Abstract
The plant hormone jasmonic acid (JA) activates host defense responses against a broad spectrum of herbivores. Although it is well established that JA controls the expression of a large set of target genes in response to tissue damage, very few gene products have been shown to play a direct role in reducing herbivore performance. To test the hypothesis that JA-inducible proteins (JIPs) thwart attack by disrupting digestive processes in the insect gut, we used a MS-based approach to identify host proteins that accumulate in the midgut of Manduca sexta larvae reared on tomato (Solanum lycopersicum) plants. We show that two JIPs, arginase and threonine deaminase (TD), act in the M. sexta midgut to catabolize the essential amino acids Arg and Thr, respectively. Transgenic plants that overexpress arginase were more resistant to M. sexta larvae, and this effect was correlated with reduced levels of midgut Arg. We present evidence indicating that the ability of TD to degrade Thr in the midgut is enhanced by herbivore-induced proteolytic removal of the enzyme's C-terminal regulatory domain, which confers negative feedback regulation by isoleucine in planta. Our results demonstrate that the JA signaling pathway strongly influences the midgut protein content of phytophagous insects and support the hypothesis that catabolism of amino acids in the insect digestive tract by host enzymes plays a role in plant protection against herbivores.
Keywords: jasmonic acid, proteomics, arginase, threonine deaminase, plant-insect interaction
Higher plants have evolved numerous defensive mechanisms to cope with the threat of phytophagous insects. One common strategy used by species throughout the plant kingdom is the induced expression of foliar compounds that negatively affect herbivore performance (1, 2). This form of plant immunity requires the accumulation of jasmonic acid (JA), which is synthesized from linolenic acid in response to tissue damage (3). JA and its bioactive derivatives and precursors powerfully activate the transcription of a large set of target genes (4, 5). Significant insight into the role of jasmonates in regulating plant defense responses has come from the characterization of mutants that are impaired in the biosynthesis or perception of JA (6, 7). A current challenge in the study of JA-regulated defenses is to determine which transcriptional responses play a direct role in thwarting herbivore feeding, growth, and reproduction.
The response of tomato (Solanum lycopersicum) to mechanical wounding and herbivory has been widely used as a model system in which to study the mechanism of induced resistance (1, 6, 8, 9). The most studied JA-inducible proteins (JIPs) in tomato and other solanaceous plants are proteinase inhibitors (PIs), which are expressed rapidly and systemically in response to wounding (1, 10). Upon consumption of induced tissues by the herbivore, these proteins bind to and inhibit digestive proteases in the insect gut. The negative effect of PIs on herbivore performance is thought to result from a compensatory response by the insect to hyperproduce digestive proteinases, which, in turn, leads to depletion of essential amino acids and reduced growth (11). Decreased growth of herbivores on induced plants cannot be explained solely by the action of PIs but rather involves multiple host compounds that exert a combination of toxic, antinutritive, and antifeedant effects (12, 13). Wound-inducible polyphenol oxidases that covalently modify dietary proteins, for example, work together with PIs to reduce the nutritional quality of ingested plant tissue (14, 15).
The objective of this study was to use a MS-based approach to identify tomato JIPs that accumulate in and interfere with digestive processes in the midgut of Manduca sexta (tobacco hornworm) larvae. We demonstrate that several JIPs significantly alter the protein content of the M. sexta midgut, and that two such proteins, arginase and threonine deaminase (TD), act in the gut to deplete amino acids that are required for insect growth. These findings indicate that host plant enzymes that metabolize essential nutrients in the insect digestive tract play a role in plant resistance to herbivore attack.
Materials and Methods
Biological Materials. Solanum lycopersicum (formerly Lycopersicon esculentum) cv. Castlemart was used as the WT for all experiments, except where otherwise noted. The 35S::Prosystemin (35S::PS), and jasmonic acid-insensitive1 (jai1) lines were backcrossed at least five times to the WT (cv. Castlemart). Plant growth conditions were previously described (16-18). M. sexta eggs were obtained from the Department of Entomology, North Carolina State University (Raleigh, NC). Hatched larvae were reared on artificial diet (Carolina Biological Supply, Burlington, NC) for 4-6 d before transfer to 6-week-old plants. Midguts and frass were obtained from fourth to fifth instar larvae that were actively feeding at time of harvest. Larvae were frozen in liquid nitrogen and stored at -20°C until further use. Frozen larvae were transected on dry ice behind the fourth pair of abdominal appendages and behind the second pair of thoracic appendages. The integument and midgut were dissected to obtain the midgut content.
Liquid Chromatography-Tandem MS (LC-MS/MS)-Based Identification of Midgut Proteins. Total midgut content was ground in liquid nitrogen to fine powder and extracted with 100 mM Tris buffer (pH 7.5) containing 1 mM EDTA, 1% (vol/vol) 2-mercapto-ethanol, and 0.1 mM PMSF. Extracts were centrifuged at 20,000 × g for 10 min. Sixty micrograms of total protein was separated by gel electrophoresis through a 4% SDS-polyacrylamide stacking gel (1.5 cm) and ≈1 cm into a 12% resolving gel. Gels were stained with Coomassie blue, and the protein-stained region of the gel was excised. Proteins within the gel piece were reduced and alkylated (19), followed by digestion with trypsin. Extracted peptides were analyzed with a Proteome X 2.0 LTQ-Fourier transform online multidimensional LC-MS/MS system (Thermo Electron, San Jose, CA). Briefly, the peptides from the ingel digest were buffer-exchanged in a large-volume OMIX C18 SPE pipette tip (Varian) and loaded onto a 100 × 0.32-mm strong cation exchange (SCX) Biobasic column (Thermo Electron). Peptides were sequentially eluted with five NH4Cl steps (0, 20, 60, 250, and 500 mM) from the SCX column onto a 150-μm × 10-cm Biobasic C18 column (Thermo Electron). Peptides were eluted from the C18 column over 50 min with a gradient of 5% B to 80% B (mobile phase A = 0.1% formic acid, mobile phase B = 95% acetonitrile, 0.1% formic acid) at a flow rate of 200 nl/min.
Peptides eluting from the C18 column were directly sprayed into a Thermo Electron LTQ-Fourier transform (FT) MS mass spectrometer. The six most abundant ions in each FT survey scan (100,000 resolution, 3-ppm minimum mass accuracy) were subjected to low-energy collision-induced dissociation, and the resulting fragments were analyzed in the linear ion trap portion of the instrument. The X!-tandem algorithm (20, 21) was used to search MS/MS spectra against 2,614 S. lycopersicum protein sequences found in GenBank as of June 28, 2005. Identifications were considered positive if the protein probability score was P ≤ 0.01 (22).
Enzyme Assays. Midgut protein extracts were prepared as described above. Measurements of protein concentration and assays of arginase activity were conducted as described (23). TD assays were performed according to Sharma and Mazumder (24). Crude protein extracts used for Ile inhibition assays were desalted on a Sephadex G-25 column (Amersham Pharmacia Biosciences) equilibrated with 100 mM Tris·HCl (pH 7.5). l-Ile was added to the assay buffer (150 mM Tris·HCl, pH 9.0/10 mM l-Thr/12 mM KCl) before the addition of enzyme extract, and TD activity was then measured.
Amino Acid and Ammonia Analysis. Levels of free amino acids and ammonia were determined simultaneously with the Waters AccQTag procedure. Isolated midgut content was ground in liquid nitrogen. The frozen powder (≈200 mg) was transferred to a 1.5-ml Eppendorf tube and extracted with 1 ml of a 1:1 mixture of chloroform and water. l-nor-Leu was added as an internal standard. After centrifugation at 20,000 × g for 10 min, the supernatant was diluted 10-fold with H2O and filtered through a 0.45-μm filter (Millipore). Samples (20 μl) were derivatized and analyzed by HPLC as described by the manufacturer. HPLC was performed with a Waters LC system equipped with a model 600 pump, a 2475 fluorescence detector, and a 717-plus autosampler.
Construction of 35S::ARG Transgenic Plants. The tomato ARG2 cDNA (23) was amplified by PCR with the primer set of 5′-TCCCCCGGGGGAGGTTCTTGTAGTAAACAA-3′ and 5′-CGAGCTCGGGGAGGCAAGTTTACAGATAAT-3′. The resulting 1,206-bp PCR product was cloned into the SmaI and SacI sites of binary vector pBI121 (Clontech). This construct, which places ARG2 under the control of the cauliflower mosaic virus 35S promoter, was introduced into tomato (cv. Microtom) cotyledon explants by Agrobacterium-mediated transformation (18). Two lines (35S::ARG#28 and 35S::ARG#39) that showed the highest arginase activity (>200 μmol/mg protein per hr) in healthy unwounded leaves were selected for further analysis. Insect-feeding assays were conducted either with plants from the T2 generation that were homozygous for the transgene (Fig. 3B, experiments b and c) or with plants from the T1 generation that were segregating for the transgene (Fig. 3B, experiments a and d). The presence of the 35S::ARG transgene in all plants was confirmed by PCR with the primer set of 35S-1 (5′-CCTTCGCAAGACCCTTCCTCTAT-3′) and ARG2-S2 (5′-GACATCAGCACCAAGGATATCA-3′), which generated a 1,023-bp product. Both homozygous and hemizygous plants exhibited high constitutive levels of arginase activity (data not shown).
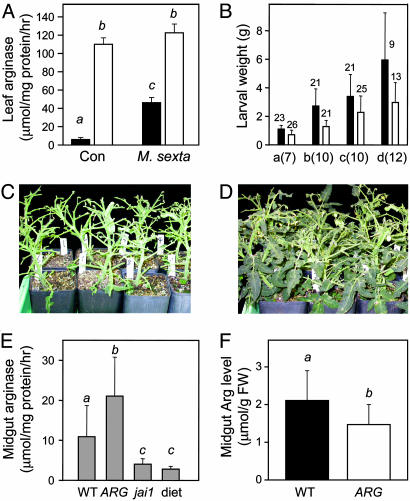
Fig. 3.
Overexpression of arginase in tomato leaves depletes Arg availability in the M. sexta midgut and increases host resistance. (A) Arginase activity in leaves of unwounded control (Con) and M. sexta-damaged (10 days postchallenge) WT (filled bar) and 35S::ARG (open bar) plants. (B) Results of four (a-d) independent feeding trials of M. sexta on WT (filled bar) and 35S::ARG (open bar) plants. Numbers in parentheses indicate the duration (days) of each trial. Significant differences in larval weights (P < 0.01) were observed in all four trials. The number of larvae in each data set is indicated above the bar. Photograph of WT (C) and 35S::ARG (D) plants after feeding by M. sexta larvae for 10 days. (E) Arginase activity in midgut extracts from larvae reared on WT, 35S::ARG (ARG), and jai1 plants, or on artificial diet (diet). (F) Arg accumulation in midguts from larvae reared on WT and 35S::ARG plants. Data in E and F show the mean ± SD (n = 10 larvae). Italicized letters denote significant differences (unpaired Students's t test) at P < 0.05.
Results
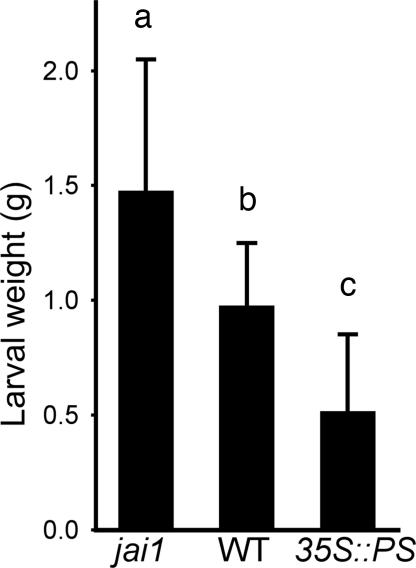
Identification of JA-Inducible Host Proteins in the Insect Midgut. To determine the contribution of the JA signaling pathway to the resistance of tomato to its natural predator, M. sexta, we measured the weight gain of larvae that were reared on WT plants and various mutants altered in JA signaling. The weight gain of larvae grown on WT plants was significantly less (P < 0.001) than that of larvae reared on the jai1-1 mutant that harbors a defective COI1 gene and, as a consequence, is insensitive to JA (16) (Fig. 1). Conversely, larvae performed significantly better (P < 0.001) on WT plants than on a transgenic line (35S::PS), in which defensive JIPs are constitutively expressed as a result of overexpression of prosystemin, a positive regulator of the JA signaling pathway (17, 25, 26). These host genotype-specific differences in larval performance presumably reflect the combined effects of all JA-regulated defenses and provide a starting point for identifying specific phytochemicals that contribute to induced resistance.
Fig. 1.
The JA signaling pathway reduces herbivore performance on tomato. M. sexta larvae were grown on jai1, WT, and 35S::PS plants for 7 days, after which larval weights were determined. Data show the mean ± SD of at least 23 larvae per host genotype. Lowercase letters denote significant differences (unpaired Students's t test) at P < 0.001.
We hypothesized that induced resistance of tomato involves JIPs other than PIs that accumulate in and interfere with the insect digestive system. To test this idea, LC-MS/MS was used to perform a nonbiased survey of the midgut protein content of larvae that were grown on WT, 35S::PS, and jai1 plants. Approximately 67 tomato proteins were confidently identified (P < 0.01) in at least one of the midgut samples (Table 2, which is published as supporting information on the PNAS web site). Among the 25 host proteins identified in 35S::PS midguts, eight (32%) met the following criteria for identification as JIPs: genes encoding the proteins were previously shown to be induced by wounding or JA, and the proteins were not detected in midguts from jai1-reared larvae. Five of these JIPs satisfied the additional criterion of being identified in WT-reared larvae (Table 1). The extent of amino acid sequence coverage and the number of spectral counts obtained for a given protein by LC-MS/MS are strongly correlated with protein abundance (27). Based on this consideration and the normalization of spectral counts to a reference protein (plastocyanin) found in all midgut samples, it was apparent that JIPs were among the most abundant leaf proteins in the midgut of insects fed on induced plants (Tables 1 and 2). Thus, the JA signaling pathway in tomato strongly influences the dietary protein content of M. sexta larvae that feed on this host plant.
Table 1. Summary of JIPs in tomato that were identified in the midgut of M. sexta larvae grown on different host genotypes.
| Spectral counts, percent amino acid coverage
|
|||||
|---|---|---|---|---|---|
| Protein ID | GenBank accession no. | Mr | 35S::PS | WT | jai1 |
| JIPs | |||||
| TD | gi|100257 | 64.9 | 159, 50.9 | 73, 29.6 | ND |
| Leucine aminopeptidase A | gi|2492529 | 60.2 | 111, 71.6 | 49, 40.6 | ND |
| Cathepsin D inhibitor | gi|9581827 | 24.2 | 46, 60.5 | 30, 43.2 | ND |
| Trypsin inhibitor-like | gi|1362094 | 25.2 | 28, 32.9 | 23, 32.4 | ND |
| Arginase2 | gi|54648782 | 36.9 | 51, 57.1 | 2, 7.1 | ND |
| Cysteine protease inhibitor | gi|3452387 | 29.0 | 13, 27.6 | ND | ND |
| Proteinase inhibitor II | gi|31088242 | 16.3 | 4, 25.6 | ND | ND |
| Proteinase inhibitor I | gi|82105 | 12.6 | 2, 14.4 | ND | ND |
| Reference protein | |||||
| Plastocyanin | gi|130271 | 17.0 | 10, 42.9 | 18, 38.2 | 12, 40.0 |
LC–MS/MS was used to identify tomato proteins in the midgut of M. sexta larvae reared on either WT, prosystemin-overexpressing (35S::PS), or jai1 plants. The number of spectral counts (i.e., the number of times the mass spectrometer detected a peptide corresponding to a particular protein) and the percent of the full-length protein sequence that was covered by the peptides (percent amino acid coverage) are shown. Data for plastocyanin (a non-JA-regulated protein) is also shown as an example of a relatively abundant protein that was detected in all three midgut samples. The complete list of tomato proteins identified in each midgut sample is shown in Table 2. ND, not detected.
It could be argued that the tomato proteins identified in the M. sexta midgut are derived from pieces of intact leaf tissue that were not completely degraded by gut digestive enzymes. To address this possibility, we used SDS/PAGE to assess the accumulation of the chloroplast-encoded large subunit of ribulose-1,5 bisphosphate carboxylase-oxygenase (Rubisco) in intact leaves and larval midguts. The results showed that the level of Rubsico accumulation in intact leaves was much greater than that in the midgut protein extracts (Fig. 5, which is published as supporting information on the PNAS web site). This finding provides evidence that bulk tomato protein present in the midgut extracts was exposed to digestive enzymes (e.g., proteases) and thus must have been released from plant cells and organelles before preparation of the extract.
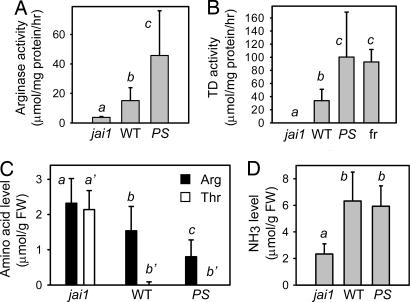
The identified midgut JIPs included several PIs; leucine aminopeptidase A (LAP-A); arginase, which degrades Arg to urea and ornithine; and TD, which metabolizes Thr to α-ketobutyrate and NH3 (Table 1). Because Arg and Thr are dietary requirements for M. sexta and most other phytophagous insects (28), subsequent work was focused on testing the hypothesis that arginase and TD act in the insect gut to deplete these nutrients. Midgut extracts from larvae reared on jai1 plants or artificial diet contained low levels of arginase activity and no detectable TD activity (Fig. 2 A and B; data not shown). Significantly higher levels of both enzyme activities were found in midgut extracts from larvae grown on WT and 35S::PS plants. Thus, arginase and TD retain activity in the M. sexta midgut. High levels of TD activity were also found in the frass of larvae grown on induced but not on jai1 plants (Fig. 2B; data not shown), indicating that this enzyme is remarkably stable in the lepidopteran digestive tract.
Fig. 2.
JA-regulated arginase and TD are active in the M. sexta digestive tract. (A) Arginase and (B) TD activity in midgut extracts from larvae that were grown on jai1, WT, and 35S::PS (PS) plants. Also shown in B is TD activity in frass (fr) from WT-reared larvae. (C) Arg (filled bar), Thr (open bar), and (D) NH3 levels in midgut extracts from the same larvae used in A and B. Data show the mean ± SD of 10 larvae per plant genotype. Italicized letters denote significant differences (unpaired Students's t test) at P < 0.05.
The high-pH optimum of tomato arginase (23) and TD (29) suggested that they are metabolically active in the alkaline environment of the M. sexta midgut. Consistent with this hypothesis, increased arginase and TD activity in midgut extracts was associated with reduced levels of free Arg and Thr in the same extracts (Fig. 2C). We also found that midgut Thr levels were inversely proportional to the level of NH3, which is a product of the TD reaction (Fig. 2D). This observation supports the idea that Thr depletion in the midgut of WT- and 355::PS-reared larvae results from the action of TD. Taken together, the results indicate that active forms of JA-regulated arginase and TD catabolize essential amino acids in the larval digestive tract.
Overexpression of Arginase Increases the Resistance of Tomato to M. sexta Larvae. We used a transgenic approach to determine whether increased expression of foliar arginase is sufficient to affect midgut Arg levels and larval performance. Transgenic plants that overexpress the tomato ARG2 cDNA (23) under the control of the cauliflower mosaic virus 35S promoter were generated by Agrobacterium-mediated transformation. The constitutive level of arginase activity in unwounded leaves of selected 35S::ARG lines far exceeded that in herbivore-damaged WT leaves (Fig. 3A). High arginase activity in these plants did not result in obvious morphological or reproductive phenotypes nor did it significantly alter the level of Arg in 35S::ARG leaves (data not shown). In four independent feeding trials conducted with two 35S::ARG lines, the average weight of larvae grown on transgenic plants was significantly less than that of larvae reared on WT plants (Fig. 3B; P < 0.01). It was also apparent that larvae consumed more foliage from WT than 35S::ARG plants (Fig. 3 C and D). Arginase activity in midgut extracts from 35S::ARG-reared larvae was significantly greater than that in WT-reared larvae, and this activity was associated with reduced levels of midgut Arg (Fig. 3 E and F). These findings indicate that foliar arginase ingested by M. sexta larvae depletes midgut Arg and reduces larval growth.
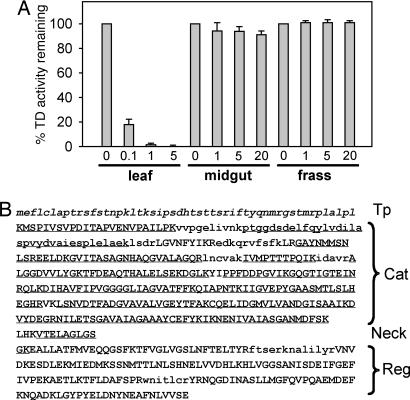
The Midgut Form of Tomato TD Is Insensitive to Feedback Inhibition by Isoleucine. In plants and microorganisms, TD catalyzes the committed step in the biosynthesis of isoleucine (Ile). The enzyme contains an N-terminal catalytic domain and a C-terminal regulatory domain and is subject to negative feedback regulation by Ile (30, 31). The high level of midgut TD activity in larvae grown on induced plants suggested that the regulatory properties of this chloroplastic enzyme might be altered in a way that enhances Thr degradation in the presence of high concentrations of Ile. To test this idea, we compared the sensitivity of leaf-, midgut-, and frass-derived forms of TD to Ile. TD activity in methyl-JA-treated leaves was strongly inhibited by Ile (Fig. 4A). In midgut and frass extracts, however, the enzyme activity was not inhibited by concentrations of Ile up to 20 mM. A likely explanation for this phenomenon came from LC-MS/MS data showing that amino acid sequence coverage of the midgut form of TD mapped exclusively to the N-terminal catalytic domain and the short “neck” region that connects the regulatory and catalytic domains (Fig. 4B). Failure to detect peptide fragments within the regulatory domain did not result from an intrinsic property of tomato TD, because LC-MS/MS analysis of the protein isolated from tomato tissues identified peptides that spanned >85% of this domain (Fig. 4B). The most straightforward interpretation of these results is that the regulatory domain of TD is proteolytically cleaved from the catalytic domain during the ingestion or digestion of leaf material, resulting in an enzyme that efficiently degrades Thr in the insect gut without being inhibited through feedback by Ile.
Fig. 4.
Ingested tomato TD is insensitive to negative feedback regulation by isoleucine. (A) TD activity in methyl-JA treated leaves (leaf), midguts of WT-reared larvae (midgut), and frass from WT-reared larvae (frass) was measured in the absence (0) or presence of different concentrations (mM) of Ile. Data were normalized to the amount of activity observed in the absence of Ile (100%) and show the mean ± SD of three independent experiments. (B) Complete amino acid sequence of tomato TD (gi 100257). Based on the 3D structure of Escherichia coli TD (30), amino acid sequences corresponding to the N-terminal catalytic (Cat) domain, C-terminal regulatory (Reg) domain, and the short “neck” region that connects the two domains of the homologous tomato enzyme are indicated. LC-MS/MS analysis of tomato flower TD identified amino acid sequences indicated by uppercase letters. Underlined letters (uppercase and lowercase) denote the sequence that was identified in the midgut enzyme. The chloroplast targeting peptide (Tp) (29) is shown by lowercase italicized letters.
Discussion
The rapid growth rate of lepidopteran larvae depends on the efficient acquisition and utilization of essential amino acids that must be acquired from plant tissues (32). Based on our results, we propose that induced defenses in tomato exploit this nutritional vulnerability through the synergistic action of PIs and a suite of enzymes that catabolize amino acids in the midgut. The theory that low nutrient quality can evolve as a plant defense has, until recently (33, 34), been largely discounted in favor of the view that plant antiherbivore defense is mediated by secondary metabolites (35). Our results highlight the importance of midgutactive host enzymes as a defensive strategy and allow us to suggest that the defensive function of arginase and TD is related to their ability to impair the acquisition of essential amino acids. An alternative, although not mutually exclusive, possibility is that arginase and TD reduce herbivore performance by producing metabolites that exert toxic or antifeedant effects. For example, ammonia produced by TD in the alkaline environment of the lepidopteran midgut is expected to exist predominantly in the unionized form (NH3) that is highly toxic to numerous biological processes (36).
The high-pH optimum of arginase and TD appears to be uniquely suited for defense against insects with an alkaline digestive tract. The exopeptidase LAP-A, which was among the most abundant JIPs identified in the midgut of larvae grown on induced plants (Table 1), also functions at high pH (37). These observations support the previous suggestion by Gu et al. (37) that LAP-A serves a defensive function in the lepidopteran gut. Although it remains to be determined whether LAP-A disrupts digestive physiology, it is noteworthy that the enzyme efficiently liberates Arg (in addition to leucine) from the N-terminal end of polypeptides (37). Thus, it is possible that LAP-A and arginase act synergistically in the gut to catabolize protein-derived Arg, which is an important determinant for optimal growth of leaf-eating insects (12).
The ability of JA to promote massive changes in gene expression and resistance to a broad range of herbivores raises the possibility that JA-based defenses involve midgut-active defensive proteins whose activity is tailored to herbivores with different gut physiochemistries. The “shotgun” proteomics approach that we used should be useful for identifying proteins that perform this role in other plant-insect interactions. The approach is grounded in the idea that host defense proteins are enriched in the midgut as a consequence of their increased abundance in induced leaves and/or increased resistance to gut proteases. Our identification of tomato JIPs that accumulate in the midgut was facilitated by the use of mutants that are affected in JA signaling. However, the detection of these proteins in larvae reared on WT plants (Table 1) indicates that such mutants are not a prerequisite for applying the approach to other plant-herbivore interactions. It is also noteworthy that our protocol for matching MS data to specific gene products involved data searches against a limited set (2,614 protein sequences) of tomato proteins. Thus, it is likely that additional tomato JIPs that accumulate in the gut of M. sexta or other insect pests of tomato remain to be discovered.
The notion that plant proteins differentially accumulate in the insect midgut is supported by our finding that Rubisco, the most abundant soluble protein in tomato leaves, was identified by MS in midguts from jai1-reared larvae but not from larvae grown on induced plants (Table 2). Whereas Rubisco is a major nutritional source of bulk amino acids for phytophagous insects (38), gut-accumulating JIPs that are resistant to gut proteases likely provide limited nutritional value as a source of amino acids. In this context, the well documented down-regulation of Rubisco by JA (39) may represent a plant defense strategy to limit the nutritional value of induced plant tissues. Additional work is needed to determine how proteins such as TD, which we found to be highly active in the midgut and frass, escape proteolytic destruction in the digestive tract.
Based on our data, we suggest that enzymes involved in primary metabolism may play an important role in the evolution of plant defenses. Selective pressure imposed by lepidopteran herbivores on Solanum species may have guided the evolution of defensive forms of arginase and TD from preexisting enzymes that fulfill other roles (i.e., amino acid metabolism) in plant growth and development. A number of different mechanisms could account for this evolution. In the case of TD, limited proteolysis converts the Ile-sensitive form of the enzyme, which participates in Ile biosynthesis in planta, to an Ile-insensitive enzyme that efficiently degrades Thr in the insect digestive tract. This proposed defensive role for tomato TD is consistent with the enzyme's extraordinarily high expression in flowers (29) and JA-induced leaves (16, 40). Increasing evidence of proleolytic activation of defensive proteins in the insect gut (41, 42) indicates that this phenomenon may be more common than previously realized. The idea that plant enzymes can exert antinutritional effects on phytophagous insects by perturbing amino acid homeostasis in the digestive tract may be extended to other classes of plant-derived nutrients, including lipids, carbohydrates, and vitamins. Systematic identification of enzymes from plants or other organisms that metabolize these nutrients may assist the development of new methods for insect control.
Supplementary Material
Acknowledgments
We acknowledge Matthew Larson (Michigan State University Genomic Technology Support Facility) for assistance with the bioinformatic analysis of LC-MS/MS data. We thank Sheng Yang He and Maeli Melotto for helpful suggestions during the course of the research. This research was supported in part by Michigan Life Science Corridor Grant 085P1000466 and U.S. Department of Energy Grant DE-FG02-91ER20021 (to G.A.H.).
Author contributions: H.C., C.G.W., and G.A.H. designed research; H.C., J.A.K., and B.S.P. performed research; H.C., C.G.W., J.A.K., B.S.P., and G.A.H. analyzed data; and G.A.H. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: JA, jasmonic acid; JIP, JA-inducible protein; TD, threonine deaminase; PI, proteinase inhibitor; LC-MS/MS, liquid chromatography tandem MS; LAP-A, leucine aminopeptidase A.
See Commentary on page 18771.
References
- 1.Gatehouse, J. A. (2002) New Phytol. 156, 145-169. [DOI] [PubMed] [Google Scholar]
- 2.Kessler, A., Halitschke, R. & Baldwin, I. T. (2004) Science 305, 665-668. [DOI] [PubMed] [Google Scholar]
- 3.Schaller, F., Schaller A. & Stintzi A. (2005) J. Plant Growth Regul. 23, 179-199. [Google Scholar]
- 4.Reymond, P., Bodenhausen, N., Van Poecke, R. M., Krishnamurthy, V., Dicke, M. & Farmer, E. E. (2004) Plant Cell 16, 3132-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halitschke, R. & Baldwin, I. T. (2005) J. Plant Growth Regul. 23, 238-245. [Google Scholar]
- 6.Howe, G. A. (2005) J. Plant Growth Regul. 23, 223-237. [Google Scholar]
- 7.Devoto, A. & Turner, J. G. (2005) Physiol. Plant. 123, 161-172. [Google Scholar]
- 8.Ryan, C. A. (2000) Biochim. Biophys. Acta 1477, 112-121. [DOI] [PubMed] [Google Scholar]
- 9.Kennedy, G. G. (2003) Annu. Rev. Entomol. 48, 51-72. [DOI] [PubMed] [Google Scholar]
- 10.Ryan, C. A. (1990) Annu. Rev. Phytopathol. 28, 425-449. [Google Scholar]
- 11.Broadway, R. M. & Duffey, S. S. (1986) J. Insect. Physiol. 32, 827-833. [Google Scholar]
- 12.Broadway, R. M. & Duffey, S. S. (1988) J. Insect. Physiol. 34, 1111-1117. [Google Scholar]
- 13.Duffey, S. S. & Stout, M. J. (1996) Arch. Insect. Biochem. Physiol. 32, 3-37. [Google Scholar]
- 14.Duffey, S. S. & Felton, G. W. (1991) Am. Chem. Soc. Symp. Ser. 449, 166-197. [Google Scholar]
- 15.Constabel, C. P., Bergey, D. R. & Ryan, C. A. (1995) Proc. Natl. Acad. Sci. USA 92, 407-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, L., Zhao, Y., McCaig, B. C., Wingerd, B. A., Wang, J., Whalon, M. E., Pichersky, E. & Howe, G. A. (2004) Plant Cell 16, 126-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, C., Williams, M. M., Loh, Y. T., Lee, G. I. & Howe, G. A. (2002) Plant Physiol. 130, 494-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, C., Schilmiller, A. L., Liu, G., Lee, G. I., Jayanty, S., Sageman, C., Vrebalov, J., Giovannoni, J. J., Yagi, K., Kobayashi, Y. & Howe, G. A. (2005) Plant Cell 17, 971-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shevchenko, A., Wilm, M., Vorm, O. & Mann, M. (1996) Anal. Chem. 68, 850-858. [DOI] [PubMed] [Google Scholar]
- 20.Craig, R. & Beavis, R. C. (2004) Bioinformatics 20, 1466-1467. [DOI] [PubMed] [Google Scholar]
- 21.Craig, R. & Beavis, R. C. (2003) Rapid Commun. Mass. Spectrom. 17, 2310-2316. [DOI] [PubMed] [Google Scholar]
- 22.Eriksson, J. & Fenyo, D. (2004) J. Proteome Res. 3, 32-36. [DOI] [PubMed] [Google Scholar]
- 23.Chen, H., McCaig, B. C., Melotto, M., He, S. Y. & Howe, G. A. (2004) J. Biol. Chem. 279, 45998-46007. [DOI] [PubMed] [Google Scholar]
- 24.Sharma, R. J. & Mazumder, R. (1970) J. Biol. Chem. 245, 3008-3014. [PubMed] [Google Scholar]
- 25.McGurl, B., Orozco-Cardenas, M., Pearce, G. & Ryan, C. A. (1994) Proc. Natl. Acad. Sci. USA 91, 9799-9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bergey, D. R., Howe, G. A. & Ryan, C. A. (1996) Proc. Natl. Acad. Sci. USA 93, 12053-12058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Old, W. M., Meyer-Arendt, K., Aveline-Wolf, L., Pierce, K. G., Mendoza, A., Sevinsky, J. R., Resing, K. A. & Ahn, N. G. (2005) Mol. Cell Proteomics 4, 1487-1502. [DOI] [PubMed] [Google Scholar]
- 28.Dadd, R. H. (1985) in Comprehensive Insect Physiology, Biochemistry, and Pharmacology, eds. Kerkut, G. A. & Gilbert, L. I. (Pergamon, New York), pp. 313-390.
- 29.Samach, A., Hareven, D., Gutfinger, T., Ken-Dror, S. & Lifschitz, E. (1991) Proc. Natl. Acad. Sci. USA 88, 2678-2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gallagher, D. T., Gilliland, G. L., Xiao, G. Y., Zondlo, J., Fisher, K. E., Chinchilla, D. & Eisenstein, E. (1998) Structure (London) 6, 465-475. [DOI] [PubMed] [Google Scholar]
- 31.Garcia, E. L. & Mourad, G. S. (2004) Plant Mol. Biol. 55, 121-134. [DOI] [PubMed] [Google Scholar]
- 32.Neal, J. J. (1996) Arch. Insect. Biochem. Physiol. 32, 55-64. [Google Scholar]
- 33.Berenbaum, M. R. (1995) J. Chem. Ecol. 21, 925-940. [DOI] [PubMed] [Google Scholar]
- 34.Felton, G. W. (1996) Arch. Insect. Biochem. Physiol. 32, 107-130. [Google Scholar]
- 35.Fraenkel, G. S. (1959) Science 129, 1466-1470. [DOI] [PubMed] [Google Scholar]
- 36.Visek, W. J. (1984) J. Dairy Sci. 67, 481-498. [DOI] [PubMed] [Google Scholar]
- 37.Gu, Y. Q., Holzer, F. M. & Walling, L. L. (1999) Eur. J. Biochem. 263, 726-735. [DOI] [PubMed] [Google Scholar]
- 38.Johnson, K. S. & Felton, G. W. (1996) Arch. Insect. Biochem. Physiol. 32, 85-105. [Google Scholar]
- 39.Reinbothe, S., Mollenhauer, B. & Reinbothe, C. (1994) Plant Cell 6, 1197-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Samach, A., Broday, L., Hareven, D. & Lifschitz, E. (1995) Plant J. 8, 391-406. [DOI] [PubMed] [Google Scholar]
- 41.Ferreira-DaSilva, C. T., Gombarovits, M. E. C., Masuda, H., Oliveira, C. M. & Carlini, C. R. (2000) Arch. Insect. Biochem. Physiol. 44, 162-171. [DOI] [PubMed] [Google Scholar]
- 42.Wang, J. H. & Constabel, C. P. (2004) Planta 220, 87-96. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.