Abstract
Engineering drought -resistant crop plants is a critically important objective. Overexpression of the vacuolar H+-pyrophosphatase (H+-PPase) AVP1 in the model plant Arabidopsis thaliana results in enhanced performance under soil water deficits. Recent work demonstrates that AVP1 plays an important role in root development through the facilitation of auxin fluxes. With the objective of improving crop performance, we expressed AVP1 in a commercial cultivar of tomato. This approach resulted in (i) greater pyrophosphate-driven cation transport into root vacuolar fractions, (ii) increased root biomass, and (iii) enhanced recovery of plants from an episode of soil water deficit stress. More robust root systems allowed transgenic tomato plants to take up greater amounts of water during the imposed water deficit stress, resulting in a more favorable plant water status and less injury. This study documents a general strategy for improving drought resistance of crops.
Keywords: root development, biotechnology, water deficit stress, tomato
Overcoming food shortages through self-reliance and sustainable agriculture is a major goal worldwide. To meet this challenge, it will be necessary to increase the productivity of land already under cultivation and to regain the use of arable land lost to scarce water supplies (1, 2). Transgenic crops engineered to tolerate some measure of drought could significantly increase yields for many developing countries and help to alleviate an increasingly imminent threat of famine (3).
Our increasing knowledge of drought stress adaptation processes has been key to engineering plants with improved tissue tolerance to dehydration (4–7). However, these strategies do not necessarily translate into improved productivity under drought conditions. Engineering plants with drought avoidance characteristics, i.e., that reduce the negative impact of soil water deficits on productivity by maintaining a more favorable plant water status, has been limited (8). Some naturally occurring drought-tolerant plant species seem to employ deep and dense root systems to maximize water uptake (9–15). Despite their obvious role in water uptake, roots have not been targeted in genetic engineering strategies to improve crop performance under drought conditions (16).
We have shown that overexpression of the H+-pyrophosphatase (H+-PPase) AVP1 results in salt and water stress-tolerant Arabidopsis plants (17). The tolerance was initially explained by an enhanced uptake of ions into their vacuoles. Presumably, the greater AVP1 activity in vacuolar membranes provides increased vacuolar H+ to drive the secondary active uptake of toxic (i.e., sodium) and nontoxic ions into the vacuole. The resulting decline in vacuolar osmotic potential may trigger water uptake, permitting plants to survive under conditions of low soil water potentials (18). Significantly, further characterization of these AVP1-overexpressing plants revealed a dramatic enhancement of their root development, with obvious implications for their ability to withstand drought (19). Moreover, root development in avp1-1 loss-of-function mutants was impaired (19). These results suggest that the H+-PPase AVP1 is a potential target for genetic engineering of root systems in agriculturally important crop plants.
To test whether the water stress resistance phenotype triggered by the overexpression of AVP1 in Arabidopsis (17) could provide a more universal strategy to improve the performance of crops under water deficit conditions, we engineered plants of a commercial cultivar of tomato (Lycopersicon esculentum) to ectopically express the Arabidopsis AVP1 H+-PPase. Here, we present data that show that transgenic tomatoes expressing the Arabidopsis H+-PPase are indeed more resistant than controls to imposed soil water deficits. We report the ability to engineer the root development of an agriculturally important crop to reduce plant damage due to water deficit stress conditions.
Methods
Plant Material, Transformation, and Growth Conditions. Tomato (Lycopersicon esculentum Mill. cultivar Money Maker) transformation was performed by means of the Agrobacterium-mediated transformation method using cotyledon and hypocotyl explants as described (20). Agrobacterium tumefaciens strain GV3101, with either the pGR395 (CaMV35S::AVP1D) or the pPZP212 expression vector (17), was used for this study. The pRG395 plasmid was generated by cloning the AVP1D gene downstream of a tandem repeat of the 35S promoter of Cauliflower mosaic virus as described (17). Of note, the AVP1D gene is the E229D gain-of-function mutant of the AVP1 gene that has a coordinated increase of both PPi hydrolytic activity and PPi-dependent H+-translocation (21). T1 XAVP1D plants were screened on 100 mg/liter kanamycin selection medium and then transferred to soil. Segregation analysis on T2 seeds from self-pollinated T1 XAVP1D plants were carried out on 100 mg/liter kanamycin selection medium, and homozygous T2 XAVP1D lines were selected to use in all of the experiments reported in this study.
DNA Isolation and Southern Blot Analysis. Tomato genomic DNA was extracted from leaf tissue harvested from primary plants and their progeny as described (22). All further DNA steps and blotting were done by using standard conditions (23). The BglII fragment containing the AVP gene from pGR209 was used as a probe (17).
Membrane Isolation and Western Blots. Tonoplast-enriched fractions were isolated from root tissues of hydroponically grown tomato plants (see below). Protein separated by 10% SDS/PAGE was immunoblotted with antibodies raised against a keyhole limpet hemocyanin-conjugated peptide (CTKAADVGADLVGKIE) corresponding to the PPi-binding site of the Arabidopsis H+-PPase that is conserved among all of the reported plant H+-PPases (24). The original design of this antibody was reported by Rea et al. (25). Quantification of the intensity of the H+-PPase in the Western blots was performed by using Bio-Rad quantity one quantitation software.
Transport Assays. Preparation of membrane vesicles, H+-pump activity, and Ca2+ uptake were performed on 6-week-old plants grown in hydroponic conditions (26, 27). Vacuolar membrane fractions were isolated from tomato root tissue by purification of the microsomal membrane fraction through a two-step sucrose gradient (28). Only the V-ATPase inhibitor bafilomycin, a marker of tonoplast, inhibited H+ pump activity measured in vesicles in this fraction, whereas inhibitors of other membrane-localized H+-ATPases did not inhibit H+ pump activity (data not shown), indicating that the tonoplast fraction isolated from the tomato root tissue had no significant contamination from other membrane fractions. Hydrolytic activity of V-type H+-ATPase and vacuolar H+-PPase was determined by measuring the release of inorganic phosphate (Pi) essentially as described (29). Time-dependent 10 μM 45CaCl2 uptake measurements into membrane vesicles were performed as described (26).
Soil Water Deficit Experiments. Seeds from T2 homozygous vector-control and XAVP1D expressing lines were germinated on Murashige and Skoog (MS) inorganic salts, 3% (wt/vol) sucrose, MS vitamins, and 100 mg/liter kanamycin. Positive candidates were selected after 15 days and transferred to small pots containing sand, where a constant supply of a diluted MS solution was added every day for 3 weeks. After this time, the plants were repotted into 1-gallon pots filled with Metro-Mix 500 soil. The plants were regularly watered to field capacity and fertilized on a weekly basis with 1 gram per pot of 20:20:20 fertilizer (Scotts). The temperature of the greenhouse was maintained within a range of 25°C to 30°C. In a typical soil water deficit stress experiment, plants were grown for a period of 5 weeks in soil and watered regularly to field capacity; then the soil was allowed to dry by withholding water. The length of the stress varied depending on the climatic conditions at the time and location of the experiment. However, in every experiment, the water was withheld until all plants showed severe drought stress symptoms (i.e., visible loss of turgor and wilting) (see Fig. 3A). Seven independent experiments were performed with vector controls and T2 XAVP1D lines. Three of them took place at the greenhouse facilities of College Station, Texas, and four at the Agricultural Biotechnology greenhouse of the University of Connecticut.
Fig. 3.
Control and transgenic tomato plants respond differently to an imposed soil water deficit. Ten-week-old, well watered plants were subjected to imposed water stress by withholding irrigation. In each panel, images of six control plants are shown on the left and three plants each of lines XAVP1D-1 (white arrows) and XAVP1D-2 (green arrows) are shown on the right. (A) Photographs were taken after 13 days of stress. Pots were irrigated to field capacity on day 13, and photographs were taken 1 day (B) and 4 days (C) after rewatering.
Plant Water Relations. In all cases, leaflets from mature, nonsenescing leaves were used. Leaf water potential was measured periodically during water stress regimes at the same time of day (9–10 a.m.) by using a PMS (Corvallis, OR) pressure chamber. During some stress regimes, plant water uptake (which occurred over the time period between leaf water potential measurements) was estimated by weighing the pots; a method used previously to calculate (long-term) transpirational water use of container-grown tomato plants (30). Care was taken to avoid soil loss from pots during the stress regimes. Plastic netting was placed within the pots before filling with potting mix, and the soil level in all pots was kept ≈5 cm below the rim. Relative water content was calculated by measuring the fresh, rehydrated (overnight incubation at 4°C with leaflet petioles in distilled water), and dry (80°C for a minimum of 2 days) weights of leaflets. The leaf osmotic potential was measured by using a Wescor (Logan, UT) 5500 osmometer. Pressure/volume isotherms (31) were generated by measuring the leaf water potential and relative water content of leaflets (≈20 per plant) that had been rehydrated overnight, and then before measurement, left to dehydrate under a small fan on a bench at room temperature for varying times.
Results
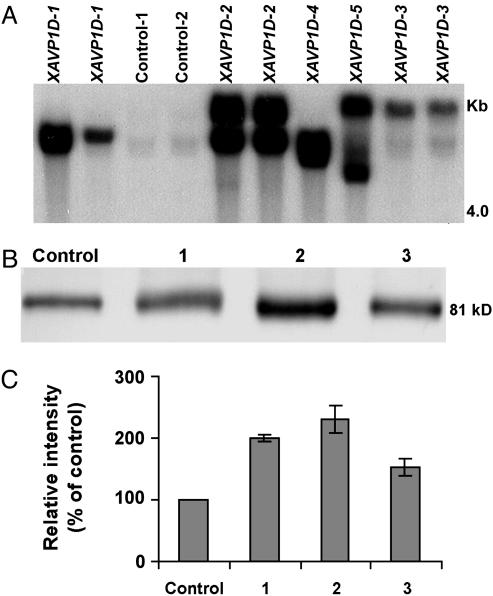
The E229D gain-of-function mutant (AVP1D) of the Arabidopsis H+-PPase AVP1 has been shown to coordinately increase both PPi hydrolytic activity and PPi-dependent H+-translocation when expressed in yeast (21). A construct containing the Arabidopsis AVP1D gene was introduced into the genome of Lycopersicon esculentum (cv. Moneymaker). Fourteen independent transgenic lines transformed with AVP1D (XAVP1D) were generated. After antibiotic selection, we randomly selected 10 transgenic lines and confirmed by Southern blot analysis that these lines contained the AVP1D expression vector (data not shown). The stable integration and transmission of the 35S::AVP1D chimera in the genome of T2 XAVP1D tomatoes was confirmed by Southern blot analysis (Fig. 1A). Of note, the lines we have termed XAVP1D-1 and XAVP1D-3 seem to contain single insertions, whereas line XAVP1D-2 had more than one integration event (Fig. 1 A). The T1 seeds of these three lines showed a segregation pattern of 3:1 for the kanamycin resistance marker gene consistent with a single insertion in lines XAVP1D-1 and XAVP1D-3 and the cosegregation of two copies of the AVP1D gene in line XAVP1D-2 (data not shown). To obtain homozygous T2 XAVP1D lines, segregation analysis on T2 seeds from self-pollinated T1 XAVP1D plants was carried out on 100 mg/liter kanamycin selection medium, and five homozygous T2 XAVP1D lines were obtained. Three (XAVPD1-1, -2, and -3) of five homozygous T2 XAVP1D lines showing a low copy number (either one or two copies) of the AVP1D gene were selected and further characterized. Western blot analysis of H+-PPase protein levels in tonoplast (vacuolar membrane)-enriched fractions isolated from roots of control and XAVP1-1, -2, and -3 tomatoes confirmed the ectopic expression of the Arabidopsis AVP1D proton pump in the transgenic lines (Fig. 1B). The antibody used was raised against a keyhole limpet hemocyanin-conjugated peptide corresponding to the PPi-binding site of the Arabidopsis H+-PPase that is conserved among all of the reported plant H+-PPases (32); thus, the expected cross reaction with the endogenous tomato H+-PPase was observed.
Fig. 1.
Molecular characterization of AVP1D expression in transgenic tomato. (A) Southern blot analysis indicates the absence of the transgene from control plants, and the presence of the 35S::AVP1D construct in genomic DNA of the transgenic tomato plants. In all cases, genomic DNA (10 μg) was digested with EcoRI, separated on a 0.9% agarose gel by electrophoresis, and probed with a BglII fragment of the AVP1D ORF. Results are shown for two replicate plants for each of the controls, and five independently generated transgenic lines (XAVP1D-1, -2, -3, -4, and -5). (B) Western blot analysis of H+-PPase protein expression in control and transgenic tomato. Size-fractionated protein (20 μg protein per lane) from tonoplast-enriched vesicles prepared from root tissue of vector-transformed (control) and XAVP1D-1 (blot 1), -2 (blot 2), and -3 (blot 3) lines was probed with an antibody immunoreactive to both the endogenous, and the recombinant H+-PPase. Results are shown for one representative experiment with a control plant and three transgenic lines (see A); this experiment was repeated a total of four times. Migration of molecular mass markers indicated that the single immunoreactive band in each lane had a mass of ≈81 kDa (i.e., the deduced mass of the endogenous and recombinant H+-PPases). (C) Quantification of relative H+-PPase protein levels in control (Control) and transgenic plants (lines 1–3 as above). Pooled results (means ± SE) are presented for four experiments similar to that shown in B; data are presented as protein levels in transgenic lines relative to that found in the control plants.
Measurements of the relative intensity of four independent Western blots of transgenic lines XAVP1D-1, XAVP1D-2, and XAVP1D-3, and the control are shown in Fig. 1C. Plants representing these transgenic lines had 200%, 230%, and 150% greater levels of H+-PPase protein in their root tonoplast membrane fractions, respectively, as compared with controls (Fig. 1C). These results are consistent with the expression of AVP1D in the transgenic tomato lines.
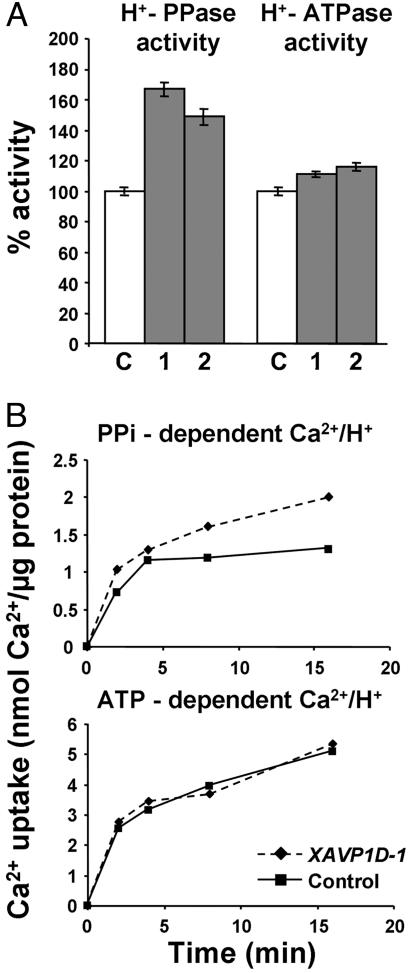
Biochemical and transport studies confirmed the functional expression of the recombinant protein in the transgenic tomato lines. Measurements of tonoplast H+-PPase hydrolytic activity from roots of two representative XAVP1D lines (XAVP1D-1 and XAVP1D-2) and control plants are shown in Fig. 2A. Results indicate that transgenic lines have a mean 56% increase in activity as compared with the control plants. Measurement of V-type H+-ATPase activity demonstrate that there was no significant change in activity of this other tonoplast enzyme in the transgenic lines compared with the control plants (Fig. 2 A). Further characterization of differences in H+-PPase activity between control and XAVP1D plants is shown in Fig. 2B. Kinetics of PPi- and ATP-dependent 45Ca2+ uptake into vacuolar membrane vesicles from transgenic (XAVP1D-1 and XAVP1D-2) and control plants were monitored. A proton gradient across the membrane vesicles (acid inside) was generated by activation of either the PPi-dependent H+-PPase or the ATP-dependent H+-ATPase. H+/Ca2+ antiport activity was measured in the presence of the Ca2+-ATPase inhibitor vanadate and in the presence and absence of the protonophore FCCP. The enhanced PPi-dependent 45Ca2+ uptake capacity displayed by the XAVP1D lines resembled that reported for the Arabidopsis AVP1OX lines (17). That is, the PPi-dependent 45Ca2+ uptake was 31% greater in vesicles from the XAVP1D lines than the control, whereas ATP-dependent (H+-ATPase-energized) 45Ca2+ uptake was unchanged by AVP1D expression (Fig. 2B).
Fig. 2.
Transgenic tomato plants have increased H+-PPase activity. (A) H+-PPase and H+-ATPase hydrolytic activity was determined from purified vacuolar membrane vesicles from root tissue of two lines of transgenic plants (bar 1 = XAVP1D-1 and bar 2 = XAVP1D-2) and control plants (bar C). Results (means ± SE of four to five replications) are shown as percentage of the control activity. (B) Ca2+/H+ antiport activity into vacuolar membrane vesicles from root tissue of XAVP1D-1 and control plants. A time course of 10 μM 45CaCl2 uptake was measured after energization by either ATP (H+-ATPase dependent) or PPi (H+-PPase-dependent). Uptake was performed in the presence of the Ca2+-ATPase inhibitor vanadate. Net results are shown after subtraction of background values (uptake in the presence of the protonophore FCCP).
Drought stress phenotypes were assayed by imposing a 13-day water deficit stress period, after which both control and transgenic plants were adversely affected (Fig. 3A). However, the transgenic plants demonstrated recovery after relief of the water deficit stress that was not evident in controls (Fig. 3 B and C). These transgenic plants did not display deleterious phenotypes (e.g., reduced vegetative growth, flower set, fruit yield) during normal growth and development (data not shown). One possible deleterious consequence of H+-PPase overexpression could be the accumulation of toxic metals in the fruit of transgenic plants. We evaluated fruit cation contents of control and XAVP1D plants and found no significant difference in levels of Pb2+, Mo2+, Mn2+, Cd2+, Zn2+, Cu2+, Fe2+, or Ca2+ (data not shown). Further studies were undertaken to identify the physiological basis for the differences in recovery from the imposed water deficit stress shown by control and transgenic plants.
The water status of control and XAVP1D plants was followed as soil water content decreased during a stress regime (Fig. 4). Transgenic plants maintained greater leaf water potentials from day 4 onward compared with control plants (Fig. 4A). During the latter part of this stress cycle, the leaf water potential of XAVP1D plants was 0.2–0.3 MPa greater than control plants at the same day of stress. Relative water content was also monitored during the stress regime shown in Fig. 4A. This second method of monitoring plant water status also indicated that XAVP1D plants maintained a more favorable water balance during the latter part of the stress episode (data not shown). Enhanced plant water status during the latter part of the stress episode is also visually evident, as shown by the improved growth of XAVP1D plants compared with controls at day 5 of the imposed stress (Fig. 4B).
Fig. 4.
Transgenic plants maintain greater leaf water potentials and take up greater amounts of water during imposed soil water deficits. Sets of control and transgenic plants were subjected to water deficit stress as described in the legend of Fig. 3. In all cases, pots were irrigated to field capacity on day one. (A) Leaf water potentials of four control plants and a total of eight transgenic plants (representing lines XAVP1D-1, XAVP1D-2, and XAVP1D-5 from Fig. 1) were monitored during the imposed stress. Results are shown as means ± SE of leaf water potential measured in control and (pooled) transgenic plants. (B) Photograph of representative control and XAVP1D plants taken on day 5 of the imposed stress. (C) Water uptake of control and transgenic plants during an imposed period of soil water deficit. The experiment shown in A was repeated with a second set of control and transgenic plants (representing lines XAVP1D-1 and XAVP1D-2); in this case, four transgenic and four control plants were tested. Water uptake measurements are shown for periods of the stress during which transgenic plants displayed significantly greater leaf water potentials (i.e., on days 5, 6, and 8, as was the case during the stress regime shown in A). The leaf water potentials with corresponding standard error values (in MPa) of control and transgenic plants measured on the day that water uptake was measured (i.e., on day 5, day 6, and day 8) are shown above the bars representing water uptake values.
One possible explanation for the maintenance of greater leaf water potentials (Fig. 4A) and enhanced plant performance (Fig. 4B) could be that stomata closure was greater in XAVP1D plants, thus restricting water usage during the stress. However, gas exchange analysis (data not shown) indicated that stomata conductance was similar in both sets of plants throughout the course of the water stress. An alternative explanation is that water uptake during the later part of the stress episode was greater in these transgenic plants. To test this hypothesis, leaf water potential and water uptake were both monitored during another imposed stress regime (Fig. 4C). The leaf water potential decline and the differences in leaf water potential between control and transgenic plants were similar to that demonstrated in the experiment shown in Fig. 4A. As shown in Fig. 4C, water uptake was determined to be significantly greater in transgenic plants as compared with control plants between days 3 and 5 (i.e., by 14%), days 5 and 6 (by 75%), and days 6 to 8 (by 45%). The greater degree of water uptake by transgenic plants occurred concomitantly with the maintenance of less negative leaf water potentials in this experiment (Fig. 4C, see values above each bar).
One adaptive physiological mechanism allowing for enhanced plant survival during periods of soil water deficits is leaf osmotic adjustment (31, 33–35). In prior work with Arabidopsis-overexpressing AVP1, greater solute accumulation in leaves was identified as one basis for altered response to water deficit (17). Therefore, we examined leaf osmotic adjustment in response to soil water deficits in control and transgenic tomato plants. Leaf osmotic adjustment was evaluated in rehydrated leaf tissue (31) of control and XAVP1D plants at the end of a water deficit stress regime by two methods. Neither measurements of osmotic potential with an osmometer (31) nor pressure/volume curve analysis (33) showed significant differences between the controls and the XAVP1D plants (data not shown).
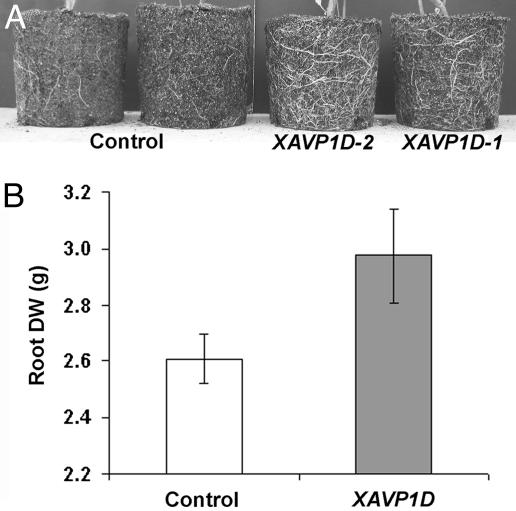
The water deficit recovery phenotype may be due in part to increased root growth in the AVP1-expressing plants. Results presented in Fig. 5 are consistent with this explanation for the enhanced performance displayed by transgenic plants in response to imposed soil water deficits. Visual observations (Fig. 5A) are in agreement with measurements of root dry weight (Fig. 5B); both indicated that transgenic tomato plants had significantly more extensive root systems. Root system biomass was also monitored on plants after exposure to an episode of soil water deficit as described in Fig. 4; XAVP1D plants were again found to have larger root systems than controls (data not shown).
Fig. 5.
Ectopic expression of the H+-PPase results in larger root systems. (A) Intact roots along with potting mix were removed from pots, exposing the root systems of 10-week-old, well watered plants. The intact root systems are shown for two representative control plants and two transgenic plants (representing lines XAVP1D-1 and XAVP1D-2). (B) Potting mix was removed from the intact root system of plants before the stress (as in A) by rinsing with water. The washed root mass was dried at 80°C for 1 week before measurement of dry weight (DW). Means (±SE) are shown for control plants (n = 6) and transgenic lines (n = 9) XAVP1D-1, XAVP1D-2, and XAVP1D-3 (three plants per line).
Discussion
Work presented here demonstrates that AVP1D expression increases root growth, water uptake, leaf water potentials, and plant survival under soil water deficit. Increased H+-PPase expression in tomato plants recapitulates the phenotypes seen in transgenic Arabidopsis lines (see below) and thus allows the engineering of root growth in agriculturally important crops for enhanced performance under soil water deficit conditions.
In some plant species, deeper and/or more extensive root systems are associated with increased drought resistance (9–15). A more extensive root system allows for water uptake to occur from a greater volume of the soil during periods of growth on limited soil water, thus reducing the extent of plant dehydration (9–15).
The development of a more robust root system in the AVP1D-expressing plants may provide the morphological and/or physiological basis for enhanced performance of plants exposed to low soil water conditions, resulting in less severe symptoms than those observed in controls (Figs. 3, 4, 5). Indeed, our transgenic plants maintained ≈0.2–0.3 MPa greater leaf water potentials than controls during the latter portion of the imposed soil water deficit stress regimes (Fig. 4). Haupt-Herting and Fock (36) evaluated soil water deficit effects on photosynthesis and physiology of Moneymaker tomato (the cultivar used in our studies) grown under similar conditions, and noted that a leaf water potential of –0.9 MPa could be considered a mild stress whereas moderate stress occurred at –1.3 MPa: i.e., a 0.4-MPa differential. Therefore, the 0.2- to 0.3-MPa difference in leaf water potential between control and transgenic plants as the leaf water potential declined below –0.9 MPa could be considered “physiologically relevant” in this context (see also Fig. 4B).
AVP1D expression certainly increases root growth in tomato to help facilitate improved water deficit recovery. Consistent with these findings, AVP1-overexpressing Arabidopsis lines also develop more robust root systems than wild type (19). In contrast, Arabidopsis avp1-1-null mutants have severely disrupted root development. The characterization of these gain- and loss-of-function AVP1 mutants revealed that AVP1 plays an important role in organ development through facilitating the auxin fluxes that regulate organogenesis (19). It is tempting to speculate that a similar mechanism may be responsible for the enhanced root development displayed by the AVP1D-expressing tomato plants, but further characterization is needed for conclusive evidence.
Crop plant species vary in their ability to undergo leaf osmotic adjustment in response to water deficits (37, 38). For example, in Arabidopsis, the up-regulation of the H+-PPase resulted in greater solute accumulation in leaves and enhanced root development, and both phenotypes had a positive impact on the plant performance under a water deficit stress (17, 19). Significantly, our data show that the ectopic expression of the Arabidopsis H+-PPase in tomato influenced only root development but had no effect in leaf osmotic adjustment. These results are consistent with the behavior of tomato reported by Torrecillas et al. (38), who found only a modest degree of leaf osmotic adjustment in response to soil water deficits.
We conclude that increasing H+-PPase expression in plants enhances root system development and that this phenotype can help confer a significant degree of water deficit stress resistance. This approach should be generally applicable to a range of agriculturally important plants and may be a general mechanism by which crops can be engineered to improve yields under water deficit conditions.
Acknowledgments
We thank T. Shigaki for comments and C. Andersen for editing. This work was supported by National Research Initiative of the U.S. Department of Agriculture (USDA) Cooperative State Research, Education and Extension Grant 2001-35100-10772 and Storrs Agricultural Experimental Station Hatch; by a University of Connecticut Research Foundation grant (to R.A.G.); by USDA Cooperative State Research, Education and Extension Grant 2004-34402-14768 (Designing Foods for Health) (to S.P.); by the USDA/Agricultural Research Service under Cooperative Agreement 58-6250-6001; by National Science Foundation Grant 0209777 (to K.D.H.); and by National Science Foundation Grant 00675 (to G.A.B.).
Author contributions: G.A.B., K.D.H., and R.A.G. designed research; S.P., J.L., J.K.P., G.A.B., H.Y., S.U., J.M., and R.A.G. performed research; K.D.H. and R.A.G. contributed new reagents/analytic tools; S.P., J.L., J.K.P., G.A.B., H.Y., S.U., K.D.H., and R.A.G. analyzed data; and J.K.P., G.A.B., K.D.H., and R.A.G. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviation: H+-PPase, H+-pyrophosphatase.
References
- 1.Herrera-Estrella, L. (1999) Proc. Natl. Acad. Sci. USA 96, 5978–5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Serrano, R. & Gaxiola, R. (1994) Crit. Rev. Plant Sci. 13, 121–138. [Google Scholar]
- 3.Thomson, J. A. (2002) J. Nutr. 132, 3441S–3442S. [DOI] [PubMed] [Google Scholar]
- 4.Chaves, M. M. & Oliveira, M. M. (2004) J. Exp. Bot. 55, 2365–2384. [DOI] [PubMed] [Google Scholar]
- 5.Penna, S. (2003) Trends Plant Sci. 8, 355–357. [DOI] [PubMed] [Google Scholar]
- 6.Vinocur, B. & Altman, A. (2005) Curr. Opin. Biotechnol. 16, 123–132. [DOI] [PubMed] [Google Scholar]
- 7.Zhang, J. Z., Creelman, R. A. & Zhu, J.-K. (2004) Plant Physiol. 135, 615–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laporte, M. M., Shen, B. & Tarczynski, M. C. (2002) J. Exp. Bot. 53, 699–705. [DOI] [PubMed] [Google Scholar]
- 9.Chaves, M. M., Pereira, J. S., Maroco, J., Rodrigues, M. L., Ricardo, C. P. P., Osorio, M. L., Carvalho, I., Faria, T. & Pinheiro, C. (2002) Ann. Bot. 89, 907–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costa e Silva, F., Shvaleva, A., Maroco, J. P., Almeida, M. H., Chaves, M. M. & Pereira, J. S. (2004) Tree Physiol. 24, 1165–1172. [DOI] [PubMed] [Google Scholar]
- 11.Ober, E. S. & Sharp, R. E. (2003) J. Exp. Bot. 54, 813–824. [DOI] [PubMed] [Google Scholar]
- 12.Pinheiro, H. A., DaMatta, F. M., Chaves, A. R. M., Loureiro, M. E. & Ducatti, C. (2005) Ann. Bot. 96, 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharp, R. E. & LeNoble, M. E. (2002) J. Exp. Bot. 53, 33–37. [PubMed] [Google Scholar]
- 14.Sharp, R. E., Poroyko, V., Hejlek, L. G., Spollen, W. G., Springer, G. K., Bohnert, H. J. & Nguyen, H. T. (2004) J. Exp. Bot. 55, 2343–2351. [DOI] [PubMed] [Google Scholar]
- 15.Tschaplinski, T. J., Tuskan, G. A., Gebre, G. M. & Todd, D. E. (1998) Tree Physiol. 18, 653–658. [DOI] [PubMed] [Google Scholar]
- 16.Beeckman, T. (2004) Trends Biotechnol. 22, 379–381. [DOI] [PubMed] [Google Scholar]
- 17.Gaxiola, R. A., Li, J., Undurraga, S., Dang, L. M., Allen, G. J., Alper, S. L. & Fink, G. R. (2001) Proc. Natl. Acad. Sci. USA 98, 11444–11449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaxiola, R. A., Fink, G. R. & Hirschi, K. D. (2002) Plant Physiol. 129, 967–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, J., Yang, H., Peer, W. A., Richter, G., Blakeslee, J. J., Bandyopadhyay, A., Titapiwantakun, B., Undurraga, S., Khodakovskaya, M., Richards, E. L., et al. (2005) Science 310, 121–125. [DOI] [PubMed] [Google Scholar]
- 20.Park, S. H., Morris, J. L., Park, J., Hirschi, K. D. & Smith, R. H. (2003) J. Plant Physiol. 160, 1253–1257. [DOI] [PubMed] [Google Scholar]
- 21.Zhen, R. G., Kim, E. J. & Rea, P. A. (1997) J. Biol. Chem. 272, 22340–22348. [DOI] [PubMed] [Google Scholar]
- 22.Paterson, A. H., Brubaker, C. L. & Wendel, J. F. (1983) Plant Mol. Biol. Rep. 11, 122–127. [Google Scholar]
- 23.Park, S. H., Pinson, S. R. M. & Smith, R. H. (1996) Plant Mol. Biol 32, 1135–1148. [DOI] [PubMed] [Google Scholar]
- 24.Maeshima, M. (2000) Biochim. Biophys. Acta 1465, 37–51. [DOI] [PubMed] [Google Scholar]
- 25.Rea, P. A., Kim, Y., Sarafian, V., Poole, R. J., Davies, J. M. & Sanders, D. (1992) Trends Biochem Sci. 17, 348–353. [DOI] [PubMed] [Google Scholar]
- 26.Cheng, N. H., Pittman, J. K., Barkla, B. J., Shigaki, T. & Hirschi, K. D. (2003) Plant Cell 15, 347–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirschi, K. D., Korenkov, V. D., Wilganowski, N. L. & Wagner, G. J. (2000) Plant Physiol. 124, 125–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vera-Estrella, R., Barkla, B. J., Bohnert, H. J. & Pantoja, O. (2004) Plant Physiol. 135, 2318–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams, L. E., Nelson, S. J. & Hall, J. L. (1990) Planta 182, 540–545. [DOI] [PubMed] [Google Scholar]
- 30.Holbrook, N. M., Shashidhar, V. R., James, R. A. & Munns, R. (2002) J. Exp. Bot. 53, 1503–1514. [PubMed] [Google Scholar]
- 31.Gupta, S. & Berkowitz, G. A. (1987) Plant Physiol. 85, 1040–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim, E. J., Zhen, R. G. & Rea, P. A. (1994) Proc. Natl. Acad. Sci. USA 91, 6128–6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta, S. A., Berkowitz, G. A. & Pier, P. A. (1989) Plant Physiol. 89, 1358–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morgan, J. M. (1984) J. Exp. Bot. 31, 655–665. [Google Scholar]
- 35.Radin, J. W. (1983) in Limitations to Efficient Water Use in Crop Production, ed. Taylor, H. M. (Am. Soc. Agron., Madison, WI), pp. 267–276.
- 36.Haupt-Herting, S. & Fock, H. P. (2002) Ann. Bot. 89, 851–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perez-Alfocea, F., Estan, M. T., Caro, M. & Guerrier, G. (1993) Physiol. Plant. 87, 493–498. [Google Scholar]
- 38.Torrecillas, A., Guillaume, C., Alarcon, J. J. & Ruiz-Sanchez, M. C. (1995) Plant Sci. 105, 169–176. [Google Scholar]