Abstract
In addition to its role in cancer, the c-Myc oncoprotein controls many normal cellular processes as a consequence of its function as a basic helix–loop–helix leucine zipper transcription factor. Determining which of the myriad genes under c-Myc control are relevant for these various roles is thus a major challenge. mt-mc1 is a direct c-Myc target gene whose overexpression recapitulates multiple c-Myc phenotypes, including transformation. Using transcriptional profiling, we now show that MT-MC1-overexpressing myeloid cells misregulate a total of 47 distinct transcripts, a large proportion of which are involved in signal transduction and/or cancer. Analysis of these genes reveals a consensus promoter structure consisting of multiple, often closely spaced c-Myc binding sites and three additional Wilm's tumor and Egr1-like motifs. More than one-third of MT-MC1 target genes are also clustered on six cancer-associated chromosomal loci. Most surprisingly, all of the transcripts examined also are regulated by c-Myc. Finally, an estrogen receptor–MT-MC1 fusion protein was used to establish that all examined transcripts were regulated directly by the chimeric protein. Our results thus indicate that MT-MC1 target genes largely comprise a subset of those regulated by c-Myc. We propose that the properties imparted by MT-MC1 are the result of its control of a small and select c-Myc target gene population.
Keywords: CCL6, DNA microarray, gene profiling, IL-3, Max
The basic helix–loop–helix leucine zipper transcription factor, c-MYC, is deregulated in diverse human cancers (1). The causes of c-Myc overexpression include chromosomal translocations, gene amplification, noncoding region mutations affecting c-Myc mRNA stability, and the deregulation of transcription factors controlling C-MYC gene promoter activity (1). c-Myc binds to several hundred genomic loci harboring consensus c-Myc binding sites, termed E-boxes, resulting in the transcriptional activation of their adjacent genes (2, 3). The degree to which these genes are deregulated is dictated by the levels of c-Myc, its affinity for its cognate E-boxes, the cell type, and by the levels of other basic helix–loop–helix leucine zipper proteins that compete for c-Myc's obligate heterodimerization partner, Max (3). Additionally, a large number of genes are down-regulated by c-Myc. The means by which this is achieved, however, is more varied than for positively regulated targets and appears to involve an inhibitory interaction between c-Myc and other transcription factors, such as Miz-1 and YY-1 (4, 5).
As might be anticipated from such global transcriptional alterations, the c-Myc phenotype is complex. In addition to promoting transformation and tumorigenesis in a variety of cell types, c-Myc overexpression affects growth rate, cell size, cell cycle progression, morphology, susceptibility to various apoptotic stimuli, differentiation, and genomic instability (6–15). Thus, a major challenge is to determine which of the myriad c-Myc target genes contributes to the individual phenotypes of c-Myc and how this is accomplished at the molecular level.
To date, the roles of only a small number of c-Myc targets in mediating specific phenotypes have been investigated. For example, ornithine decarboxylase, HMG-I/Y, and Hsp90A are transforming; telomerase is immortalizing; cdk4 and serine hydroxymethyl-transferase promote cell cycle progression and accelerated proliferation; and cyclin B1 induces genomic instability (10, 16–21). Although overexpression tends to recapitulate only a single c-Myc-like property, several examples have been reported in which individual target genes can impart additional phenotypes. For example, ornithine decarboxylase over-expression also enhances susceptibility to certain apoptotic stimuli and cdk4 can also cooperate with activated Ras oncogenes to transform primary cells (22, 23).
mt-mc1, a direct, positive c-Myc target gene (24, 25), encodes a 188-aa nuclear protein with weak similarity to certain DNA-binding proteins and helicases. MT-MC1 is unique in that its overexpression can recapitulate multiple c-Myc phenotypes. Among these phenotypes are in vitro transformation and in vivo tumorigenesis, the promotion of genomic instability, alteration of cellular morphology, inhibition of differentiation, and increased apoptosis in response to growth factor deprivation (25). Several of these properties appear not to require the cooperation of other deregulated c-Myc target genes or even c-Myc itself, because they can be mimicked in c-Myc-null fibroblasts (26).
MT-MC1 also regulates some c-Myc target genes, thus suggesting a potential means by which the former protein might orchestrate the complex c-Myc phenotype (25, 26). However, because these analyses were performed with only a small number of genes, the extent and nature of this regulation remains largely undefined. Nonetheless, the findings imply that MT-MC1 may impart multiple c-Myc-like properties to diverse cell types by deregulating its own target gene repertoire, which overlaps that of c-Myc.
We have now used transcriptional profiling to obtain a more comprehensive appraisal of MT-MC1 target genes. DNA microarrays were used to evaluate the differential expression of these genes in myeloid cells constitutively expressing MT-MC1. With this approach, we have identified 47 genes whose expression is deregulated by >2-fold as a result of MT-MC1 overexpression. Further characterization of a subset of these genes shows them to be direct transcriptional targets for MT-MC1 and c-Myc. Thus, MT-MC1 appears to participate in a novel form of regulation of other c-Myc target genes, which may explain its unique ability to impart multiple c-Myc-like phenotypes.
Materials and Methods
Cell Culture. 32Dcl3 murine myeloid cells that overexpress c-Myc (32D-c-Myc cells) or MT-MC1 (32D-MT-MC1 cells) have been previously described, as has the control cell line (32D-neo) that was transfected with the empty parental plasmid (24, 25). 32D-MT-MC1del (2–32) cells express a mutant form of MT-MC1 missing 31 aa from its N terminus (see Plasmid Constructs below). Each cell line consisted of a pooled population of several hundred clones derived by stable electroporation with linearized plasmid DNA. Overexpression of the proteins of interest was confirmed by immunoblotting.
Microarray and Quantitative Real-Time (QRT)-PCR Analyses. Isolation of total RNA was performed as described in refs. 24–26. The preparation of cRNA was performed by the PittArray DNA Microarray Facility of the Genomics and Proteomics Core Laboratories at the University of Pittsburgh Medical Center and is described in more detail in Supporting Materials and Methods, which is published as supporting information on the PNAS web site. cRNAs were hybridized overnight to triplicate murine genome MG-U74Av2 GeneChips (Affymetrix, Santa Clara, CA) and further analyzed using microarray suite 5.0 and dchip 1.3 software (27). Identified gene information and accession numbers were confirmed by searches of the LocusLink database (www.ncbi.nlm.nih.gov/LocusLink). Genes were grouped into functional categories based on their known molecular and/or biological functions denoted in LocusLink. For QRT-PCR, we used 50 ng of total input RNA in a SYBR Green-based assay (QuantiTect, Qiagen, Valencia, CA) according to the directions of the supplier. Oligonucleotide primers for select MT-MC1 target genes were chosen to span intron–exon boundaries and were designed with the primer 3 program. Primers were synthesized by Integrated DNA Technologies (Coralville, IA) and are listed in Table 1, which is published as supporting information on the PNAS web site. Reverse transcription and PCR amplification and analyses were performed on a LightCycler 2 (Roche Diagnostics) under the conditions recommended by the supplier. All analyses used lightcycler 4 relative quantification software. Standard errors on all samples were generally <2%. Cτ values for each reaction were normalized to those for GAPDH reactions included in each set of QRT-PCR runs and are expressed relative to values obtained with RNAs from 32D-neo cells. Additional methods and analyses are provided in Supporting Materials and Methods.
Computational Analysis of MT-MC1 Target Gene Promoters and Chromosomal Localizations. For the purpose of this study, “promoter region” was defined as the 3 kb of contiguous sequence upstream and 1 kb downstream of the transcription start site, which was computationally identified with the footer internet tool (28). Briefly, the coding sequence of each gene was used in a blast search against the mouse RefSeq collection (29). We assumed that the first nucleotide of the longest cDNA identified in this manner corresponded to, or lay only a short distance from, the actual transcription start site. The promoters were then analyzed with the program consensus (30, 31) for identification of common DNA patterns. Lengths of 6–15 bp were tested, because this range includes most of the known transcription factor binding sites.
The chromosomal localizations of MT-MC1-regulated genes were performed by using the LocusLink database. The potential involvement of human syntenic loci in cancer was investigated using the Mitelman Database of Chromosome Aberrations in Cancer from the Cancer Genome Anatomy Project (http://cgap.nci.nih.gov/Chromosomes/Mitelman).
Plasmid Constructs. Expression vectors for murine c-Myc and myc-epitope-tagged MT-MC1 are been described in refs. 10, 24, and 25. The construction of other vectors is described in Supporting Materials and Methods.
Results
Transcriptional Profiling of 32D-MT-MC1 Cells. In 32D cells, the enforced expression of either c-Myc or MT-MC1 imparts genomic instability and increased sensitivity to apoptotic stimuli (24, 25). Further mimicking of the c-Myc phenotype by MT-MC1 includes the acquisition of anchorage-independent growth and tumorigenic behavior by Rat1a fibroblasts, morphological reversion of c-Myc-null fibroblasts, and the inhibition of chemically induced differentiation of Friend murine erythroleukemia cells (25, 26).
To gain a better understanding of the molecular underpinnings of MT-MC1's wide-ranging biological effects, and to relate this to the c-Myc phenotype, we performed a microarray analysis that compared the transcriptional profiles of early passage, diploid, log-phase 32D-MT-MC1 cells to those of 32D-neo control cells. Strict criteria were applied for the identification of differentially expressed transcripts, including the requirement that they register in three independent experiments and that at least a 2-fold differential expression ratio be observed on each occasion. The raw data for these transcripts (Affymetrix in excel format), is shown in Data Set 1, which is published as supporting information on the PNAS web site.
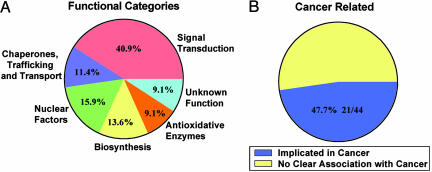
As seen in Table 2, which is published as supporting information on the PNAS web site, 47 transcripts met the above criteria. Of these transcripts, 33 (70%) were up-regulated (range 2.1- to 16.3-fold) and 14 (30%) were down-regulated (range 2.2- to 9.6-fold). Only seven transcripts (15%) encoded proteins of unknown or uncertain function. Of the 40 transcripts with established function, 18 (45%) could be broadly categorized as being involved in signal transduction. The remaining proteins could be classified into four other functional categories involving subcellular trafficking and transport, nuclear maintenance, biosynthesis, and oxidative or other stress responses. Of the 44 proteins with known or uncertain function, 21 (48%) have been implicated in cancers and 7 (16%) have been identified as c-Myc targets in previous microarray studies performed in fibroblasts (Fig. 1).
Fig. 1.
Functional categories of MT-MC1 target genes. (A) Of the 47 differentially expressed genes listed in Table 2, 40 encoded proteins of known function can be classified into five broad categories. (B) Of these proteins, 48% have also been previously implicated in cancer.
Taken together, the above results indicate that the de novo biological properties of 32D-MT-MC1 myeloid cells are associated with the deregulation of a number of genes, particularly those whose products are involved in signal transduction and neoplastic pathways. In keeping with our previous observations (25, 26), a significant number of these genes have also been previously identified as c-Myc targets.
Analysis of MT-MC1-Responsive Promoters. To search for common regulatory elements, the sequences of all 47 MT-MC1-regulated transcripts listed in Table 2 were retrieved from public databases and used to identify their respective promoters as described in Materials and Methods. We successfully identified the promoters for 39 of the 47 genes (83%), although 3 were incomplete at their 5′ ends. The remaining eight promoters contained large sections of incomplete and/or ambiguous sequences.
Because of the above described relationship between certain MT-MC1 and c-Myc target genes, we first asked whether the promoter sequences contained the consensus palindromic c-Myc E-box element CACGTG, the alternate E-box CATGTG, or its reverse complement CACATG. As seen in Fig. 2, standard string search analysis showed that 35 of 39 promoters contained multiple E-box elements and the remaining promoters contained a single such site. As a group, up-regulated genes harbored a significantly greater number of E-boxes than did down-regulated genes (average, 4.3 sites per gene vs. 2.8 sites per gene; P = 0.01, Student's t test). In the former case, E-box elements also often occurred in clusters of more than three within 1 kb of one another (for example, Ccl6, Aqp9, Lmo2, Piga, and Pik3ca).
Fig. 2.
Analysis of MT-MC1-responsive gene flanking sequences. Promoters for 39 of the 47 MT-MC1-regulated genes are shown, with small black arrows indicating the presumed start of transcription initiation (See Fig. 6 for the actual sequences analyzed and the location of each consensus motif). Those designated in green indicate promoters up-regulated by MT-MC1, whereas those designated in red are down-regulated. We initially screened for consensus c-Myc binding sites of the sequence CACGTG (red boxes) or for alternate E-boxes of the sequence CATGTG and its reverse complement (CACATG) (yellow arrows). The genomic sequences were then further analyzed with the program consensus (30) to identify other common sites. Three additional common consensus motifs [A, (C/G)(A/T)GGGGGC(A/T); B, GGGGTGGGG; and C, CCTCTGCCTCC] were identified and are depicted here in blue, lavender, and green, respectively, with arrows indicating their 5′→3′ orientation.
We next used the consensus program (30) to identify other common elements shared among the aligned promoters. Lengths between 6 and 15 bp were tested, because this range includes the vast majority of known transcription factor binding sites. The output of consensus consists of a number of top-scoring patterns organized in the form of count matrices, which is the most popular form of transcription factor binding preference representation (31). After manual examination of the results for elimination of subpatterns, we identified three consensus motifs, all bearing resemblance to early growth response 1 (EGR1) or Wilm's tumor 1 suppressor-binding sites according to the corresponding patterns in the transfac 6.0 database (www.gene-regulation.com/pub/databases.html#transfac) (32). Motifs A and B are related to the EGR1 site (the first only in its middle six nucleotides), whereas motif C represents a suboptimal pattern of Wilm's tumor 1. Each of these consensus motifs was present in each of the 39 promoters (see Fig. 6, which is published as supporting information on the PNAS web site, for the precise locations of all of the above sites in each promoter). Together, these data provide a picture of the consensus MT-MC1-regulated gene as containing A, B, and C motifs, as well as a significantly greater number of E-boxes than predicted in the case of positively regulated MT-MC1 targets.
Partial Loss of MT-MC1 Function Correlates with Misregulation of a Subset of Target Genes. Deletion analysis of MT-MC1 has revealed that its various functions map to distinct regions of the protein (D.E.C., K.R.R., and E.V.P., unpublished data). To determine whether loss of one or more of these properties might also be associated with an inability to regulate some of the above MT-MC1 target genes, we examined the behavior of an N-terminal deletion mutant [MT-MC1-del(2–32)]. As seen in Fig. 3A, this mutant protein was expressed in 32D cells at levels comparable with those of full-length MT-MC1. Consistent with our original observations using GFP-tagged full-length MT-MC1 (25), both proteins also localized primarily to the nucleus (Fig. 3B). Furthermore, the del(2–32) mutant was as effective as full-length MT-MC1 in promoting the acquisition of tetraploidy (Fig. 3C). In contrast, 32D-del(2–32) cells were severely compromised in their ability to enhance apoptosis after the removal of IL-3 (Fig. 3D). These findings indicate that the N terminus of MT-MC1 is dispensable for nuclear localization and the promotion of genomic instability but is required for the acceleration of apoptosis after the removal of IL-3.
Fig. 3.
Differential function and target gene regulation by a truncated form of MT-MC1. (A) Stable expression in 32D cells of myc-epitope-tagged, full-length MT-MC1 and a deletion mutant lacking amino acids 2–32. (Upper) The results of a whole-cell lysate Western analysis, indicating that the two MT-MC1 proteins were expressed at equivalent levels in 32D cells but not in control 32D-neo cells. For the full-length and deleted proteins, two distinct bands are seen, likely representing different posttranslationally modified forms. (Lower) The same blot probed with a monoclonal antibody against actin, which was used as a control for protein loading. (B) Subcellular localization of MT-MC1 proteins. 32D-MT-MC1 and 32D-MT-MC1-del(2–32) cells were fractionated into nuclear and cytoplasmic components. (Top) Western analyses for MT-MC1. (Middle and Bottom) Western analyses for caspase 3 (Middle) and acetylated histone H3 (AcH H3) (Bottom), which were used as controls for proteins localizing to the cytoplasmic and nuclear compartments, respectively. (C) Induction of genomic instability by MT-MC1 proteins. The indicated cell lines were carried continuously in culture for 6 months. At the end of this period, nuclei were stained with propidium iodide and the fraction of pseudotetraploid cells was determined by flow cytometry as described in refs. 24 and 25. (D) Effects of MT-MC1 proteins on apoptosis. The indicated cell lines were deprived of IL-3 and then serially examined for viability as described in refs. 24 and 25. The results show the average of three experiments ± SE. (E) Gene expression profiles of 32D cell lines. QRT-PCR analyses were performed on 12 of the genes listed in Table 2. Each QRT-PCR analysis was performed in triplicate and normalized to GAPDH transcripts whose levels were determined in parallel. The normalized levels of individual transcripts in each of the cell lines are expressed relative to those in control 32D-neo cells.
To independently confirm the differential expression of the transcripts that were identified by the above microarray experiments and to determine whether differential gene expression could account for the defect in 32D-del(2–32) cells, we performed QRT-PCR analyses on a select subset of the transcripts listed in Table 2. We also concurrently assessed the expression of these genes in 32D-c-Myc cells whose biological behavior is closely mimicked by MT-MC1 (25, 26). As shown in Fig. 3E, all 12 of the analyzed transcripts showed differential expression in 32D-MT-MC1 cells compared with 32D-neo cells, thus confirming the results of the original microarray analyses. In 8 of 12 cases, 32D-del(2–32) cells showed a >4-fold change in transcript levels compared with 32D-MT-MC1 cells. Thus, the altered biological activity of this MT-MC1 mutant correlated with altered target gene expression. Surprisingly, all 12 transcripts were also altered in 32D-c-Myc cells. In some cases, however, the nature and/or extent of regulation differed from that seen in 32D-MT-MC1 cells. For example, transcripts for Aldh1a1, which were down-regulated ≈10-fold in 32D-MT-MC1 cells, were down-regulated by nearly 250-fold in 32D-c-Myc cells. In contrast, transcripts for the chemokine Ccl6, which were up-regulated 15.8-fold by MT-MC1, were down-regulated 120-fold by c-Myc. Thus, we conclude that MT-MC1 target genes are also targets for c-Myc, although the two proteins often regulate these genes differently.
Frequent Chromosomal Clustering of MT-MC1 Target Genes. In determining the chromosomal location for each of the murine genes listed in Table 2, we noted that 16 (34%) mapped to six chromosomal loci ranging from 0.32 to 13 megabases (Mb) (Table 3, which is published as supporting information on the PNAS web site). For example, the genes for CXCR4, Daf2, and Daf1 mapped to within 2 Mb of one another on chromosome 1. In another case, the genes for Gp1ba, Spag7, Ccl9, and Ccl6 mapped to within 13 Mb on chromosome 11. In virtually all cases, the syntenic regions for these genes in humans are associated with a variety of acute and chronic leukemias, carcinomas, and other types of cancers.
Many MT-MC1 Genes Are Direct Targets. Unlike c-Myc, which regulates transcription by directly binding to DNA or by binding to and inhibiting other positively acting transcription factors (2–5), the mechanism(s) by which MT-MC1 regulates target gene expression remain(s) unknown, which complicates the use of methods such as chromatin immunoprecipitation to demonstrate its direct DNA binding because no consensus DNA-binding site for MT-MC1 has been identified. The fact that MT-MC1-responsive genes are often found in tightly grouped clusters (Table 3) further raised the possibility of long-range control of gene expression (33–36).
With these concerns in mind, we created a 32D cell line that expressed myc-epitope-tagged MT-MC1 fused at its N terminus to the hormone-binding domain of a 4-hydroxytamoxifen (4-HT)-sensitive estrogen receptor (Fig. 4A). Similar vectors have been previously used to demonstrate the direct nature of gene regulation by c-Myc and other transcription factors after the blockade of de novo protein synthesis by cycloheximide (CHX) (2, 37). In the absence of 4-HT, 32D-ER™-MT-MC1 cells (ER™, estrogen receptor responsive to tamoxifen) showed higher basal rates of apoptosis in response to IL-3 withdrawal than did control cells, thus indicating that the fusion protein was somewhat “leaky.” However, the extent of apoptotic death increased nearly 3-fold in the presence of 4-HT (Fig. 4B). Thus, the fusion protein is at least partially regulatable by 4-HT, and its effect can be quantified.
Fig. 4.
Direct regulation of MT-MC1 target gene expression by MT-MC1. (A) Expression of ER™-MT-MC1. 32D cells were stably transfected with a vector encoding a fusion protein consisting of the hormone-binding domain of a mutant form of the estrogen receptor responsive to tamoxifen and myc-epitope-tagged MT-MC1. 32D-neo cells were stably transfected with the empty pcDNA3.1(+) vector and served as controls. Both cells lines were selected for G-418-resistant clones, which were then pooled. Equivalent amounts of total cell lysate from each cell line were then subjected to immunoblotting and probed with monoclonal antibodies directed against the myc-epitope (Upper) or tubulin (Lower) as a control for protein loading. (B) Apoptosis of each of the cell lines in response to IL-3 withdrawal. Logarithmically growing 32D-neo or 32D-ER™-MT-MC1 cells were washed three times in IL-3-free medium and then resuspended in the same medium with or without 250 nM 4-HT. The percent of apoptotic cells in each group was then determined 20 h later (24, 25). Each point depicts the average of triplicate determinations ± SE. (C) QRT-PCR results. RNA from each of the following cell lines (50 ng) was used to quantify the expression of the genes listed in Fig. 3E by QRT-PCR: 1, 32D-Neo+CHX plus 4-HT; 2, 32D-ER-MT-MC1 plus CHX; and 3, 32D-ER-MT-MC1+CHX plus 4-HT. The absolute level of each transcript is expressed relative to that in 32D-neo cells that had been exposed to CHX only. Each value represents the average of triplicate determinations. Deviations from the average were <2% is all cases (data not shown).
We next blocked de novo protein synthesis in both cell lines with CHX in the absence or presence of 4-HT, harvested total RNAs 8 h later, and measured MT-MC1 target transcripts by QRT-PCR. The addition of 4-HT to control cells did not significantly alter the levels of any of the transcripts relative to those in cells treated with CHX only (data not shown). As seen in Fig. 4C, in the absence of 4-HT, 32D-ER™-MT-MC1 cells showed altered transcript levels, which was consistent with the fusion protein being moderately active and thus providing an explanation for the cells' higher rate of apoptosis (Fig. 4B). However, even greater changes in transcript levels were observed in the concurrent presence of 4-HT. Thus, despite some basal 4-HT-independent activity of ER™-MT-MT-MC1, it can be further activated by the hormone. This increased target gene deregulation occurring in the absence of protein synthesis indicated that MT-MC1's ability to alter target gene expression is at least in part, direct.
Discussion
Among the myriad of c-Myc target genes (17, 18, 21, 38–40), a small number are capable of transforming established or primary cells. Among these is MT-MC1, which is highly up-regulated in response to c-Myc overexpression in 32D myeloid cells (24, 25). That MT-MC1 can recapitulate multiple c-Myc phenotypes in several different cell lines, including c-Myc-null fibroblasts (25, 26), suggests that it occupies a unique and central position in the c-Myc transformation pathway.
We have addressed five questions pertaining to the mechanisms by which MT-MC1 alters cellular behavior. (i) What is the transcriptional profile imparted by MT-MC1? (ii) Into what general functional categories can MT-MC1 target genes be placed? (iii) Do MT-MC1 target genes share common structural and/or organizational features? (iv) Is the regulation of MT-MC1 target genes direct? (v) What is the relationship between MT-MC1 targets and those regulated by c-Myc?
First, to answer questions i and ii, MT-MC1 regulated transcripts encode a striking number of products involved in signal transduction and/or cancer (Fig. 1 and Table 2). Second, given that the growth rates and cell-cycle profiles of early passage 32D-neo, 32D-MT-MC1, and 32D-c-Myc cells are indistinguishable under the normal growth conditions used here (refs. 24 and 25 and our unpublished observations), it seems highly unlikely that such factors account for the differential transcript levels among these cell lines.
Third, the promoter regions of MT-MC1-regulatable genes show remarkable consistencies with regard to the number and positioning of E-boxes. It is of interest that genes found to be negatively regulated by MT-MC1 contained, on average, nearly the same number of E-boxes as would have been predicted from random occurrence (2.8 sites per promoter observed vs. 2.9 sites per promoter predicted), whereas genes positively regulated by MT-MC1 contained a significantly greater number of these sites (4.3 sites per promoter, P = 0.01). This difference suggests that, as is the case for c-Myc targets, the regulation of positive MT-MC1 target genes might be E-box-dependent, whereas negative targets might be E-box-independent.
In every case, three additional motifs were also identified in each promoter. Two of these motifs bore various degrees of similarity to EGR1 binding sites and one bore homology to the Wilm's tumor 1 suppressor binding site. Therefore, it is of interest that a recent in silico survey of c-Myc target gene promoters has shown that Wilm's tumor binding sites are overrepresented, suggesting functional links between these transcription factors (41). It may also be significant that the EGR1 and Wilm's tumor 1 binding sites are often recognized by both factors (42). Clearly, it will be of interest for future studies to determine the basis of regulation of these genes by MT-MC1 and to identify the functional contributions of the above discussed conserved elements.
On a larger scale, over one-third of MT-MC1-responsive genes localize to chromosomal loci of between 0.32 and 13 megabases in size. In many metazoans, genes whose products control specific pathways or which otherwise require coordinated regulation, often cluster to common chromosomal regions subject to long-distance control (33–36, 43, 44). That so many MT-MC1 target genes appear to be similarly arranged, together with the finding that their loci are commonly disrupted in human cancer (Table 3), suggests their involvement in common functions.
Fourth, we have shown that all of the examined transcripts can be largely regulated independently of de novo protein synthesis and in a manner that is qualitatively similar to that seen after constitutive expression of unmodified MT-MC1 (Fig. 4C). Thus, although we cannot state that MT-MC1's effects on gene expression are entirely direct in nature, its ability to further promote apoptosis and alter the expression of MT-MC1 target transcripts by 4-HT in the absence of de novo protein synthesis argues that much, if not all, of the effects of MT-MC1 on gene expression and biological readout are direct. At present, these studies do not permit an assessment of whether these changes occur as a result of MT-MC1 binding to DNA or its interaction with other DNA-binding proteins.
Finally, it was surprising that all MT-MC1 target genes that were examined also showed significant regulation by c-Myc, although not necessarily to the same extent or in the same direction (Fig. 3E). For example, transcripts of the Aldh1a1 gene were significantly down-regulated in 32D-MT-MC1 and 32D-c-Myc cells; however, the extent of the down-regulation varied by >20-fold between the two lines. In contrast, and as previously shown (45), Ccl6 transcripts were down-regulated by c-Myc but up-regulated by MT-MC1. Similar differences were seen with transcripts for Ccl9 and Fcgr2b. Despite these exceptions, the majority of transcripts were regulated similarly and, in some cases, remarkably so. These findings suggest that the ability of MT-MC1 to recapitulate so many c-Myc phenotypes may relate to its regulation of a critical subset of c-Myc target genes (Fig. 5). Although the overlap of c-Myc and MT-MC1 transcriptional targets appears substantial, we have not attempted to comprehensively define it here. Thus, it remains possible that MT-MC1 may have unique transcriptional targets in addition to those it shares with c-Myc, which may be particularly true in other cell types for which such comparisons have not yet been made.
Fig. 5.
Model for the regulation of the c-Myc phenotype. Overexpression of c-Myc leads to the direct transcriptional activation or repression of a large number of target genes (step A), including MT-MC1 (step B). MT-MC1 can also directly regulate a subset of c-Myc target genes either independently of or in cooperation with c-Myc (step C) (26). The coordinated regulation of these target genes generates the various c-Myc phenotypes (step D). It remains possible that MT-MC1 also regulates non-c-Myc target genes (step E), although these genes have yet to be identified.
Nonetheless, our finding that 32D-neo and 32D-MT-MC1 cells differ by the expression of only 47 genes argues that MT-MC1's transcriptional repertoire is small, highly selective, and largely focused on a subset of c-Myc targets. Indeed, the combination of previously described c-Myc target genes (Table 2) and the ones described in the present work (Fig. 3E), permit us to say that, at a minimum, 17 (36%) of 47 MT-MC1 target genes are also regulated by c-Myc. Because every MT-MC1-regulated transcript examined to date is also regulated by c-Myc (Fig. 3E), it seems quite likely that significant additional overlap exists. Given the known ability of MT-MC1 to recapitulate a large number of c-Myc phenotypes (25, 26), these results suggest that many of the key regulatory genes directing these properties may be contained within the MT-MC1 target subset.
In summary, we have shown that MT-MC1, a direct transcriptional target of c-Myc, promotes the positive and negative regulation of numerous target genes in myeloid cells. The majority of these genes are involved in signal transduction and cancer and display remarkable similarities in terms of the placement of E-box motifs and other conserved sites. Furthermore, every gene examined appeared to be directly regulated by MT-MC1 and displayed significant responses to c-Myc overexpression. These findings point to a novel form of c-Myc target gene regulation by one of its own targets and provide a molecular basis for understanding precisely how MT-MC1 is able to recapitulate such a broad range of c-Myc phenotypes.
Supplementary Material
Acknowledgments
We thank Martin Eilers (Universität Marburg, Marburg, Germany) for supplying pBabePuro-c-Myc-ER™, Deborah Hollingshead of the PittArray core facility, and Tunda Hidvegi for advice on the use of dchip software. This work was supported by National Institutes of Health Grants CA078259 and CA105033 (to E.V.P.) and Institutional Training Grant in Pediatric Research T32HD 042987 (to D.E.C.).
Author contributions: K.R.R., D.E.C., and E.V.P. designed research; K.R.R., D.E.C., and E.V.P. performed research; K.R.R., D.E.C., D.L.C., and P.V.B. analyzed data; and D.E.C. and E.V.P. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CHX, cycloheximide; EGR1, early growth response factor 1; ER™, estrogen receptor responsive to tamoxifen; QRT, quantitative real-time; 4-HT, 4-hydroxytamoxifen.
References
- 1.Nesbit, C. E., Tersak, J. M. & Prochownik, E. V. (1999) Oncogene 18, 3004–3016. [DOI] [PubMed] [Google Scholar]
- 2.Fernandez, P. C., Frank, S. R., Wang, L., Schroeder, M., Liu, S., Greene, J., Cocito, A. & Amati, B. (2003) Genes Dev. 17, 1115–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levens, D. L. (2003) Genes Dev. 17, 1071–1077. [DOI] [PubMed] [Google Scholar]
- 4.Oster, S. K., Ho, C. S., Soucie, E. L. & Penn, L. Z. (2002) Adv. Cancer Res. 84, 81–154. [DOI] [PubMed] [Google Scholar]
- 5.Wanzel, M., Herold, S. & Eilers, M. (2003) Trends Cell Biol. 13, 146–150. [DOI] [PubMed] [Google Scholar]
- 6.Prochownik, E. V. & Kukowska, J. (1986) Nature 322, 848–850. [DOI] [PubMed] [Google Scholar]
- 7.Stern, D. F., Roberts, A. B., Roche, N. S., Sporn, M. B. & Weinberg, R. A. (1986) Mol. Cell. Biol. 6, 870–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prochownik, E. V., Kukowska, J. & Rodgers, C. (1988) Mol. Cell. Biol. 8, 3683–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prendergast, G. C. (1999) Oncogene 18, 2967–2987. [DOI] [PubMed] [Google Scholar]
- 10.Yin, X. Y., Grove, L., Datta, N. S., Katula, K., Long, M. W. & Prochownik, E. V. (2001) Cancer Res. 61, 6487–6493. [PubMed] [Google Scholar]
- 11.Fest, T., Mougey, V., Dalstein, V., Hagerty, M., Milette, D., Silva, S. & Mai, S. (2002) Oncogene 21, 2981–2990. [DOI] [PubMed] [Google Scholar]
- 12.Iritani, B. M., Delrow, J., Grandori, C., Gomez, I., Klacking, M., Carlos, L. S. & Eisenman, R. N. (2002) EMBO J. 21, 4820–4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vafa, O., Wade, M., Kern, S., Beeche, M., Pandita, T. K., Hampton, G. M. & Wahl, G. M. (2002) Mol. Cell 9, 1031–1044. [DOI] [PubMed] [Google Scholar]
- 14.Schorl, C. & Sedivy, J. M. (2003) Mol. Biol. Cell 14, 823–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maines, J. Z., Stevens, L. M., Tong, X. & Stein, D. (2004) Development (Cambridge, U.K.) 131, 775–786. [DOI] [PubMed] [Google Scholar]
- 16.Holtta, E., Auvinen, M., Paasinen, A., Kangas, A. & Andersson, L. C. (1994) Biochem. Soc. Trans. 22, 853–859. [DOI] [PubMed] [Google Scholar]
- 17.Hermeking, H., Rago, C., Schuhmacher, M., Li, Q., Barrett, J. F., Obaya, A. J., O'Connell, B. C., Mateyak, M. K., Tam, W., Kohlhuber, F., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 2229–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wood, L. J., Mukherjee, M., Dolde, C. E., Xu, Y., Maher, J. F., Bunton, T. E., Williams, J. B. & Resar, L. M. (2000) Mol. Cell. Biol. 20, 5490–5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newbold, R. F. (2002) Mutagenesis 17, 539–550. [DOI] [PubMed] [Google Scholar]
- 20.Nikiforov, M. A., Chandriani, S., O'Connell, B., Petrenko, O., Kotenko, I., Beavis, A., Sedivy, J. M. & Cole, M. D. (2002) Mol. Cell. Biol. 22, 5793–5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teng, S. C., Chen, Y. Y., Su, Y. N., Chou, P. C., Chiang, Y. C., Tseng, S. F. & Wu, K. J. (2004) J. Biol. Chem. 279, 14649–14655. [DOI] [PubMed] [Google Scholar]
- 22.Packham, G. & Cleveland, J. L. (1995) Curr. Top. Microbiol. Immunol. 194, 283–290. [DOI] [PubMed] [Google Scholar]
- 23.Haas, K., Staller, P., Geisen, C., Bartek, J., Eilers, M. & Moroy, T. (1997) Oncogene 15, 179–192. [DOI] [PubMed] [Google Scholar]
- 24.Nesbit, C. E., Tersak, J. M., Grove, L. E., Drzal, A., Choi, H. & Prochownik, E. V. (2000) Oncogene 19, 3200–3212. [DOI] [PubMed] [Google Scholar]
- 25.Yin, X., Grove, L., Rogulski, K. & Prochownik, E. V. (2002) J. Biol. Chem. 277, 19998–20010. [DOI] [PubMed] [Google Scholar]
- 26.Rothermund, K., Rogulski, K., Fernades, E., Whiting, A., Sedivy, J., Pu, L. & Prochownik, E. V. (2005) Cancer Res. 65, 2097–2107. [DOI] [PubMed] [Google Scholar]
- 27.Li, C. & Wong, W. H. (2001) Proc. Natl. Acad. Sci. USA 98, 31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corcoran, D. L., Feingold, E. & Benos, P. V. (2005) Nucleic Acids Res. 38, W442–W446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wheeler, D. L., Church, D. M., Edgar, R., Federhen, S., Helmberg, W., Madden, T. L., Pontius, J. U., Schuler, G. D., Schriml, L. M., Sequeira, E., et al. (2004), Nucleic Acids Res. 32, D35–D40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hertz, G. Z. & Stormo, G. D. (1999) Bioinformatics 15, 563–577. [DOI] [PubMed] [Google Scholar]
- 31.Stormo, G. D. (2000) Bioinformatics 16, 16–23. [DOI] [PubMed] [Google Scholar]
- 32.Matys, V., Fricke, E., Geffers, R., Gossling, E., Haubrock, M., Hehl, R., Hornischer, K., Karas, D., Kel, A. E., Kel-Margoulis, O. V., et al. (2003) Nucleic Acids Res. 31, 374–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cohen, B. A., Mitra, R. D., Hughes, J. D. & Church, G. M. (2000) Nat. Genet. 26, 183–186. [DOI] [PubMed] [Google Scholar]
- 34.Caron, H., van Schaik, B., van der Mee, M., Baas, F., Riggins, G., van Sluis, P., Hermus, M. C., van Asperen, R., Boon, K., Voute, P. A., et al. (2001) Science 291, 1289–1292. [DOI] [PubMed] [Google Scholar]
- 35.Li, Q., Peterson, K. R., Fang, X. & Stamatoyannopoulos, G. (2002) Blood 100, 3077–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spitz, F., Gonzalez, F. & Duboule, D. (2003) Cell 113, 405–417. [DOI] [PubMed] [Google Scholar]
- 37.Picard, D. (1994) Curr. Opin. Biotechnol. 5, 511–515. [DOI] [PubMed] [Google Scholar]
- 38.O'Brien, T. G., Megosh, L. C., Gilliard, G. & Soler, A. P. (1997) Cancer Res. 57, 2630–2637. [PubMed] [Google Scholar]
- 39.Shim, H., Dolde, C., Lewis, B. C., Wu, C. S., Dang, G., Jungmann, R. A., Dalla-Favera, R. & Dang, C. V. (1997) Proc. Natl. Acad. Sci. USA 94, 6658–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu, Y., Sumter, T. F., Bhattacharya, R., Tesfaye, A., Fuchs, E. J., Wood, L. J., Huso, D. L. & Resar, L. M. (2004) Cancer Res. 64, 3371–3375. [DOI] [PubMed] [Google Scholar]
- 41.Elkon, R., Zeller, K. I., Linhart, C., Dang, C. V., Shamir, R. & Shiloh, Y. (2004) Nucleic Acids Res. 32, 4955–4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rauscher, F. J., III, Morris, J. F., Tournay, O. E., Cook, D. M. & Curran, T. (1990) Science 250, 1259–1262. [DOI] [PubMed] [Google Scholar]
- 43.Roy, P. J., Stuart, J. M., Lund, J. & Kim, S. K. (2002) Nature 418, 975–979. [DOI] [PubMed] [Google Scholar]
- 44.Spellman, P. T. & Rubin, G. M. (2002) J. Biol. 1, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yi, F., Jaffe, R. & Prochownik, E. V. (2003) Cancer Res. 63, 2923–2932. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.