., Miley, M. J., Nelson, C. A., Grakoui, A., Horvath, S., Donermeyer, D. L., Kappler, J., Allen, P. M. & Fremont, D. H. (2001) J. Immunol. 166, 3345-3354. [DOI] [PubMed] [Google Scholar]
- 18.Holler, P. D., Holman, P. O., Shusta, E. V., O'Herrin, S., Wittrup, K. D. & Kranz, D. M. (2000) Proc. Natl. Acad. Sci. USA 97, 5387-5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez, P. A., Carreno, L. J., Coombs, D., Mora, J. E., Palmieri, E., Goldstein, B., Nathenson, S. G. & Kalergis, A. M. (2005) Proc. Natl. Acad. Sci. USA 102, 4824-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sykulev, Y., Cohen, R. J. & Eisen, H. N. (1995) Proc. Natl. Acad. Sci. USA 92, 11990-11992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malherbe, L., Hausl, C., Teyton, L. & McHeyzer-Williams, M. G. (2004) Immunity 21, 669-679. [DOI] [PubMed] [Google Scholar]
- 22.Foote, J. & Eisen, H. N. (2000) Proc. Natl. Acad. Sci. USA 97, 10679-10681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chlewicki, L. K., Holler, P. D., Monti, B. C., Clutter, M. A. & Kranz, D. M. (2005) J. Mol. Biol. 346, 223-239. [DOI] [PubMed] [Google Scholar]
- 24.Boder, E. T. & Wittrup, K. D. (1997) Nat. Biotechnol. 15, 553-557. [DOI] [PubMed] [Google Scholar]
- 25.Masteller, E. L., Warner, M. R., Ferlin, W., Judkowski, V., Wilson, D., Glaichenhaus, N. & Bluestone, J. A. (2003) J. Immunol. 171, 5587-5595. [DOI] [PubMed] [Google Scholar]
- 26.Garcia, K. C., Radu, C. G., Ho, J., Ober, R. J. & Ward, E. S. (2001) Proc. Natl. Acad. Sci. USA 98, 6818-6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altman, J. D., Reay, P. A. & Davis, M. M. (1993) Proc. Natl. Acad. Sci. USA 90, 10330-10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu, L. C., Tuot, D. S., Lyons, D. S., Garcia, K. C. & Davis, M. M. (2002) Nature 418, 552-556. [DOI] [PubMed] [Google Scholar]
- 29.Vidal, K., Daniel, C., Hill, M., Littman, D. R. & Allen, P. M. (1999) J. Immunol. 163, 4811-4818. [PubMed] [Google Scholar]
- 30.Kieke, M. C., Shusta, E. V., Boder, E. T., Teyton, L., Wittrup, K. D. & Kranz, D. M. (1999) Proc. Natl. Acad. Sci. USA 96, 5651-5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shusta, E. V., Kieke, M. C., Parke, E., Kranz, D. M. & Wittrup, K. D. (1999) J. Mol. Biol. 292, 949-956. [DOI] [PubMed] [Google Scholar]
- 32.Shusta, E. V., Holler, P. D., Kieke, M. C., Kranz, D. M. & Wittrup, K. D. (2000) Nat. Biotechnol. 18, 754-759. [DOI] [PubMed] [Google Scholar]
- 33.Yin, J., Mundorff, E. C., Yang, P. L., Wendt, K. U., Hanway, D., Stevens, R. C. & Schultz, P. G. (2001) Biochemistry 40, 10764-10773. [DOI] [PubMed] [Google Scholar]
- 34.Yang, J., Swaminathan, C. P., Huang, Y., Guan, R., Cho, S., Kieke, M. C., Kranz, D. M., Mariuzza, R. A. & Sundberg, E. J. (2003) J. Biol. Chem. 278, 50412-50421. [DOI] [PubMed] [Google Scholar]
- 35.Bernat, B., Sun, M., Dwyer, M., Feldkamp, M. & Kossiakoff, A. A. (2004) Biochemistry 43, 6076-6084. [DOI] [PubMed] [Google Scholar]
- 36.Krogsgaard, M., Li, Q. J., Sumen, C., Huppa, J. B., Huse, M. & Davis, M. M. (2005) Nature 434, 238-243. [DOI] [PubMed] [Google Scholar]
- 37.Holler, P. D., Chlewicki, L. K. & Kranz, D. M. (2003) Nat. Immunol. 4, 55-62. [DOI] [PubMed] [Google Scholar]
- 38.Grossman, Z. & Singer, A. (1996) Proc. Natl. Acad. Sci. USA 93, 14747-14752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sykulev, Y., Joo, M., Vturina, I., Tsomides, T. J. & Eisen, H. N. (1996) Immunity 4, 565-571. [DOI] [PubMed] [Google Scholar]
- 40.Irvine, D. J., Purbhoo, M. A., Krogsgaard, M. & Davis, M. M. (2002) Nature 419, 845-849. [DOI] [PubMed] [Google Scholar]
- 41.Krogsgaard, M. & Davis, M. M. (2005) Nat. Immunol. 6, 239-245. [DOI] [PubMed] [Google Scholar]
- 42.Stefanova, I., Dorfman, J. R. & Germain, R. N. (2002) Nature 420, 429-434. [DOI] [PubMed] [Google Scholar]
- 43.Yachi, P. P., Ampudia, J., Gascoigne, N. R. & Zal, T. (2005) Nat. Immunol. 6, 785-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.