Abstract
Several experiments have suggested that newly synthesized polypeptide chains can adopt helical structures deep within the ribosome exit tunnel. We hypothesize that confinement in the roughly cylindrical tunnel can entropically stabilize α-helices. The hypothesis is validated by using theory and simulations of coarse-grained off-lattice models. The model helix, which is unstable in the bulk, is stabilized in a cylindrical cavity provided the diameter (D) of the cylinder exceeds a critical value D*. When D < D* both the helical content and the helix–coil transition temperature (Tf) decrease abruptly. Surprisingly, we find that the stability of the α-helix depends on the number (N) of amino acid residues. Entropic stabilization, as measured by changes in Tf, increases nonlinearly as N increases. The simulation results are in quantitative agreement with a standard helix–coil theory that takes into account entropy cost of confining a polypeptide chain in a cylinder. The results of this work are in qualitative accord with most of the findings of a recent experiment in which N-dependent ribosome-induced helix stabilization of transmembrane sequences was measured by fluorescence resonance energy transfer.
Keywords: confinement effects, cylindrical pores, entropic stabilization, folding
The spectacular structure of the ribosome complex has revealed that a newly synthesized polypeptide emerges through a long, roughly cylindrical exit tunnel (1, 2). The structure of the exit tunnel, which was first observed in the mid-1980s (3, 4), has been elucidated in atomic detail using 50S ribosomes from Haloarcula marismortui (1) and Deinococcus radiodurans (2). A newly synthesized polypeptide chain traverses a distance of ≈100 Å from the peptidyl transferase center deep inside the ribosome complex to the outer surface of the ribosome. The lateral dimension of the tunnel is between 10 and 20 Å, depending on the location along the tunnel axis (1, 2). This width is just enough for a nascent polypeptide chain to form secondary structure, namely an α-helix. The possibility of secondary structure formation inside the ribosome exit tunnel has been suggested in a number of experimental studies on Escherichia coli ribosomes (5) as well as eukaryotic ribosomes (6–9). Furthermore, recent cryo-EM studies showed that a translating ribosome tunnel of E. coli expands during protein synthesis (5), which suggests that α-helix formation inside the ribosome tunnel is plausible.
Recently, Johnson and coworkers (8) used fluorescent resonance energy transfer (FRET) to show that a transmembrane sequence (TMS) adopts a compact conformation, consistent with α-helical structure, deep within the ribosome exit tunnel. The FRET efficiency decreased when the TMS emerged from the tunnel, which was interpreted as unfolding of the peptide in the bulk. It also was shown that the intratunnel FRET efficiency of a peptide with a shorter TMS, as well as of a peptide from a soluble protein, is low. Woolhead et al. (8) concluded from these observations that the ribosome-induced structure may be a complex function of the peptide–tunnel interactions and the sequence length.
Despite these insightful experiments, the mechanism of the putative formation of α-helices within the ribosome tunnel remains unclear. The tunnel walls are made mostly of rRNA, and although several ribosomal proteins penetrate into the tunnel, no apparent hydrophobic patches, low-polarity areas, or structural motifs have been identified (1, 2). Thus, it is unlikely that there are favorable interactions between the newly synthesized protein and the tunnel cavity, except perhaps at specific locations along the tunnel (6, 8, 10, 11). If this is the case, then persistent secondary structure inside the exit tunnel is merely a result of the geometry of the tunnel. Hence, α-helices may be entropically stabilized. Specific ribosome-peptide interactions may only play a secondary role in structure formation, although they can be important in recognition, signaling, and elongation processes (12–14).
In this work we test the entropic-stabilization hypothesis theoretically and answer the following questions that are in part inspired by the Johnson experiments (8):
What causes a thermodynamically unstable α-helix to fold inside the ribosome exit tunnel?
What is the role of the sequence length, i.e., the number of amino acid residues (N) in α-helix formation?
To validate the proposal that confinement causes α-helix formation within the ribosome, we perform both analytical and numerical studies of the helix–coil transition in a cylinder, which approximately mimics the geometry of the exit tunnel. Comparison of the stability of a simple off-lattice model in the bulk and in cylinders of various sizes shows a remarkable change in the helical content as the extent of confinement increases. This comparison allows us to show that the experimental observations of Woolhead et al. (8), including the role of sequence length, can be qualitatively rationalized using the entropic stabilization hypothesis.
Methods
Theory of Confinement-Induced α-Helix Formation. The thermodynamic equilibrium between the helix and the coil states of a polypeptide chain is determined by the difference in the Gibbs free energy between the two
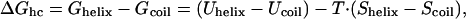 |
[1] |
where Uhelix (Ucoil) is the enthalpy of the helix (coil) state and Shelix (Scoil) is the entropy of the helix (coil) state. For simplicity, we normalize all quantities by a, the unit of length, which is set to the average Cα distance (≈3.8 Å), and calculate energy per residue. The difference between the two enthalpic terms is mainly due to the formation of hydrogen bonds between residues i and i + 4 in the helix state. If ΔGhc < 0, the helical state is thermodynamically stable, whereas for ΔGhc > 0 the coiled state is the more stable one. Confinement of the polypeptide within a cylinder with inert walls does not affect the enthalpic terms in Eq. 1. Thus, ΔGhc changes upon confinement only because of changes in the entropy of the helix and coil states. As can be seen in Fig. 1, the confinement-induced reduction in the number of allowed conformations of the coil state is far larger than that of the (less flexible) helix state. We model the change in the coil-configurational entropy, ΔScoil, using the energy cost per unit length of a confined self-avoiding random walk (SAW) (15)
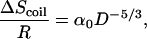 |
[2] |
where D is the cylinder diameter, α0 is a constant on the order unity, and R is the universal gas constant. For the helical state we use the results of Lamura et al. (16), who showed that the entropy cost per unit length of a helical polymer with persistence length lp, pitch p, and diameter d0 is the same as that of a semiflexible chain with an effective persistence length 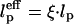 , where
, where 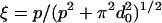 , confined in a cylinder with effective diameter Deff, Deff ≃ D – d0 for P « D and Deff ≈ D for p » D
, confined in a cylinder with effective diameter Deff, Deff ≃ D – d0 for P « D and Deff ≈ D for p » D
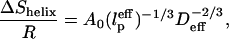 |
[3] |
where A0 ≃ 2.375 (17). In our model α-helix p ≈ 2.1 and d0 ≈ 1.3, so that ξ ≈ 0.45. Because in our simulations p is neither much larger nor much smaller than D, we set Deff ≃ D – f·d0, where 0 ≤ f ≤ 1 is a free parameter.
Fig. 1.
Confinement effects on conformational entropy of a polypeptide chain. In the coiled state, confining the peptide to a cylinder of diameter D << RF (where RF is the Flory radius) excludes many configurations shown as gray lines. On the other hand, the rigid α-helix for which D « lp (where lp is the α-helix persistence length) loses only a few conformations, which makes the loss in entropy much smaller.
Using Eqs. 2 and 3 the confinement-induced change in the energy difference between helix and coil states, ΔΔGhc, becomes
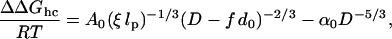 |
[4] |
where α0, f, and lp are free parameters, and T is the temperature. For an infinitely rigid α-helix, lp → ∞, we expect f → 1, and hence ΔΔGhc/RT = –α0D–5/3 for D > d0 and ΔΔGhc/RT > 0 for D ≈ d0. Thus, the helical state is entropically stabilized by confinement as long as D > D* ≈ d0. For an α-helix with a large but finite persistence length, ΔΔGhc < 0 for large D and increases sharply as D < D*. Therefore, for a peptide that is unstable in solution, cylindrical confinement may render the helix state thermodynamically stable.
The arguments given above ignore intermediates that must be included in light of the noncooperative nature of the helix–coil transition (18–20). To incorporate intermediates into our theory, we use a simple model (21) in which each residue has two possible states, helix or coil, with thermodynamic weights u and v, respectively. The overall weight (i.e., the unnormalized probability) for a particular conformation is given by the product of the individual weights. Because of the lack of interactions between residues in this model, the average helical content, i.e., the average fraction of helical residues, is just θ = u/(v + u) ≡ s/(1 + s) where s ≡ u/v. The Gibbs free energy of Eq. 4 is introduced into the model by writing sc = sbulk exp(–ΔΔGhc/RT), with sc and sbulk the values of the parameter s in confinement and in bulk, respectively. Thus one gets for the confined peptide
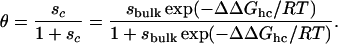 |
[5] |
Because θ is a monotonically increasing function of sc, and since ΔΔGhc < 0 for D > D*, it follows that the helical content should increase upon confinement.
Off-Lattice Simulations. We studied numerically the effect of confinement on α-helix formation by performing Langevin dynamics simulations. A coarse-grained representation of the polypeptide chain was used, in which only the positions of the Cα carbons are retained, and each residue is either hydrophobic (B) or hydrophilic (L) (22). Most simulations were conducted with an N = 16 residue sequence L(LBLLBBL)2L, but we also performed simulations with N = 9 [LLBLLBBLL], N = 30 [L(LBLLBBL)4L], and N = 47 [L(LBLLBBL)6LBLL]. The hydrogen bonds between the backbone carbonyl oxygen CO and the amine hydrogen NH groups were mimicked by using virtual moieties located between backbone α-carbons (19). The potential energy of a conformation, represented by the set of α-carbons positions {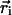 } (i = 1, 2,..., N), was E({
} (i = 1, 2,..., N), was E({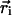 }) = VBL + VBA + VDIH + VHB + VNON + VCYL, where VBL, VBA, VDIH, VHB, and VNON are bond-length potential, bond-angle potential, dihedral angle potential, hydrogen-bond potential, nonbonded long-range potential, and cylindrical-confinement potential, respectively. The forms of the potentials VBL, VBA, VHB, and VNON are given in ref. 19. We used the dihedral angle potential VDIH
}) = VBL + VBA + VDIH + VHB + VNON + VCYL, where VBL, VBA, VDIH, VHB, and VNON are bond-length potential, bond-angle potential, dihedral angle potential, hydrogen-bond potential, nonbonded long-range potential, and cylindrical-confinement potential, respectively. The forms of the potentials VBL, VBA, VHB, and VNON are given in ref. 19. We used the dihedral angle potential VDIH
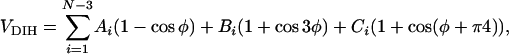 |
[6] |
where A1 = B1 = C1 = εh. The unit of energy in our simulations, εh, (≈1.2 kcal/mol), corresponds to the minimum of interaction potential of two B residues. This form of the dihedral angle potential leads to a stable four-helix bundle in the bulk (23), but an isolated α-helix is unstable (19). We chose this destabilized model to test whether confinement inside a cylinder can shift the thermodynamic equilibrium of this model polypeptide.
For all simulations in confined space, we used a confining potential with cylindrical symmetry, VCYL,
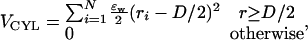 |
[7] |
where D is the cylinder diameter, and 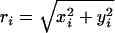 is the distance of residue i from the z axis. The strength of the confinement potential (εW) is taken as εW = 100εh. All simulation observables were normalized by the unit of length a (≈3.8 Å). Temperature was measured in units ofεh/R.
is the distance of residue i from the z axis. The strength of the confinement potential (εW) is taken as εW = 100εh. All simulation observables were normalized by the unit of length a (≈3.8 Å). Temperature was measured in units ofεh/R.
We used simulated annealing to perform simulations in the bulk (i.e., without VCYL) and in cylinders with varying D, carrying out 60 runs under each condition. Each annealing simulation was preceded by heating the polypeptide chain to T = 4 followed by a temperature quench to T = 1.2 to randomize the initial conditions. The temperature was then lowered by 0.02 every 1,000,000 time-steps, where each time-step (τ) is ≈3 ps (19). After each temperature change the system was equilibrated for 500,000 steps before collecting the averages of physical observables. Simulations were performed by using the velocity form of the Verlet algorithm at a value of friction coefficient ζ = 0.05τ–1 and a time integration step h = 0.005τ. All thermodynamic averages were calculated by using the multiple histogram method (24).
Analysis. To evaluate the state of the polypeptide chain during the simulation we calculated the number of helical dihedral angles (nHD) for each saved configuration. A dihedral angle was considered to be helical if it deviates by at most δφ ≈ 10° from its value in the helix state. The helical content θ of a conformation was defined as θ = nHD/(N – 3), where N – 3 is the total number of dihedral angles. The folding temperature Tf was determined by the condition 〈θ 〉Tf, where 〈... 〉T is the thermodynamic average at temperature T, and 〈θ 〉 is the average helical content.
Results
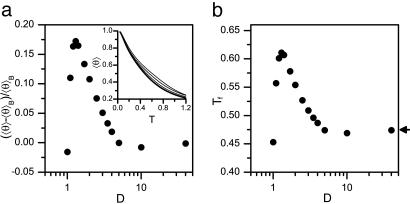
Cylindrical Confinement Induces α-Helix Formation. Comparison of the temperature dependence of 〈θ 〉 for a peptide in the bulk and in cylinders with decreasing D shows a substantial increase in 〈θ 〉 as D decreases (Fig. 2a Inset). At a fixed temperature T = 0.5, corresponding to ≈300 K, 〈θ 〉 increases by as much as 17% when going from bulk to D = 1.3 (Fig. 2a), which is essentially the native α-helix diameter. It is surprising to find such a large increase, especially because the sequence is inherently unstable in the bulk. Below D = 1.3 the helix becomes too large to fit inside the cylinder, and 〈θ 〉 decreases sharply. When D ≤ 1, α-helix formation does not occur even at very low temperatures. The folding temperature Tf exhibits a similar behavior, increasing from ≈285 K in the bulk to ≈360 K for D = 1.3 and decreasing abruptly as D decreases (Fig. 2b). Thus, our model α-helix, which does not fold in the bulk at ambient temperature (i.e., T ≈ 300 K), can readily fold when confined inside a cylinder with D > D* ≈ 1. Below D*, however, there is an abrupt destabilization of the helix as the polypeptide undergoes a helix-stretched coil transition. These results are in qualitative accord with the theoretical arguments presented above.
Fig. 2.
Confinement-induced α-helix formation. (a) Change of mean helical content 〈θ 〉 of an N = 16 helix model relative to mean helical content in the bulk 〈θ 〉B, as a function of cylinder diameter at T = 0.5 (≈300 K). For D > 1.3 〈θ 〉 is larger than in the bulk, i.e., the α-helix is stabilized by the cylinder. Below D ≈ 1.3, 〈θ 〉 decreases sharply. (Inset) Temperature dependence of 〈θ 〉 in the bulk (thick line) and in cylinders of diameter D = 3, 2, 1.4 (thin lines, bottom to top). (b) The folding temperature Tf as a function of D. Tf increases remarkably up to D ≈ 1.3. Arrow indicates folding temperature in the bulk.
Although thermodynamic stability increases upon confinement, the transition becomes less cooperative as D decreases, as observed in the more gentle decay of 〈θ 〉 with increasing temperature. This finding is consistent with previous studies of protein folding in various confining geometries (25, 26), which showed that folding cooperatively decreases in restricted spaces.
To further investigate the origin of confinement-induced α-helix formation, we analyze our simulation results by using the theoretical model (see Methods). We calculate from the simulations the distribution of the number of helical dihedral angles nHD at T = 0.5 in the bulk and for various values of D. We then fit this value to the distribution function P(nHD) of our theoretical model
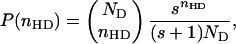 |
[8] |
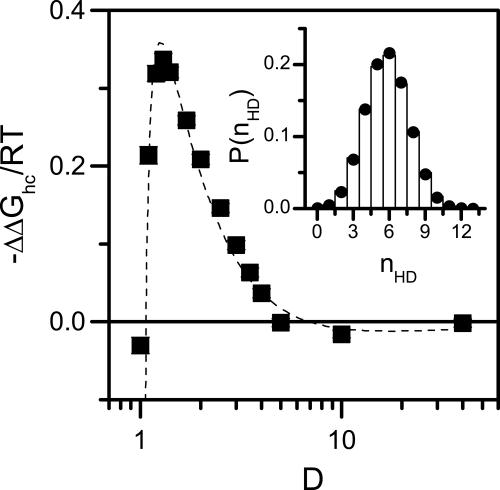
where ND = N – 3 is the total number of dihedral angles. An example of such a fit is shown in Fig. 3 Inset. From the ratio between sc for a peptide confined in a cylinder of diameter D and sbulk calculated from a simulation in the bulk we compute the free energy difference ΔΔGhc as a function of D (Fig. 3). We find that the D-dependence of ΔΔGhc is well fitted using Eq. 4. The persistence length obtained from the fit is lp = 230 ± 72. This value agrees well with a recent calculation of the elastic properties of long α-helices (27), which predicted lp ≈ 263 (=100 nm), although other authors quote a somewhat smaller number (28). Thus, entropic interactions are sufficient to account for the thermodynamic stabilization of the helical state under cylindrical confinement observed in our simulations.
Fig. 3.
Helix stabilization is entropic in nature. The change in the Gibbs free energy difference between helix and coil states, ΔΔGhc, as a function of D at T = 0.5. α-Helix formation is favored when D is larger than the diameter of the native helix (≈1.3). For smaller D the helix state cannot fit within the cylinder, and ΔGhc increases sharply. For D < 1 helix does not form even at T = 0. Dashed line is a fit to Eq. 4 with fitted parameters α0 = 3.9 ± 0.2, f = 0.778 ± 0.005, and lp = 230 ± 72. (Inset) Comparison of measured (bars) and calculated (circles) probability distributions of helical dihedral angles P(nHD) in a cylinder with D = 1.4. Circles are calculated by fitting Eq. 8 to the simulation data obtaining s = 1.28 ± 0.01.
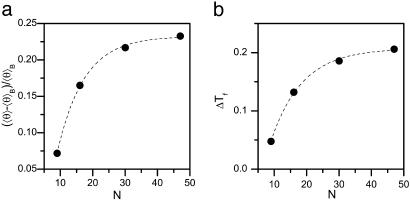
Helix Stability in a Cylinder Depends on Sequence Length. Our analytical model ignores finite-length effects, such as the increased energetic cost of α-helix formation at the chain termini. It is likely that these effects play some role in short peptides. To evaluate the role of peptide length, we performed simulations with N varying from 9 to 47 residues. For all of the peptides in the bulk, there is no apparent effect of N on 〈θ 〉. On the other hand, upon confining the peptides to a cylinder with D = 1.4, 〈θ 〉 increases with N (see Fig. 4a), indicating stronger stabilization of the helical state for longer peptides. In addition, the folding temperature Tf, which is the same for all peptides in the bulk, increases with N (Fig. 4b).
Fig. 4.
Confinement-induced α-helix formation is length-dependent. (a) The mean helical content 〈θ 〉, calculated for D = 1.4 and T = 0.5 relative to the bulk value, 〈θ 〉B, increases with N. (b) Change of folding temperature ΔTf upon confinement in a cylinder of D = 1.4 at T = 0.5. In the bulk Tf ≈ 0.48 for all peptides. Dashed lines are guides to the eye.
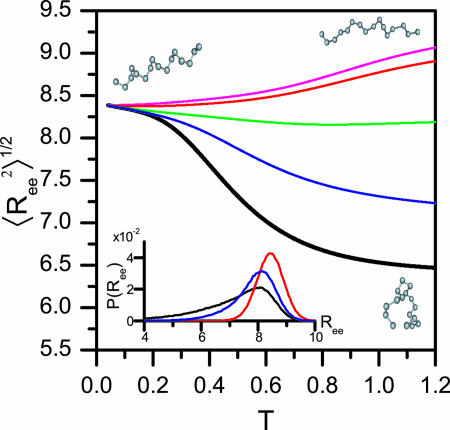
Coiled State Undergoes a Structural Transition upon Confinement. Plotting the root-mean-squared (rms) end-to-end distance 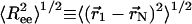 as a function of temperature in the bulk (black line) and in cylinders of different sizes (Fig. 5), we observe a confinement-induced structural transition in the coiled state even at high T. For a peptide in the bulk,
as a function of temperature in the bulk (black line) and in cylinders of different sizes (Fig. 5), we observe a confinement-induced structural transition in the coiled state even at high T. For a peptide in the bulk, 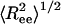 decreases the peptide unfolds. This when result is because
decreases the peptide unfolds. This when result is because 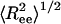 for an α-helix scales with N, the number of residues, whereas in the coiled state it scales as Nν with ν = 3/5. In contrast,
for an α-helix scales with N, the number of residues, whereas in the coiled state it scales as Nν with ν = 3/5. In contrast, 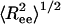 increases upon unfolding in cylindrical confinement, because unlike in the bulk the coil state can adopt only extended conformations, such as the one shown in the upper right corner of Fig. 5. This increase is clearly observed in the distributions of Ree plotted in Fig. 5 Inset. We also observe a significant decrease in the fluctuations of Ree, manifested by a decrease in its standard deviation from 1.25 in the bulk to ≈0.5 in a cylinder with D = 1.3.
increases upon unfolding in cylindrical confinement, because unlike in the bulk the coil state can adopt only extended conformations, such as the one shown in the upper right corner of Fig. 5. This increase is clearly observed in the distributions of Ree plotted in Fig. 5 Inset. We also observe a significant decrease in the fluctuations of Ree, manifested by a decrease in its standard deviation from 1.25 in the bulk to ≈0.5 in a cylinder with D = 1.3.
Fig. 5.
Structural transition upon confinement. The rms end-to-end distance 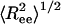 in cylindrical confinement as a function of temperature in the bulk (black line) and in cylinders of diameter D = 3 (blue), 2 (green),1.4 (red), and 1.3 (magenta). At low temperature the peptide adopts a helical conformation (drawn in the left-hand side). At the high T limit in the bulk, the peptide samples many conformations, with either small
in cylindrical confinement as a function of temperature in the bulk (black line) and in cylinders of diameter D = 3 (blue), 2 (green),1.4 (red), and 1.3 (magenta). At low temperature the peptide adopts a helical conformation (drawn in the left-hand side). At the high T limit in the bulk, the peptide samples many conformations, with either small 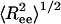 (such as the conformation drawn in bottom right corner) or with ends far away (such as the conformation drawn in top right corner). For the confined peptide, only extended conformations are observed, signifying a random coil → stretch structural transition upon confinement. (Inset) The end-to-end distance distribution function, P(Ree) in the bulk (black line) and in cylinders with diameter D = 3 (blue line) and D = 1.4 (red line). The fluctuations in the end-to-end distance are suppressed as D decreases, and the distributions sharpen.
(such as the conformation drawn in bottom right corner) or with ends far away (such as the conformation drawn in top right corner). For the confined peptide, only extended conformations are observed, signifying a random coil → stretch structural transition upon confinement. (Inset) The end-to-end distance distribution function, P(Ree) in the bulk (black line) and in cylinders with diameter D = 3 (blue line) and D = 1.4 (red line). The fluctuations in the end-to-end distance are suppressed as D decreases, and the distributions sharpen.
Conformational Sampling Decreases upon Confinement. To evaluate the effects of confinement on the helix–coil transition kinetics, we calculate the “ergodic measure” Ω for the helical content (29). The ergodic measure corresponds to the time scale of diffusion on the conformational energy landscape, projected on the chosen reaction coordinate (in this case θ). To calculate Ω we first define
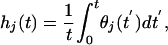 |
where θj(t) is the helical content of trajectory j at time t. By using this definition, an ergodic measure is calculated (29), and a energy-landscape diffusion coefficient dE is obtained from the asymptotic behavior
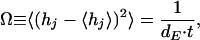 |
where the averages are performed over all trajectories.
We calculated dE for different size cylinders at T = 0.5 and normalized it by the respective quantity in the bulk, dE,bulk. The resulting diffusion coefficients are dE/dE,bulk = 2 ± 0.05 for D = 1.4, dE/dE,bulk = 1.3 ± 0.03 for D = 1.7, and dE/dE,bulk ≈ 1 for larger cylinders. Thus, we observe an increase in the time needed to cover phase-space for small cylinders, which quickly approaches the bulk time scale for cylinders larger then D ≈ 2.
Discussion
In this work, we have proposed and theoretically validated the hypothesis that cotranslational α-helix formation within the ribosome exit tunnel is entropically driven. Both our analytical arguments and numerical simulations indicate that confinement within a cylindrical space can stabilize the helical states of polypeptide chains. This stabilization effect is purely entropic in nature and is a consequence of the severe restriction of the conformational space of the coil states inside the cylinder.
The simulation results are quantitatively explained by a model for the helix–coil transition, which takes into account only the relative reduction of entropy in the coil and helix states. Confinement not only stabilizes the helical state but also has an effect on the coil state. Coil conformations that cannot be accommodated within the cylinder are disallowed, which leaves only those that make the chain much more extended even at high temperatures compared with the bulk. This structural transition is accompanied by a significant reduction in chain dynamics. The latter is evidenced by a narrowing of the end-to-end distance distribution, as well as an increase in the time scales for navigation of conformational space. These findings are consistent with an intuitive picture of confined kinetics, where entropic barriers become larger as the confining dimensions become smaller.
The length dependence of the confinement-induced α-helix formation, found in the simulations, is somewhat surprising and cannot be understood within the framework of our simple analytical model. Previous experimental studies have shown a weak length dependence of the helix–coil transition in solution (30, 31). Compared with intrinsic helical propensity and entropic stabilization in confined spaces, we expect that the dependence of helix formation on N should be a secondary effect. In our model the probability of finding each residue in a helical conformation does not significantly depend on N. The interesting outcome of the simulations under confinement is that long peptides are more likely to adopt a helical conformation upon confinement than short peptides. Thus, the simulations indicate that additional finite-size effects become relevant in confined space. The confinement-induced α-helix stabilization also depends on the helix-forming propensity of the sequence that in our model is parameterized by the dihedral-angle potential VDIH (Eq. 6); a model peptide that does not form an α-helix in bulk shows no helix formation upon confinement (data not shown). This finding may partially explain the origin of the experimental observation of sequence-dependent structure formation inside the ribosome exit tunnel.
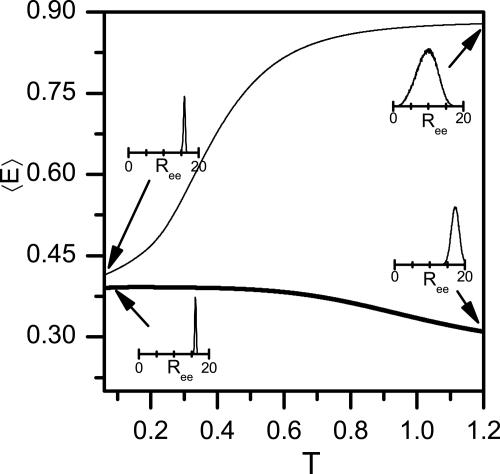
We explore the implication of our work for the experimental results, especially those of Johnson and coworkers (8), who measured the FRET efficiency between probes attached to the two ends of an α-helix forming peptide. By using the probability distribution of the end-to-end distance P(Ree), we calculated the average FRET efficiency (〈E 〉)
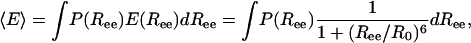 |
[9] |
where R0 is the Förster distance, i.e., the distance at which 〈E 〉= 0.5. The FRET efficiency dependence on temperature is plotted in Fig. 6 for a peptide of N = 30, using R0 = 15 (≈57 Å), which is similar to the parameter used by Woolhead et al. (8). In the bulk 〈E 〉 grows with temperature. This assumption is understood based on arguments presented above. Indeed, we showed that the rms end-to-end distance decreases upon transition from helix to coil, and because conformations with short distance contribute significantly to 〈E 〉, the latter should increase strongly. On the other hand, upon confinement the coil state is more extended than the helix state, and therefore 〈E 〉 shows a decrease at high temperatures.
Fig. 6.
Förster energy transfer for a peptide with N = 30. The average FRET efficiency 〈E 〉, calculated from the simulations according to Eq. 10, and plotted as function of temperature in the bulk (thin line) and under confinement in a cylinder of D = 1.4 (thick line). Insets show P(Ree) at the indicated points. At low temperatures both bulk and confined peptides are helical, whereas at high temperatures the bulk peptide is a random coil with a high 〈E 〉, but the confined peptide is relatively extended, with low 〈E 〉.
Our simulations also showed that short peptides are less affected by confinement. This finding implies that a peptide with a short TMS sequence (but with the same overall number of amino acids), will have a larger end-to-end distance and a lower FRET efficiency than a full TMS peptide. This conclusion is in qualitative agreement with the results of Woolhead et al. (8) for peptides within the ribosome exit tunnel. Surprisingly, however, they also found that the helical peptide, when released into solution, shows low FRET efficiency. However, our simulations, as well as simple arguments based on polymer theory, suggest that the FRET efficiency should increase in the bulk. This discrepancy calls for further experimental and theoretical work.
Although our simulations indicate that entropic stabilization can explain α-helix formation within the ribosome exit tunnel, we cannot entirely rule out a role for specific interactions, e.g., with ribosomal proteins forming part of the tunnel walls. Such interactions would explain the cross-linking experimental results of Woolhead et al. (8). Although possibly not contributing to the formation of secondary structure elements, these interactions may play a role in signaling events related to chain elongation and communication with the translocon (12–14). This interesting conjecture awaits experimental verification.
Coarse-grained model for the polypeptide chain used in this study have yielded qualitative insights into the folding of secondary and tertiary structure elements of proteins (32). For this model we find that α-helices are most stabilized in a cylindrical cavity with diameter of ≈8.3Å(= 1.3 × 3.8 Å + 2 × 1.7 Å, where we added two van der Waals radii of the Cαs), which is about the narrowest diameter found in the ribosome exit tunnel. Our coarse-grained model neglects side chains, which might change the results in two ways: First, the diameter of the α-helix will increase, but this increase is expected to only shift the curves of Figs. 2 and 3, so that the maximum effect occurs at a larger D. Second, specific amino acid side chains might contribute to further stabilization or destabilization of the helical state. Another simplification of our simulations is the use of a straight, constant-diameter cylinder to represent the ribosome exit tunnel. The simulations show, however, that the helical state is stabilized over a range of tunnel diameters, so that even as the tunnel diameter changes along its length the helix-forming propensity would not change by much. Further, the natural flexibility of α-helices [leading to bent helices such as those appearing in coiled coils (33)] is likely to accommodate tunnel bending. We also have not included solvent effects explicitly. It is possible that preferential interactions of the polypeptide chains with water in confined space can influence helix stability. Indeed, a recent work (E. J. Sorin and V. S. Pande, private communication) suggests that a helix confined to carbon nanotubes, whose surface is apolar, is destabilized as a result of confinement-induced changes in solvent entropy. It is unclear whether similar arguments apply to folding in the exit tunnel of ribosome whose surface is polar.
Conclusions
We showed in this work, using simulations of coarse-grained off-lattice peptide models, that confinement within the ribosome exit tunnel can cause the formation of secondary structure in the nascent polypeptide chain. The diameter of the exit tunnel is very close to that of an α-helix, and our simulations showed that in this case the confinement effect is maximized. We also found that longer helix-forming peptides are more stabilized by confinement than shorter ones. This finding has implications for the possible timing of cotranslational secondary structure formation within the exit tunnel, which may only initiate after a long-enough peptide was synthesized.
Our findings are in accord with recent experimental work (5–9). Consequently, the simplest proposition that long helices can be entropically stabilized in the tunnel is sufficient to rationalize many of the available experimental data, without invoking specific interactions with the tunnel walls. It will be interesting to understand the possible implications of confinement-induced secondary structure formation for cotranslational folding of proteins within the ribosome exit tunnel.
Acknowledgments
We thank A. Bashan and A. Yonath for useful discussions. This work was supported in part by U.S.-Israel Binational Science Foundation and National Science Foundation Grant NSF CHE-05-14056.
Author contributions: G.H. and D.T. designed research; G.Z. performed research; G.Z. and D.T. analyzed data; and G.Z., G.H., and D.T. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: TMS, transmembrane sequence.
References
- 1.Nissen, P., Hansen, J., Ban, N., Moore, P. B. & Steitz, T. A. (2000) Science 289, 920–930. [DOI] [PubMed] [Google Scholar]
- 2.Harms, J., Schluenzen, F., Zarivach, R., Bashan, A., Gat, S., Agmon, I., Bartels, H., Franceschi, F. & Yonath, A. (2001) Cell 107, 679–688. [DOI] [PubMed] [Google Scholar]
- 3.Milligan, R. A. & Unwin, P. N. (1986) Nature 319, 693–695. [DOI] [PubMed] [Google Scholar]
- 4.Yonath, A., Leonard, K. R. & Wittman, H. G. (1987) Science 236, 813–816. [DOI] [PubMed] [Google Scholar]
- 5.Gilbert, R. J. C., Fucini, P., Connell, S., Fuller, S. D., Nierhaus, K. H., Robinson, C. V., Dobson, C. M. & Stuart, D. I. (2004) Mol. Cell 14, 57–66. [DOI] [PubMed] [Google Scholar]
- 6.Lu, J. & Deutsch, C. (2005) Biochemistry 44, 8230–8243. [DOI] [PubMed] [Google Scholar]
- 7.Whitley, P., Nilsson, I. & von Heijne, G. (1996) J. Biol. Chem. 271, 6241–6244. [DOI] [PubMed] [Google Scholar]
- 8.Woolhead, C. A., McCormick, P. J. & Johnson, A. E. (2004) Cell 116, 725–736. [DOI] [PubMed] [Google Scholar]
- 9.Mingarro, I., Nilsson, I., Whitley, P. & von Heijne, G. (2000) BMC Cell Biol. 1, 1471-2121/1/3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakatogawa, H. & Ito, K. (2002) Cell 108, 629–636. [DOI] [PubMed] [Google Scholar]
- 11.Amit, M., Berisio, R., Baram, D., Harms, J., Bashan, A. & Yonath, A. (2005) FEBS Lett. 579, 3207–3213. [DOI] [PubMed] [Google Scholar]
- 12.Stroud, R. M. & Walter, P. (1999) Curr. Opin. Struct. Biol. 9, 754–759. [DOI] [PubMed] [Google Scholar]
- 13.White, S. H. & von Heijne, G. (2004) Curr. Opin. Struct. Biol. 14, 397–404. [DOI] [PubMed] [Google Scholar]
- 14.Baram, D. & Yonath, A. (2005) FEBS Lett. 579, 948–954. [DOI] [PubMed] [Google Scholar]
- 15.Grosberg, A. Y. & Khokhlov, A. R. (1994) Statistical Physics of Macromolecules (AIP, New York).
- 16.Lamura, A., Burkhardt, T. W. & Gompper, G. (2004) Phys. Rev. E Stat. Nonlinear Soft Matter Phys. 70, 051804. [DOI] [PubMed] [Google Scholar]
- 17.Bicout, D. J. & Burkhardt, T. W. (2001) J. Phys. A 34, 5745–5750. [Google Scholar]
- 18.Williams, S., Causgrove, T. P., Gillmanshin, R., Fang, K. S., Callender, R. H., Woodruff, W. H. & Dyer, R. B. (1996) Biochemistry 35, 691–697. [DOI] [PubMed] [Google Scholar]
- 19.Klimov, D. K., Betancourt, M. R. & Thirumalai, D. (1998) Folding Des. 3, 481–496. [DOI] [PubMed] [Google Scholar]
- 20.Baumketner, A. & Shea, J. E. (2003) Phys. Rev. E Stat. Nonlinear Soft Matter Phys. 68, 051901. [DOI] [PubMed] [Google Scholar]
- 21.Poland, D. & Scheraga, H. A. (1970) Theory of Helix-Coil Transitions in Biopolymers (Academic, New York).
- 22.Veitshans, T., Klimov, D. & Thirumalai, D. (1997) Folding Des. 2, 1–22. [DOI] [PubMed] [Google Scholar]
- 23.Guo, Z. & Thirumalai, D. (1996) J. Mol. Biol. 263, 323–343. [DOI] [PubMed] [Google Scholar]
- 24.Ferrenberg, A. M. & Swendsen, R. H. (1989) Phys. Rev. Lett. 63, 1195–1197. [DOI] [PubMed] [Google Scholar]
- 25.Takagi, F., Koga, N. & Takada, S. (2003) Proc. Natl. Acad. Sci. USA 100, 11367–11372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klimov, D. K., Newfield, D. & Thirumalai, D. (2002) Proc. Natl. Acad. Sci. USA 99, 8019–8024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choe, S. & Sun, S. X. (2005) J. Chem. Phys. 122, 244912. [DOI] [PubMed] [Google Scholar]
- 28.Chakrabarti, B. & Levine, A. J. (2005) Phys. Rev. E Stat. Nonlinear Soft Matter Phys. 71, 031905. [DOI] [PubMed] [Google Scholar]
- 29.Thirumalai, D., Mountain, R. D. & Kirkpatrick, T. R. (1989) Phys. Rev. A 39, 3563–3574. [DOI] [PubMed] [Google Scholar]
- 30.Ingwall, R. T., Scheraga, H. A., Lotan, N., Berger, A. & Katchalski, E. (1968) Biopolymers 6, 331–368. [DOI] [PubMed] [Google Scholar]
- 31.Scholtz, J. M., Qian, H., York, E. J., Stewart, J. M. & Baldwin, R. L. (1991) Biopolymers 31, 1463–1470. [DOI] [PubMed] [Google Scholar]
- 32.Thirumalai, D. & Klimov, D. K. (1999) Curr. Opin. Struct. Biol. 9, 197–207. [DOI] [PubMed] [Google Scholar]
- 33.Lupas, A. (1996) Trends Biochem. Sci. 21, 375–382. [PubMed] [Google Scholar]