Abstract
The adapter SLP-76 is essential for T cell development and function. SLP-76 binds to the src homology 3 domain of Lck in vitro. This interaction depends on amino acids 185–194 of SLP-76. To examine the role of the Lck-binding region of SLP-76 in T cell development and function, SLP-76-/- mice were reconstituted with an SLP-76 mutant that lacks amino acids 185–194. Double and single positive thymocytes from reconstituted mice were severely reduced in numbers and exhibited impaired positive selection and increased apoptosis. Peripheral T cells were also reduced in numbers, exhibited impaired phospholipase C-γ1 and Erk phosphorylation, and failed to flux calcium, secrete IL-2, and proliferate in response to T cell antigen receptor ligation. Delayed cutaneous hypersensitivity responses and Ab responses to T cell-dependent antigen were severely impaired. These results indicate that the Lck binding region of SLP-76 is essential for T cell antigen receptor signaling and normal T cell development and function.
Keywords: Lck, proline-rich region
Maturation, proliferation, and selection of thymocytes and activation of mature T cells require signals delivered by the pre-T cell antigen receptor (TCR) and TCR. Adapter proteins, which lack intrinsic enzymatic activity, are key components in this signaling. SLP-76 [src homology 2 (SH2) domain containing leukocyte-specific phosphoprotein of 76 kDa] is an adapter protein expressed predominantly in hematopoietic cells (1). SLP-76 has three distinct domains: an NH2-terminal domain, a central proline rich domain and a C-terminal SH2 domain. Phosphorylated tyrosine residues in the NH2-terminal domain bind SH2 domain-containing proteins that include the Tec-family protein tyrosine kinase Itk (IL-2 inducible T cell kinase) and the Rac/Rho guanine exchange factor Vav (2, 3). The central proline-rich domain of SLP-76 binds to src homology 3 (SH3) domain-containing proteins that include Grb2-related adaptor downstream of shc (Gads) and phospholipase C-γ1 (PLC-γ1) (4). Upon TCR stimulation, Gads recruits SLP-76 to LAT which results in translocation of SLP-76 to glycolipid enriched regions (GEMs) (5). LAT, through Grb2, interacts with Sos, a guanine nucleotide exchange factor of Ras GTPase, and may link SLP-76 to the Ras/mitogen activated protein kinase (MAPK)/extracellular signal regulated protein kinase (ERK) pathway. The C-terminal SH2 domain of SLP-76 associates with the phosphotyrosine containing adapter protein ADAP and the hematopoietic progenitor kinase 1 (HPK1) (6, 7).
SLP-76 plays an important role in T cell receptor signal transduction and T cell activation. SLP-76-deficient human leukemia Jurkat T cells (J14 cells) exhibit impaired TCR/CD3 signaling with reduced PLC-γ1 activation, calcium mobilization, ERK phosphorylation, and IL-2 production (8). SLP-76-/- mice have a complete block in thymocyte development at the CD4-CD8- double negative (DN) stage and lack peripheral T cells (9, 10). The role of SLP-76 domains and residues in reconstituting T cell development and function in SLP-76-/- mice has been examined in two independent studies (11, 12). WT SLP-76 transgene completely rescued T cell development and function in SLP-76-/- mice. An SLP-76 NH2-teminal domain deletion mutant completely failed to restore thymocyte development, suggesting a critical role of this domain in pre-TCR signaling. Mice reconstituted with a mutant where all three tyrosine molecules in the NH2-terminal domain were replaced with phenylalanine (Y3F) had partial rescue of thymic development but absent peripheral T cell function. Gads binding domain (within the proline-rich region) and C-terminal SH2 domain deletion mutants restored thymic development and peripheral T cells more efficiently than did Y3F. However, peripheral T cell function remained impaired.
p56lck (Lck) is a member of the src family of protein tyrosine kinases that is highly expressed in T cells. Lck is required for thymopoiesis, because there is a severe deficit of double positive (DP) and single positive (SP) thymocytes in Lck-/- mice. Mature TCRαβ T cells are present in reduced numbers in the periphery but are partially responsive to TCR stimulation (13). Association between SLP-76 and the SH3 domain of Lck has recently been demonstrated in vitro (14). By using a peptide competition based strategy, a 10-aa-long sequence (amino acids 185–194) in the proline-rich domain of SLP-76 upstream of the Gads binding site has been identified as a docking area for the SH3 domain of Lck. To address the functional importance of this region, we reconstituted SLP-76-/- mice with an SLP-76 deletion mutant that lacks amino acids 185–194 (SLP-76Δ185–194) and selectively fails to bind Lck.
Experimental Procedures
Cells. Jurkat T cells, SLP-76-deficient Jurkat T cell line J14 (a generous gift of A. Weiss, University of California, San Francisco), and SLP-76 transfected J14 cells were maintained in RPMI medium 1640 (supplemented with 10% FBS and 10 mM Hepes and penicillin and streptomycin) at 106 cells per ml. The SLP-76Δ185–194 mutant was generated by using the QuikChange site-directed mutagenesis kit (Stratagene). For stable transfection, SLP-76 WT and mutant constructs were subcloned in pCMVTag3A (Stratagene), which provides G418 resistance.
Immunoprecipitation and Western Blotting. SLP-76 immunoprecipitates were run on SDS/PAGE gels and Western-blotted with polyclonal rabbit Abs to SLP-76 (11), PLC-γ1 (sc-81, Santa Cruz Biotechnology) and Itk (2F11, BD Biosciences) and with mAb to Lck (BD Biosciences) and Vav (Upstate Biotechnology, Lake Placid, NY).
IL-2 Production and Measurement. Cells stimulated with plate-bound anti-CD3 mAb (5 μg/ml) or phorbol 12-myristate 13-acetate (PMA) (50 ng/ml) and ionomycin (500 ng/ml) for 48 h, and IL-2 levels in the supernatants were measured by ELISA.
Intracellular Immunofluorescence. Intracellular immunofluorescence staining for nuclear factor of activated T cells (NFAT) 1 was carried out essentially as described in ref. 15.
Transgenic Mice. Mutant SLP-76 cDNA was sequenced and subcloned in the Bam H1 site of the p1017 vector carrying the Lck proximal promoter (kindly provided by R. Perlmutter, Amgen, Thousand Oaks, CA). Mutant transgenic mice were generated essentially as described in ref. 11. Transgenic mice that express bcl-2 under the control of Eμ promoter (line 2–25) were obtained from The Jackson Laboratory (16). All animal experiments were performed in compliance with the National Institutes of Health and institutional guidelines approved by the Children's Hospital internal animal care and use committee.
Antibodies and Flow Cytometry Analysis. Streptavidin-FITC, streptavidin-phycoerythrin (PE), and streptavidin-CyChrome and mAbs and polyclonal Abs (unlabeled or labeled to FITC or PE), as well as procedures for staining cells for flow cytometry, are described in ref. 11. FITC-anti-bcl-2 mAb was from BD Biosciences. Annexin V-FITC (BioVision, Mountain View, CA) staining was performed as per the manufacturer's instructions.
Up-Regulation of CD25 and CD69 Expression, Proliferation of Splenic T Cells, and PLC-γ1, Vav, and ERK Phosphorylation. Purification of splenic T cells, up-regulation of CD25 and CD69 expression, proliferation, and PLC-γ1, Vav, and ERK phosphorylation assays were performed essentially as described in ref. 11.
Intracellular Calcium Measurements. Intracellular calcium measurements on purified T cells from SLP-76+/+ and SLP-76Δ185–194 mice were performed as described in ref. 17. Details of the procedure are provided in Supporting Text, which is published as supporting information on the PNAS web site.
Oxazolone-Induced Contact Hypersensitivity, Serum Ig Levels, and T-Dependent Antibody Responses. These were measured as described in ref. 11.
Densitometry and Statistical Analysis. Blots were scanned and analyzed by using nih image software (Version 1.62, http://rsb.info.nih.gov/nih-image/Default.html). Statistical analyses (Student's t test) were performed with prism software (Version 3.0a, Graph-Pad, San Diego).
Results
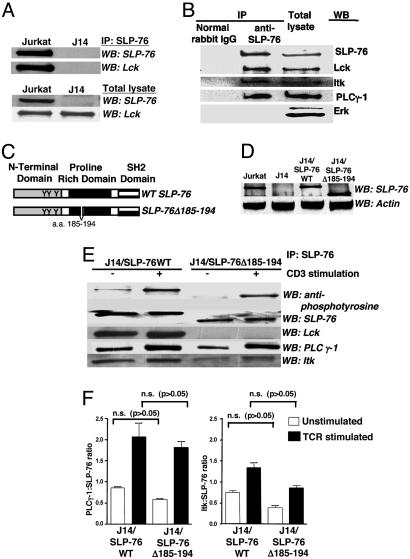
Characterization of SLP-76Δ185–194. To establish that SLP-76 and Lck interact in T cells, SLP-76 immunoprecipitates from Jurkat cells were probed for Lck. Fig. 1A shows that Lck was present in SLP-76 immunoprecipitates from WT Jurkat cells but not from the SLP-76-deficient subline J14. Lck also coimmunoprecipitated with SLP-76 in mouse splenic T cells (Fig. 1B). PLC-γ1 and Itk also associated constitutively with SLP-76 in these cells.
Fig. 1.
Interaction of SLP-76 with Lck, PLC-γ1, and Itk in Jurkat and splenic T cells. (A and B) SLP-76 associates with Lck in Jurkat cells (A) and purified splenic T cells (B). (C) WT SLP-76 and SLP-76-Δ185–194 deletion mutant used to reconstitute J14 cells and SLP-76-/- mice. (D) Levels of SLP-76 protein in Jurkat, J14, and J14 cells transfected with WT SLP-76 or SLP-76-Δ185–194. (E) Baseline and TCR induced association of Lck, PLC-γ1, and Itk with WT SLP-76 in J14 cells transfected with WT SLP-76 or SLP-76-Δ185–194. (F) Pooled results from three experiments of PLC-γ1 and Itk association with SLP-76 (expressed in arbitrary units as the densitometric intensity ratio of PLC-γ1:SLP-76 and Itk:SLP-76) in J14/SLP-76WT and SLP-76Δ185–194 cells.
To examine the role of amino acids 185–194 of SLP-76 in Lck binding, J14 cells were stably transfected with WT SLP-76 (J14/SLP-76WT) or SLP-76Δ185–194 (J14/SLP76Δ185–194) (Fig. 1C). Transfectants with TCR/CD3 surface expression comparable with that of WT Jurkat cells were selected. J14/SLP-76WT expressed SLP-76 protein of appropriate size, whereas J14/SLP76Δ185–194 cells expressed a faster-migrating SLP-76 (Fig. 1D). The level of SLP-76 protein in these cells was comparable with that in Jurkat cells. Lck was present in SLP-76 immunoprecipitates from J14/SLP-76WT cells but not in J14/SLP76Δ185–194 cells (Fig. 1E). Expression of Lck was comparable in J14/SLP-76Δ185–194 cells and J14/SLP76WT controls (Fig. 8, which is published as supporting information on the PNAS web site).
The amino acids 157–223 proline-rich region of SLP-76 is important for SH3 domain-mediated constitutive association with PLC-γ1, whereas the amino acids 184–195 and 196–208 regions of SLP-76 are important for its SH3-domain-mediated constitutive association with Itk (2, 4). Expression of PLC-γ1 and Itk was comparable in J14/SLP-76Δ185–194 cells and J14/SLP76WT controls (Fig. 8). Fig. 1 E and F shows that there was a modest, but not significant, reduction in the binding of PLC-γ1 to the mutant SLP-76 in unstimulated cells. Binding of Itk to the mutant SLP-76 was more affected, although the decrease was not statistically significant.
After TCR ligation, there is increased association of SLP-76 complex with PLC-γ1 and Itk (2, 4). TCR ligation in J14/SLP-76WT cells caused SLP-76 phosphorylation and increased association of PLC-γ1 (2.5-fold) and Itk (1.8-fold) (Fig. 1 E and F). There was no increase in the association of Lck with SLP-76 after TCR ligation (1.1-fold) (Fig. 1E). TCR ligation in J14/SLP-76Δ185–194 cells caused tyrosine phosphorylation of SLP-76Δ185–194 to a degree comparable with that observed for WT protein (Fig. 1E). The ratio of phosphorylated to nonphosphorylated SLP-76 was 15 for WT SLP-76 versus 13 for SLP76Δ185–194. Association of Lck with SLP-76Δ185–194 remained undetectable after TCR ligation. TCR ligation caused increased recruitment of PLC-γ1 (3.1-fold) and Itk (2.2-fold) to SLP-76Δ185–194. The ratios of PLC-γ1:SLP-76 and of Itk:SLP-76 in anti-CD3-stimulated J14/SLP-76Δ185–194 cells were, respectively, 88% and 72% those in anti-CD3-stimulated J14/SLP76 WT controls and were not significantly different (P > 0.05).
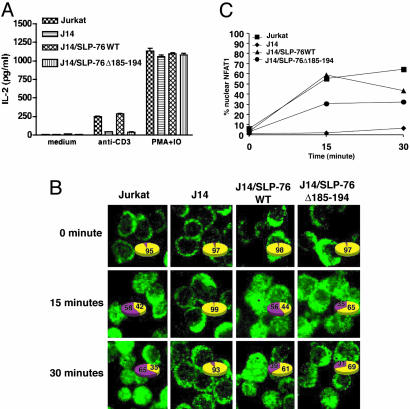
Impaired IL-2 Production and NFAT Nuclear Translocation After TCR Ligation in J14SLP-76Δ185–194 Cells. Fig. 2A shows that J14/SLP-76Δ185–194 cells were severely impaired in their ability to secrete IL-2 after anti-CD3 stimulation. In contrast, they secreted IL-2 normally in response to PMA and ionomycin, which bypass via TCR signaling.
Fig. 2.
Impaired IL-2 production and NFAT activation in SLP-76 mutant T cells. (A) IL-2 secretion by Jurkat cells stimulated with plate-bound anti-CD3 mAb or PMA and ionomycin for 48 h. (B) Cells were left unstimulated or stimulated for 15 and 30 min with anti-CD3. Cells were spun immediately onto coverslips, stained with anti-NFAT1 Abs to assess nuclear translocation of NFAT (green, NFAT1). Numbers given in the pie charts represent percent numbers of cells with cytoplasmic (yellow) and nuclear (purple) NFAT, respectively. A total of 150–300 cells were counted for each time point. The graph (C) represents the percentage of cells with nuclear NFAT1 for each of the cell lines and time points tested. Similar results were obtained in two different experiments.
NFAT is a critical factor in the activation of the IL-2 promoter (18, 19). After TCR ligation, NFAT1 is dephosphorylated by the Ca2+-dependent phosphatase calcineurin, resulting in a downward shift of its molecular weight and allowing its translocation to the nucleus (20). There was no detectable shift in the molecular weight of NFAT1 in J14/SLP-76Δ185–194 cells stimulated with anti-CD3 (Fig. 9, which is published as supporting information on the PNAS web site). Fig. 2 B and C shows that nuclear translocation of NFAT1 after anti-CD3 stimulation was impaired in J14 cells and was only partially corrected by introduction of SLP-76Δ185–194.
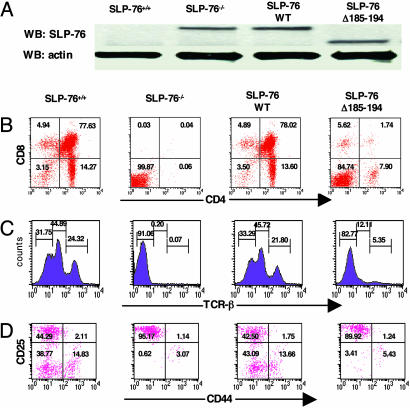
Impaired Thymocyte Development in SLP-76Δ185–194 Transgenic Mice. We introduced the SLP-76Δ185–194 transgene in the SLP-76-/- background (SLP-76Δ185–194 mice). Controls included non-transgene-bearing SLP-76+/+ mice, SLP-76-/- littermates, and the previously described SLP-76-/- mice reconstituted with an SLP-76WT transgene (SLP-76WT mice) (11). Transgenic lines that expressed SLP-76 protein in thymocytes in amounts comparable with those expressed in SLP-76+/+ mice, as assessed by Western blot (Fig. 3A), were studied.
Fig. 3.
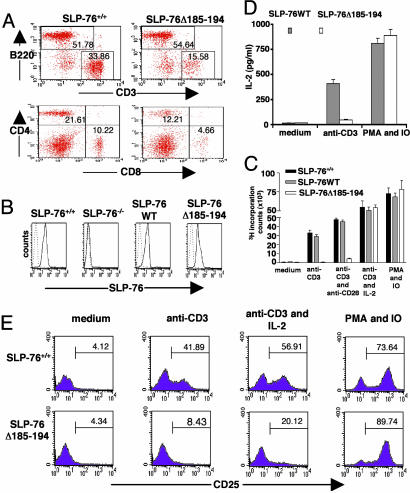
FACS analysis of thymocytes from SLP-76-/- mice reconstituted with SLP-76-Δ185–194. (A) Expression of SLP-76 in the thymocytes of mice as assessed by Western blot using rabbit anti-SLP-76 Ab. (B) Surface expression of CD4 (anti-CD4-FITC) vs. CD8 (anti-CD8-PE) on total thymocytes. (C) TCRβ expression on total thymocytes. (D) CD44 and CD25 expression on DN thymocytes. Cells were triple-stained with anti-CD44-FITC, anti-CD25-PE, and a mixture of biotin-conjugated mAbs to CD3, CD4, CD8, B220, Mac1, and Gr-1 followed by streptavidin-CyChrome. Analysis was performed on gated CyChrome-negative cells. The results are representative of at least three experiments.
SLP-76-/- mice have thymic cellularity that is <1% of that of SLP-76+/+ mice, no detectable CD4+CD8+ DP, CD4+8- SP, or CD4-8+ SP thymocytes (Fig. 3B), and no detectable TCRβ+ cells (Fig. 3 B and C). The SLP-76WT transgene completely restored thymic cellularity and restored thymocyte development with normal TCRβ expression (Fig. 3 B and C). In contrast, thymic cellularity remained severely decreased in SLP-76Δ185–194 mice (5.6 ± 3.5 × 106 vs. 110 ± 7.8 × 106 in SLP-76+/+ mice, and 118 ± 9.4 × 106 in SLP-76WT mice). DN cells accumulated in SLP-76Δ185–194 thymi, and their progression from DN to DP cells was severely impaired (Fig. 3B). There was also decreased percentage of CD4+, but not of CD8+, SP cells, and the CD4:CD8 ratio was decreased compared with WT mice. The density of TCRβ on thymocytes was decreased in SLP-76Δ185–194 mice (Fig. 3C). Similar results were obtained by using two independent lines of SLP-76Δ185–194 mice (Fig. 10, which is published as supporting information on the PNAS web site).
SLP-76-/- mice show a relative increase of the CD25+CD44- population, whereas the most mature CD25-CD44- cells are almost undetectable. This block is completely overcome by reconstitution with SLP-76 WT transgene. In contrast, it remained severe in SLP-76Δ185–194 mice with <5% of DN cells being at the CD25-CD44- (Fig. 3D).
Because in all subsequent experiments, and as previously reported (11), thymocytes and peripheral lymphocytes from SLP-76+/+ mice and mice reconstituted with the SLP-76WT transgene behaved identically, in most of the remainder of the article, we present data on only SLP-76+/+ controls.
Increased Apoptosis in DP and SP Thymocytes of SLP-76Δ185–194 Transgenic Mice. We used annexin V staining to assess apoptosis in SLP-76Δ185–194 thymocytes. There was a modest percentage of apoptotic cells in DP and CD4+ SP thymocytes in SLP-76+/+ mice (10–15%), whereas their CD8+ SP cells showed minimal apoptosis (<2%). In contrast, there was massive apoptosis in the DP compartment (≈70%) and CD4+ SP compartment (≈95%), but not in the CD8+ SP compartment (≈5%), of SLP-76Δ185–194 thymus (Fig. 11A, which is published as supporting information on the PNAS web site). A similar low percentage of apoptotic cells were obtained when CD8+TCRhi and CD8+TCRlo cells were analyzed separately (data not shown). Introduction of the antiapoptotic gene bcl-2, which can rescue thymocytes from apoptosis (16) in SLP-76Δ185–194 mice, did not rescue the increased apoptosis or defective thymocyte development (Fig. 11).
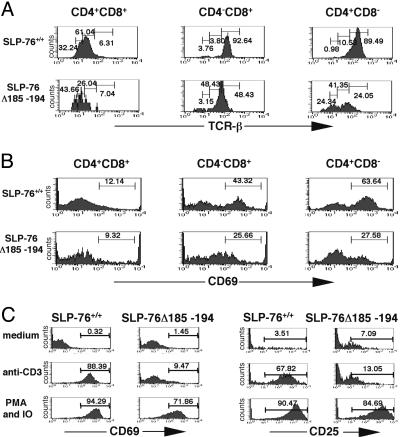
Deficient Up-Regulation of CD69 on SP Thymocytes from SLP-76Δ185–194 Mice. Positive selection of thymocytes into SP cells is accompanied by up-regulation of TCR/CD3 and CD69 expression (21, 22). TCRβ expression was decreased on DP and SP thymocytes from SLP-76Δ185–194 mice compared with SLP-76+/+ controls (Fig. 4A). CD69 expression on DP thymocytes was roughly equivalent in SLP-76Δ185–194 mice and SLP-76+/+ controls. However, up-regulation of CD69 on SP cells was impaired in SLP-76Δ185–194 mice (Fig. 4B). Furthermore, SLP-76+/+ thymocytes, but not SLP-76Δ185–194 thymocytes, up-regulated CD69 and TCRβ after anti-CD3 stimulation (Fig. 4C and data not shown). Up-regulation of these activation markers on thymocytes in response to PMA and ionomycin which bypass TCR signaling was normal.
Fig. 4.
Expression of TCRβ and CD69 on DP and SP thymocytes and induction of CD69 and CD25 expression on thymocytes after TCR ligation. Thymocytes were stained with anti-CD4-PE, anti-CD8-CyChrome, and anti-TCRβ-FITC or anti-CD69-FITC. Gated DP and SP cells were analyzed for expression of TCRβ (A) and CD69 (B). (C) Thymocytes were stimulated with plate-bound anti-CD3 (5 μg/ml) for 18 h, then stained with anti-CD69-FITC or anti-CD25-PE. The results are representative of at least three experiments.
Phenotypic and Functional Analysis of Peripheral T Cells in SLP-76Δ185–194 Mice. Spleens of SLP-76-/- mice contain no detectable CD3+, CD4+, or CD8+ cells (9). Introduction of the SLP-76WT transgene restores the numbers and percentages of splenic CD3+, CD4+, and CD8+ cells. The SLP-76Δ185–194 transgene partially restored the number and percentage of splenic CD3+ T cells to ≈1/4 to 1/2 of normal (n = 6), with a normal CD4:CD8 ratio (Fig. 5A) and a normal percentage of CD25+ cells among the CD4+ population (data not shown). The density of surface CD3 and TCRβ expression on splenic T cells from these mice was 2- to 3-fold lower than that of T cells from SLP-76+/+ controls (Fig. 5A and data not shown). T cells from spleens of mice reconstituted with SLP-76 transgenes expressed SLP-76 protein at comparable intensities (Fig. 5B).
Fig. 5.
Phenotypic and functional analysis of splenic T cells in SLP-76Δ185–194 mice. (A) Surface expression of CD3 and B220 and of CD4 and CD8 on spleen cells. (B) Expression of SLP-76 protein in purified T cells by intracellular staining with FITC-conjugated rabbit anti-SLP-76 Ab. (C) Proliferation of purified T cells to anti-CD3 (coated at 5 μg/ml) in the presence or absence of anti-CD28 (coated at 5 μg/ml) and mouse rIL-2 (20 ng/ml), and to PMA and ionomycin (IO). (D) IL-2 secretion by purified splenic T cells after stimulation with anti-CD3 and PMA and ionomycin. (E) FACS analysis of CD25 surface expression on purified splenic T cells stimulated as above. The results are representative of three experiments.
Splenic T cells from SLP-76Δ185–194 mice failed to proliferate and secrete IL-2 in response to stimulation with immobilized anti-CD3 (Fig. 5 C and D). IL-2, but not anti-CD28 mAb, corrected the proliferation failure. Anti-CD3 stimulation caused a small, but detectable increase in IL-2Rα/CD25 and CD69 expression in T cells from SLP-76Δ185–194 mice. This increase was further up-regulated by costimulation with exogenous IL-2 (Fig. 5E and data not shown). SLP-76Δ185–194 T cells proliferated, produced IL-2, and up-regulated CD25 and CD69 normally in response to PMA and ionomycin (Fig. 5 C–E).
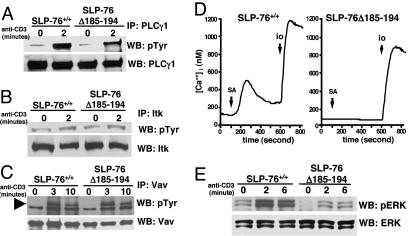
Calcium Influx and PLC-γ1 and ERK Phosphorylation. Tyrosine phosphorylation of PLC-γ1 after TCR/CD3 ligation was reduced in T cells of SLP-76Δ185–194 mice compared with SLP-76+/+ controls (Fig. 6A). This decrease was specific, because tyrosine phosphorylation of Itk (Fig. 6B) and Vav-1 (Fig. 6C), two important proximal components of the TCR-mediated signaling, was normal in these cells. Phosphorylation and activation of PLC-γ1 results in the hydrolysis of PIP2 and generation of IP3, which initiates an increase in intracellular calcium levels. T cells from SLP-76Δ185–194 mice showed no appreciable change in calcium influx after TCR/CD3 ligation (Fig. 6D). SLP-76 is also important for the phosphorylation and activation of the MAP kinase ERK (23). Both baseline and anti-CD3-induced phosphorylation of Erk1/2 was lower in SLP-76Δ185–194 T cells than in control T cells (Fig. 6E).
Fig. 6.
Phosphorylation of PLC-γ1, Itk, Vav, and Erk, and calcium flux after TCR/CD3 ligation in splenic T cells. Cells were stimulated with anti-CD3 (5 μg/ml), washed, and crosslinked with 10 μg/ml goat F(Ab′)2 anti-rat Ig. Lysates were immunoprecipitated with anti-PLC-γ1 (A), anti-Itk (B), or anti-Vav1 (C) Abs, run on 10% SDS/PAGE gels, probed with antiphosphotyrosine (pTyr) mAb 4G10, and reprobed with anti-PLC-γ1, anti-Itk, or anti-Vav Abs as loading controls. (D)Ca2+ mobilization: Purified T cells preloaded with fura-2 acetoxymethyl ester were stimulated with biotinylated anti-CD3 and crosslinked with streptavidin (SA) then later stimulated with ionomycin (io) in 2 mM Ca2+ Ringer's solution. (E) Erk phosphorylation. Lysates from purified spleen T cells stimulated with crosslinked anti-CD3 as described in A were probed with phospho-ERK-specific Ab (pErk) and reprobed with Erk-specific Ab. The results shown are representative of three experiments for A–C and two experiments for D.
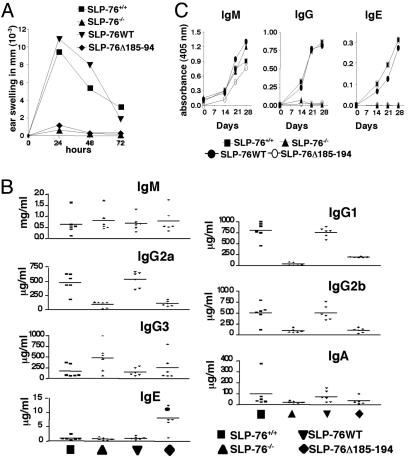
In Vivo T Cell Effector Responses, Serum Ig Levels, and Antibody Response to T-Dependent Antigen. SLP-76-/- mice completely failed to exhibit ear swelling in response to sensitization and challenge with the hapten oxazolone, had significantly decreased serum levels of the T dependent isotypes IgG1, IgG2a, IgG2b, and IgE, and failed to mount Ab responses to the T-dependent antigen ovalbumin (OVA). All of these deficiencies were corrected by introduction of the SLP-76WT transgene (11). SLP-76Δ185–194 mice had no appreciable ear swelling response to oxazolone (Fig. 7A). Their serum IgG1, IgG2a, and IgG2b remained significantly decreased (Fig. 7B), and they made only an IgM response with no detectable IgG or IgE responses to OVA (Fig. 7C). Surprisingly, their serum IgE levels were significantly elevated.
Fig. 7.
Hapten-induced contact hypersensitivity, serum immunoglobulins, and Ab response to OVA. (A) Groups of four mice were sensitized with oxazolone and challenged with hapten (right ear) or ethanol (left ear). The results represent the difference in thickness between hapten- and vehicle-challenged ears. (B) Serum Ig isotypes were determined on samples from 6- to 8-week-old mice (n = 6 per group). (C) Anti-OVA IgM, IgG1, and IgE Abs in the serum of mice immunized i.p. with 100 μg of OVA in alum (n = 6 per group). For clarity, SDs are not shown, but they were uniformly <10% of the mean.
Discussion
Our studies demonstrate that SLP-76 and Lck constitutively interact in T cells and that the region of SLP-76 involved in Lck binding is essential for T cell development and function.
Constitutive association of SLP-76 and Lck in Jurkat T cells required amino acids 185–194 of SLP-76 (Fig. 1E), in agreement with the previous finding that an amino acids 185–194 peptide inhibits the binding of SLP-76 to GST-SH3 Lck fusion protein (14). In contrast, constitutive association of PLC-γ1 with SLP-76Δ185–194 was not significantly decreased, although amino acids 185–194 partially overlap with a minimal amino acid sequence of SLP-76 reported to be essential for binding PLC-γ1 by Gonen et al. (24). However, in a more recent study, SH3 domain of PLC-γ1 was found to associate with SLP-76 through at least three distinct sites (25). More importantly, after TCR ligation, there was little difference in the association of PLC-γ1 with SLP-76Δ185–194 compared with WT SLP-76 control (Fig. 1F). Because increased association of PLC-γ1 with SLP-76 after TCR ligation depends on the formation of a complex between SLP-76, Gads, and LAT, which recruits PLC-γ1, this finding suggests that the formation of this complex is essentially intact in cells that express the SLP-76Δ185–194 mutant.
Deletion of amino acids 185–194 diminished, albeit not significantly, the constitutive association of Itk with SLP-76 (Fig. 1E), consistent with the observation that amino acids 184–195 peptide partially inhibits the interaction of SLP-76 with GST-SH3 Itk (2). The association of Itk and SLP-76Δ185–194 after TCR ligation, which involves recruitment of Itk mediated by interaction of its SH2 domain with phosphorylated Y145 residue of SLP-76, was not significantly diminished (Fig. 1F), suggesting that the inducible recruitment of Itk to the SLP-76 mutant was reasonably intact.
TCR signaling was impaired in J14/SLP-76Δ185–194 cells, as evidenced by impaired NFAT1 nuclear translocation and by IL-2 production (Fig. 2). Because the association of PLC-γ1 and Itk with SLP-76 after TCR ligation, which is important for the transmission of the TCR signal, was well preserved in J14/SLP-76Δ185–194 cells, it is unlikely that the decreased constitutive association of Itk with the mutant accounts for the defective TCR signaling in these cells. Rather, the results suggest that Lck binding to SLP-76 may be important for TCR signaling. However, it is possible that the Δ185–194 deletion caused a conformational change and/or compromised as yet unknown interactions that impaired SLP-76 function. With the exception of loss of Lck binding, we did not detect differences in the pattern of proteins pulled down from Jurkat T cells by WT and mutant SLP-76 GST fusion proteins, or in the intracellular localization of the WT and mutant proteins (unpublished observations). It has been reported that reconstitution of J14 cells with an SLP-76 mutant lacking amino acids 177–212 resulted in no impairment of TCR signaling (24). It is possible that small differences in the deletion mutant used and differences in techniques may account for these results. In our study, we have confirmed the results obtained in reconstituted J14 cells in SLP-76-/- mice reconstituted with the deletion mutant.
The SLP-76 mutant was severely deficient in its ability to reconstitute thymic development in SLP-76-/- mice. Thymic cellularity, CD25-CD44--mature DN, DP, and SP cells were all severely decreased in SLP-76Δ185–194 mice (Fig. 3). Because transition from CD25+CD44--immature to CD25-CD44--mature DN cells and maturation of DN into DP cells require signaling via the pre-TCR, these results suggest that the Lck binding region of SLP-76 is important for signaling via the pre-TCR. Furthermore, thymic selection was affected, because SP cells had decreased expression of TCRβ and of the activation marker CD69, which are normally up-regulated upon maturation of DP into SP cells (Fig. 4 A and B), and up-regulation of CD69 and CD25 on thymocytes after in vitro crosslinking of TCR/CD3 was severely defective (Fig. 4C). These results suggest that the Lck binding region of SLP-76 is also critical for TCR signaling in thymocytes. There was increased thymocyte apoptosis in SLP-76Δ185–194 mice (Fig. 11 A). This finding may reflect the failure of TCR engagement to deliver survival signals. TCR activation of Ras leading to Erk activation plays an important role in positive selection and thymocyte survival (26).
The percentage of CD3+ cells in spleens and lymph nodes was decreased in SLP-76Δ185–194 mice (Fig. 5A). T cells from these mice had decreased CD25 and CD69 expression after ligation of TCR/CD3 and completely failed to secrete IL-2 and proliferate in response to anti-CD3 (Fig. 5 C–E). This defect is unlikely to be simply due to their decreased TCR/CD3 expression. Up-regulation of CD25 and CD69 expression and correction of the proliferative defect by IL-2 suggest that residual signaling via TCR/CD3 occurs in these cells.
PLC-γ1 phosphorylation after TCR ligation is thought to depend on the activity of several kinases, which include ZAP-70, Lck, and Tec kinases, and on SLP-76 (8, 27). PLC-γ1 phosphorylation was impaired in T cells from SLP-76Δ185–194 mice. In contrast, ZAP-70, Itk, and Lck phosphorylation was intact (Fig. 6B and unpublished observations). Lck is known to phosphorylate and activate Itk directly (28). SLP-76 may serve as a scaffold that brings together Lck, Itk, and PLC-γ1. This arrangement may allow Lck and Itk to synergize in phosphorylating PLC-γ1. Disruption of Lck binding to SLP-76, and possibly the modest decrease in Itk association with SLP-76Δ185–194, may have resulted in impaired phosphorylation of PLC-γ1. In a recent study, PLC-γ1 phosphorylation was reported to be normal in J14 cells reconstituted with an SLP-76 mutant that lacks amino acids 177–212 (24), which contrasts with our findings with primary T cells from SLP-76Δ185–194 mice. The discrepancy may be due to technical differences such as the level of expression of the mutant proteins and the source of cells used.
Calcium influx was abolished in T cells from SLP-76Δ185–194 mice (Fig. 6D). This abolition cannot simply be explained by their lower level of expression of TCR/CD3, because we had previously observed that T cells from SLP-76-/- mice reconstituted with an SLP-76 SH2 domain deletion mutant (that have comparable low expression of TCR/CD3) flux calcium almost normally (11). Activation of the MAP kinase Erk was decreased in T cells from SLP-76Δ185–194 mice (Fig. 6E). This decrease may be due to decreased generation of diacylglycerol and subsequent impaired activation of the Ras exchange factor RasGRP and of the Ras/Raf/Erk pathway (29).
T cell-dependent functions in vivo were severely impaired in SLP-76Δ185–194 mice. These mice failed to develop contact hypersensitivity response to oxazolone (Fig. 7A), provide help for isotype switching (Fig. 7B), and mount an IgG or IgE Ab response to the T-dependent antigen OVA (Fig. 7C). The elevated serum IgE levels in these mice may be a consequence of impaired T cell activation, because it has been proposed that weak TCR signals promote the development of IL-4-secreting Th2 cells that drive IgE switching (30).
The phenotype of the SLP-76-/- mice reconstituted with an SLP-76 N-terminal domain deletion mutant is similar to that of SLP-76-/- mice. The phenotype of SLP-76Δ185–194 mice is more severe than that of SLP-76Y3F (12) mice, which exhibit higher numbers of thymocytes and a less impaired DP-to-DN transition. In turn, the Y3F phenotype is more severe than that of SLP-76ΔGads mice, which in turn is more severe than that of SLP-76ΔSH2 mice (11, 12). This observation suggests the following hierarchy in SLP-76 function: N-terminal domain → Lck binding region → Y3F → Gads binding region → SH2 domain.
Supplementary Material
Acknowledgments
We thank Tatyana Sannikova for excellent assistance with animal care, Dr. Miguel A. de la Fuente and Dr. Alex Kettner for useful discussions, Dr. Paul Bryce for help in CHS experiments, and Ms. Haifa Jabara and Dr. Hans Oettgen for critically reviewing the manuscript. This work was supported by U.S. Public Health Service Grants AI-35714 (to R.S.G.), AI-40127 (to A.R.), and AI-054933 (to S.F.).
Author contributions: L.K. and R.S.G. designed research; L.K. and S.F. performed research; L.K. contributed new reagents/analytic tools; L.K., S.F., A.R., and R.S.G. analyzed data; and L.K. and R.S.G. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: DP, double positive; OVA, ovalbumin; NFAT, nuclear factor of activated T cells; PE, phycoerythrin; PLC-γ1, phospholipase C-γ1; PMA, phorbol 12-myristate 13-acetate; SH2, src homology 2; SH3, src homology 3; SP, single positive; SLP-76, SH2 domain containing leukocyte-specific phosphoprotein of 76 kDa; TCR, T cell antigen receptor.
References
- 1.Jackman, J. K., Motto, D. G., Sun, Q., Tanemoto, M., Turck, C. W., Peltz, G. A., Koretzky, G. A. & Findell, P. R. (1995) J. Biol. Chem. 270, 7029-7032. [DOI] [PubMed] [Google Scholar]
- 2.Bunnell, S. C., Diehn, M., Yaffe, M. B., Findell, P. R., Cantley, L. C. & Berg, L. J. (2000) J. Biol. Chem. 275, 2219-2230. [DOI] [PubMed] [Google Scholar]
- 3.Wu, J., Motto, D. G., Koretzky, G. A. & Weiss, A. (1996) Immunity 4, 593-602. [DOI] [PubMed] [Google Scholar]
- 4.Yablonski, D., Kadlecek, T. & Weiss, A. (2001) Mol. Cell. Biol. 21, 4208-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu, S. K., Fang, N., Koretzky, G. A. & McGlade, C. J. (1999) Curr. Biol. 9, 67-75. [DOI] [PubMed] [Google Scholar]
- 6.Motto, D. G., Ross, S. E., Wu, J., Hendricks-Taylor, L. R. & Koretzky, G. A. (1996) J. Exp. Med. 183, 1937-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liou, J., Kiefer, F., Dang, A., Hashimoto, A., Cobb, M. H., Kurosaki, T. & Weiss, A. (2000) Immunity 12, 399-408. [DOI] [PubMed] [Google Scholar]
- 8.Yablonski, D., Kuhne, M. R., Kadlecek, T. & Weiss, A. (1998) Science 281, 413-416. [DOI] [PubMed] [Google Scholar]
- 9.Pivniouk, V., Tsitsikov, E., Swinton, P., Rathbun, G., Alt, F. W. & Geha, R. S. (1998) Cell 94, 229-238. [DOI] [PubMed] [Google Scholar]
- 10.Clements, J. L., Yang, B., Ross-Barta, S. E., Eliason, S. L., Hrstka, R. F., Williamson, R. A. & Koretzky, G. A. (1998) Science 281, 416-419. [DOI] [PubMed] [Google Scholar]
- 11.Kumar, L., Pivniouk, V., de la Fuente, M. A., Laouini, D. & Geha, R. S. (2002) Proc. Natl. Acad. Sci. USA 99, 884-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Myung, P. S., Derimanov, G. S., Jordan, M. S., Punt, J. A., Liu, Q. H., Judd, B. A., Meyers, E. E., Sigmund, C. D., Freedman, B. D. & Koretzky, G. A. (2001) Immunity 15, 1011-1026. [DOI] [PubMed] [Google Scholar]
- 13.Molina, T. J., Kishihara, K., Siderovski, D. P., van Ewijk, W., Narendran, A., Timms, E., Wakeham, A., Paige, C. J., Hartmann, K. U., Veillette, A., et al. (1992) Nature 357, 161-164. [DOI] [PubMed] [Google Scholar]
- 14.Sanzenbacher, R., Kabelitz, D. & Janssen, O. (1999) J. Immunol. 163, 3143-3152. [PubMed] [Google Scholar]
- 15.Wang, D. Z., McCaffrey, P. G. & Rao, A. (1995) Ann. N.Y. Acad. Sci. 766, 182-194. [DOI] [PubMed] [Google Scholar]
- 16.Strasser, A., Harris, A. W. & Cory, S. (1991) Cell 67, 889-899. [DOI] [PubMed] [Google Scholar]
- 17.Grynkiewicz, G., Poenie, M. & Tsien, R. Y. (1985) J. Biol. Chem. 260, 3440-3450. [PubMed] [Google Scholar]
- 18.Jain, J., Miner, Z. & Rao, A. (1993) J. Immunol. 151, 837-848. [PubMed] [Google Scholar]
- 19.Rooney, J. W., Sun, Y. L., Glimcher, L. H. & Hoey, T. (1995) Mol. Cell. Biol. 15, 6299-6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feske, S., Okamura, H., Hogan, P. G. & Rao, A. (2003) Biochem. Biophys. Res. Commun. 311, 1117-1132. [DOI] [PubMed] [Google Scholar]
- 21.Bendelac, A., Matzinger, P., Seder, R. A., Paul, W. E. & Schwartz, R. H. (1992) J. Exp. Med. 175, 731-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swat, W., Dessing, M., von Boehmer, H. & Kisielow, P. (1993) Eur. J. Immunol. 23, 739-746. [DOI] [PubMed] [Google Scholar]
- 23.Cantrell, D. (1996) Annu. Rev. Immunol. 14, 259-274. [DOI] [PubMed] [Google Scholar]
- 24.Gonen, R., Beach, D., Ainey, C. & Yablonski, D. (2005) J. Biol. Chem. 280, 8364-8370. [DOI] [PubMed] [Google Scholar]
- 25.Jia, C. Y., Nie, J., Wu, C., Li, C. & Li, S. S. (2005) Mol. Cell. Proteomics 4, 1155-1166. [DOI] [PubMed] [Google Scholar]
- 26.Swan, K. A., Alberola-Ila, J., Gross, J. A., Appleby, M. W., Forbush, K. A., Thomas, J. F. & Perlmutter, R. M. (1995) EMBO J. 14, 276-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carpenter, G. & Ji, Q. (1999) Exp. Cell Res. 253, 15-24. [DOI] [PubMed] [Google Scholar]
- 28.Heyeck, S. D., Wilcox, H. M., Bunnell, S. C. & Berg, L. J. (1997) J. Biol. Chem. 272, 25401-25408. [DOI] [PubMed] [Google Scholar]
- 29.Ebinu, J. O., Stang, S. L., Teixeira, C., Bottorff, D. A., Hooton, J., Blumberg, P. M., Barry, M., Bleakley, R. C., Ostergaard, H. L. & Stone, J. C. (2000) Blood 95, 3199-3203. [PubMed] [Google Scholar]
- 30.Tao, X., Constant, S., Jorritsma, P. & Bottomly, K. (1997) J. Immunol. 159, 5956-5963. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.