Abstract
Recent studies have established distinctive serum polypeptide patterns through mass spectrometry (MS) that reportedly correlate with clinically relevant outcomes. Wider acceptance of these signatures as valid biomarkers for disease may follow sequence characterization of the components and elucidation of the mechanisms by which they are generated. Using a highly optimized peptide extraction and matrix-assisted laser desorption/ionization–time-of-flight (MALDI-TOF) MS–based approach, we now show that a limited subset of serum peptides (a signature) provides accurate class discrimination between patients with 3 types of solid tumors and controls without cancer. Targeted sequence identification of 61 signature peptides revealed that they fall into several tight clusters and that most are generated by exopeptidase activities that confer cancer type–specific differences superimposed on the proteolytic events of the ex vivo coagulation and complement degradation pathways. This small but robust set of marker peptides then enabled highly accurate class prediction for an external validation set of prostate cancer samples. In sum, this study provides a direct link between peptide marker profiles of disease and differential protease activity, and the patterns we describe may have clinical utility as surrogate markers for detection and classification of cancer. Our findings also have important implications for future peptide biomarker discovery efforts.
Introduction
Recent scientific advances, including sequencing of the genome (1) and new approaches to modeling complex biological systems (2) may ultimately lead to improved anticancer therapies. However, at this time, the best anticancer strategies still rely on early detection followed by close monitoring for early relapse so that therapies can be appropriately adjusted (3). There is optimism, however, that advances in genomics and proteomics may more readily lead to new and improved approaches in molecular diagnostics, capable of classifying patients into subgroups based on their predicted response to individual treatments (4, 5). Appropriate biomarker-based screens should be minimally invasive and reproducible. A simple blood or urine test that detects molecules specific to tumor tissues would be ideal. In addition, screening technology must be sufficiently sensitive to detect early cancers but specific enough to classify individuals without cancer as being free of disease (3).
While genes contain hereditary information, including genetic predisposition to cancer and other diseases, it is their products that confer the actual phenotypes of living organisms and, in the case of disease, normal versus pathological states. Since there are many posttranslational events that can modify biological structure, function, and degradation of proteins, the knowledge of genes alone does not even begin to describe the full complexity of biological systems. From a screening perspective, it is also mostly the proteins that are secreted or otherwise released from tissues into the bloodstream (6, 7). Yet, despite an intensive search during the past decade(s), only a very small number of identified cancer biomarkers, all plasma proteins (e.g., prostate-specific antigen [PSA], carcinoembryonic antigen [CEA], cancer antigen 125 [CA125], and thyroglobulin), have proven clinically useful, often in combination with other diagnostic tools, for the prognosis of response to therapy, relapse, and survival and for defining the rate of progression and monitoring of treatment, but they have been less useful for broad-based population screening (8, 9). Those proteins are typically present in plasma or serum at subnanomolar concentrations and require individual immunoassays for detection and quantitation (10, 11). New and improved cancer biomarkers and facile detection methods are clearly in order but have so far eluded discovery and implementation. Even the most recent approaches, using identity-based proteomics that involve digesting (e.g., with trypsin) complex protein mixtures into peptides for mass spectrometric (MS) analysis, have yet to translate into any practical applications, largely because of insufficient instrumental dynamic range and because the elaborate fractionation procedure coupled to multiple MS runs to detect low-abundant tryptic peptides precludes processing statistically relevant sample numbers (12).
As cancer involves the transformation and proliferation of altered cell types that produce high levels of specific proteins and enzymes such as proteases, e.g., PSA and prostate-specific membrane antigen (PSMA) (13, 14), it not only modifies the array of existing serum proteins (the serum proteome; ref. 6, 7) but also their metabolic products, i.e., peptides (the serum peptidome). It is well established that human serum contains thousands of proteolytically derived peptides (15–17), yet it remains unclear to date whether this complex peptidome may provide a robust correlate of some biological events occurring in the entire organism. As advances in MS now permit the display of hundreds of small- to medium-sized peptides using only microliters of serum (17, 18), several recent reports have advocated the use of MS-based serum peptide profiling to determine qualitative and quantitative patterns, often referred to as signatures or barcodes, that indicate the presence/absence of diseases such as cancer (19–24). However, this work has come under intense criticism as growing evidence has indicated that uncontrolled variables related to both clinical and analytical chemistry and/or signal processing artifacts may have tainted the published results (12, 25–29). Skepticism was further fueled by the use of low-grade MS equipment in these analyses, which precluded comprehensive, high-resolution read-outs, and because the identities of only a few putative markers have been established so far (30–32). The proof of the potential value of this new approach will be in the ability of several laboratories to independently show that the highly discriminatory peptides have the same amino acid sequences. To date, this has not been done.
Working toward this stated goal, we have previously developed an automated procedure for the simultaneous measurement of peptides in serum that utilizes magnetic, reverse-phase beads for analyte capture and a matrix-assisted laser desorption/ionization–time-of-flight (MALDI-TOF) MS read-out (18, 29). This system is more sensitive than surface capture on chips (33), as spherical particles have larger combined surface areas and therefore higher binding capacity than small-diameter spots. Coupled to high-resolution MS and MS/MS, hundreds of peptides have been detected in a single droplet of serum, many of which can be readily identified without further fractionation. The automation element facilitates throughput and ensures reproducibility. To round out the system, we have also developed a minimal entropy-based algorithm that simplifies and improves alignment of spectra and subsequent statistical analysis (29). With these tools in hand, we now sought to determine if selected patterns of serum peptides with known sequences can (a) separate cancer from noncancer, (b) distinguish among different types of solid tumors, and (c) allow class prediction with an independent validation set.
To this end, we have used visual inspection of spectral overlays, peptide ion relative intensity comparisons, and statistical analysis to sort through hundreds of features obtained by rigorous peptide profiling of 106 serum samples from patients with advanced prostate cancer or bladder or breast cancer and from healthy controls to identify several that are most predictive of outcome. We show that reduction in the number of key peptides to only a few (i.e., the signatures) that were easily recognized between samples did not adversely affect class predictions. MS/MS-based sequence identification of 61 signature peptides indicated that all were breakdown products, many related, of abundant proteins in the blood. By correlating the proteolytic patterns with disease groups and controls, we show that exoprotease activities superimposed on the ex vivo coagulation and complement-degradation pathways contribute to generation of not only cancer-specific but also cancer type–specific serum peptides. Our study therefore provides a direct link between peptide marker profiles of disease and differential protease activity. The patterns we describe may have clinical utility as surrogate markers for detection and classification of cancer.
Results
Unsupervised analysis of 651 peptide ion signals from MS-based serum profiling differentiates 3 types of cancer and controls.
We analyzed the serum peptide profiles of 73 patients with advanced prostate (n = 32), breast (n = 21), and bladder (n = 20) cancer, as well as 33 control sera from healthy volunteers, all collected at our institution using a single standard clinical protocol (29). Age distribution, gender, and clinical characteristics are provided in Supplemental Table 1 (supplemental material available online with this article; doi:10.1172/JCI26022DS1). Sample handling after collection was uniform, involving 2 freeze-thaw cycles to accomplish initial storage and subsequent aliquoting for peptide extraction and MS analysis (29). All 106 serum samples were processed fully automatically (i.e., peptides extracted on magnetic beads coated with C8 phase, washed, eluted, mixed with matrix, and deposited on the MALDI target plate) as a single batch, using a customized robot liquid handler followed within 1 hour by automated MALDI-TOF MS analysis (see Supplemental Methods). System reproducibility was verified on the same day by analysis, computer alignment, and visual comparison of 12 reference samples/spectra (see Supplemental Methods) as described (18, 29). Samples from patients with different cancers and from control individuals were then randomly distributed during processing and analysis. Processed spectra (see Supplemental Methods) were aligned using the custom entropycal program (29) and a total of 651 distinct mass/charge (m/z) values resolved in the 700–15,000 Da range. A spreadsheet (peak list) containing the normalized intensities (i.e., signal intensities, after baseline subtraction, were divided by the total ion current of the corresponding spectrum and multiplied by a scaling factor of 107) of all 651 peaks for each of the 106 samples was then taken for unsupervised, average-linkage hierarchical clustering using standard correlation. This resulted in clear, distinct patterns that differentiate disease from control as well as different types of solid tumor cancers in binary and multiclass comparisons (Figure 1).
Figure 1. Unsupervised hierarchical clustering and principal component analysis of MS-based serum peptide profiling data derived from 3 groups of cancer patients and healthy controls.
(A) Serum samples from healthy volunteers and patients with advanced prostate, bladder, and breast cancer were prepared following the standard protocol. The 4 groups were randomized before automated solid-phase peptide extraction and MALDI-TOF MS. Spectra were processed and aligned using the Qcealign script (see Supplemental Methods). A peak list containing normalized intensities of 651 m/z values for each of the 106 samples was generated. Numbers indicate the number of patients and controls analyzed in the respective groups. (B) Unsupervised, average-linkage hierarchical clustering using standard correlation as a distance metrics between each cancer group and the control in binary format. The entire peak list (651 × 106) was used. Columns represent samples; rows are m/z peaks (i.e., peptides). Dendrogram colors follow the color coding scheme of A. The heat map scale of normalized ion intensities is from 0 (green) to 200 (red) with the midpoint at 100 (yellow). (C) Hierarchical clustering of the 3 cancer groups plus controls (as in B). (D) Principal component analysis (PCA) of the 3 cancer groups plus controls. Color coding is as in A. The first 3 principal components, which account for most of the variance in the original data set, are shown.
Feature selection yields a 68–peptide ion signature that separates the 3 clinical groups and controls.
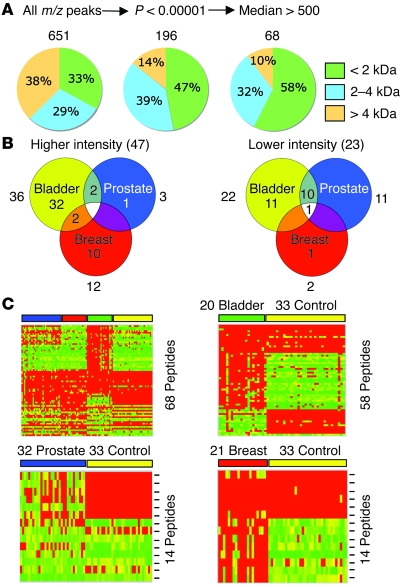
Anticipating future clinical development of this technology, we felt that correlations between patient samples involving 651 features would be difficult at different times and locations. Thus, a feature selection was performed using discriminant analysis to identify the most distinguishing peaks. A Mann-Whitney U test for each of the 3 cancer groups individually versus the control selected 196 peaks with a multiple comparison corrected P value of less than 1 × 10–5 for at least 1 type of cancer (Figure 2A). This number was further reduced to 68 by applying a threshold to the median ion intensities of each individual peak within a sample cohort (Figure 2A and Supplemental Table 2). The threshold was set high enough to select only robust peaks in the spectra with intensities that would permit MALDI MS/MS-based tandem MS sequencing and to exclude closely positioned neighboring peaks or “shoulders.” An m/z peak was selected if this criterion was met in at least 1 of the cancer groups or the control (see Supplemental Table 2). When feature selection was repeated using a multiclass Kruskal-Wallis test (adjusted P < 1 × 10–5) and the same median intensity threshold as above, 214 and 67 peaks were selected (data not shown). The majority of selected peaks corresponded to peptides with molecular mass less than 2,000 Da; most peptides with a mass of greater than 4,000 Da were removed (Figure 2A and Supplemental Table 2). Spectra from all samples were then color coded and overlaid to visually inspect the 68 peaks for correct assignment, degree of separation, and overall difference between cancer and control. Examples are shown in Figure 3. Forty-seven m/z peaks had higher ion intensities in 1 (or more) of the cancer groups, and 23 m/z peaks had lower intensities (Figure 2B). Interestingly, 2 were up in 1 type of cancer and down in another. Of the 68 peaks, 14 had biomarker (up or down) potential for prostate cancer (1 unique; 13 shared), 14 (11 unique) for breast cancer, and 58 (43 unique) for bladder cancer (Figure 2, B and C). The results, when represented in the form of heat maps in Figure 2C, indicated that data reduction (by ~90%) did not adversely affect the separation of the clinical groups. The results also illustrated that cancer-specific serum peptide signatures are not likely just indicators of a nonspecific inflammatory condition, such as arthritis or infection, in addition to cancer but are specific enough to distinguish different types of cancer from each other and from controls without cancer.
Figure 2. Feature selection and comparative analysis of serum peptide profiling data derived from 3 groups of cancer patients and healthy controls.
(A) The peak list was subjected to a Mann-Whitney U test for each individual cancer versus the control. Only peaks with adjusted P values of less than 0.00001 were passed through a second filter (median peak intensity > 500 units); a peak was selected if it passed the threshold in 1 cancer or in the control. (B) Venn diagrams show the number of peptides that passed both feature selection steps. The numbers shown outside the diagrams indicate the total number of peptides of a specific cancer group that were either up (Higher intensity) or down (Lower intensity). (C) Heat maps compare the selected features of the 3 cancer groups with controls in multiclass and binary formats. Columns represent samples (per group); rows are m/z peaks (not in numerical order). Peptides used in each binary comparison are the sum of those specifically higher and lower in each cancer group; the multiclass heat map contains the combined, nonredundant number of peptides. The multiclass, bladder, and breast heat map scales of normalized intensities range from 0 (green) to 500 (red) with the midpoint at 250 (yellow); those of the prostate map are from 0 (green) to 2,000 (red), with the midpoint at 1,000 (yellow).
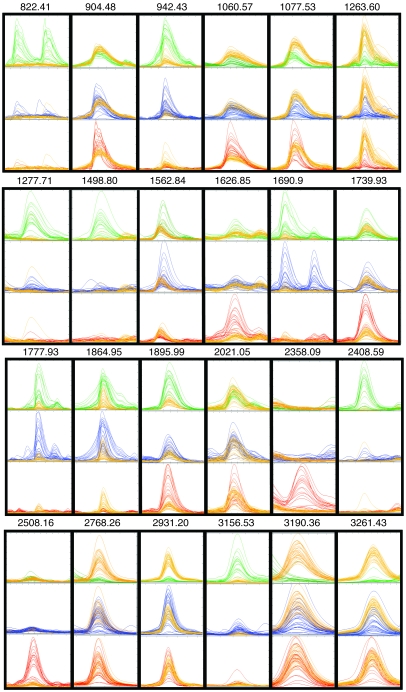
Figure 3. MALDI-TOF mass spectral overlays of selected peaks derived from serum peptide profiling of 3 groups of cancer patients and healthy controls.
Spectra were obtained, aligned, and normalized as described in Methods and were displayed using the mass spectra viewer. Peptide ions have been selected to illustrate group-specific differences in normalized intensities, except for 2021.05, which is provided here as an example of the vast majority of peptide ions with intensities that were not statistically different between any 2 groups. The 24 overlays (not to scale) each show a binary comparison for all spectra from either the bladder cancer (n = 20; green), prostate cancer (n = 32; blue), or breast cancer patient group (n = 21; red) versus the control group (n = 33; yellow). They are arrayed so that an identical mass range window is shown for each of the 3 binary comparisons in which spectral intensities have been normalized and scaled to the same size. The monoisotopic mass (m/z) is shown for each peptide ion peak.
Serum peptide signatures consist of a small but discrete set of sequence clusters.
Of the 68 selected peptides, 46 were positively identified by MALDI-TOF/TOF (Figure 4) and MALDI-Q/TOF MS/MS analysis and database searches (Figure 5). Note that the m/z values listed in Figure 5 are monoisotopic and therefore smaller than the corresponding average isotopic values listed in Supplemental Table 1. Interestingly, all but a few peptide sequences clustered into sets of overlapping fragments lined up within each group at either the C or N terminal end and with ladder-like truncations at the opposite ends. In fact, some sequence assignments had below-threshold scores (see Supplemental Methods) but could nonetheless be unequivocally assigned as the precursor ion mass and selected fragment ion masses (b or y) matched a particular rung in the ladder, taking into account whether the limited CID patterns were in agreement with established rules (34) of preferential peptide bond cleavage (e.g., Xaa-Pro or Asp/Glu-Xaa) and the putative sequence. Furthermore, 23 additional peptides outside the original group of 78 could also be matched to certain sequence clusters by hypothesis-driven, targeted MS/MS analysis. Fifteen of those had significant discriminant analysis adjusted P values (< 0.0002) for at least 1 cancer type but typically lower ion intensities (Figure 6). Two others (2553 and 2021; Figures 5 and 6) displayed very high but similar MS ion intensities across all cancer groups and the control with adjusted P values > 0.04 and can therefore be regarded as quasi-internal controls. Six more peptides (Figures 5 and 6) that fit into the clusters were randomly observed in samples of the cancer and control groups and had neither discriminant nor internal control value. The finding that the majority of peptide sequences obtained here collapsed into 10 or 11 clusters wasn’t entirely surprising in view of a recent finding that more than 250 of the most abundant plasma peptides are derived from some 20 serum proteins, also in largely overlapping clusters (17). It should be noted that we used an unbiased approach to identify marker peptides in which the peptides were selected first on the basis of discriminant analysis and then sequenced. This approach, commonly referred to as ion mapping, can be taken using any type of MS platform (35, 36).
Figure 4. MALDI-TOF/TOF MS/MS identification of serum peptide 2305.
0 as a fragment of complement C4a. Peptides from a serum sample of a breast cancer patient were extracted and analyzed by MS and the ion of choice selected for MS/MS analysis, as described in Supplemental Methods. The fragment ion spectrum shown here was taken for a Mascot MS/MS ion search of the human segment of the NR database (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Protein) and retrieved a sequence, GLEEELQFSLGSKINVKVGGNS ([MH]+ = 2305.19; Δ = 4 ppm), with a Mascot score of 38. b and y fragment ion series are indicated together with the limited sequences (arrows at top). Note that y ions originate at the C terminus and that the sequence therefore reads backwards (see direction of the arrows).
Figure 5. Serum peptide signatures for advanced prostate, bladder, and breast cancer.
Selected peptides identified by MALDI-TOF/TOF MS/MS are listed in clusters (ladders) of overlapping sequences, including 46 of the initial signature group of 68 (Figure 2 and Supplemental Table 2). m/z values are monoisotopic. Twenty-three additional peptides were positively matched to the existing clusters by hypothesis-driven, targeted MS/MS analysis. Overall, 61 entries had clear marker potential (adjusted P < 0.0002; Figure 6) for at least 1 cancer type and are color-coded blue (prostate cancer), green (bladder cancer), or red (breast cancer). Resulting signatures for the 3 cancers consist of 26 (prostate), 50 (bladder), and 25 (breast) peptide ions. Color-coded peptides have either higher (no filled circles) or lower (filled circles) differential ion intensities in a particular cohort of cancer samples compared with controls. C3f (m/z = 2021.05) and 1 member of the fibrinogen α cluster (m/z = 2553.01) gave comparable ion signals in all patient groups and control sera (see Figure 3, 2021, and Figure 6) and therefore represent effective internal standards (yellow). Six peptides (pink) were randomly observed. Residues in brackets were not experimentally observed but are shown to either indicate putative full-length sequences of the founder peptides and/or the positions of trypsin-like cleavage sites (Arg/Lys–Xaa).
Figure 6. Serum peptide signatures for advanced prostate, bladder, and breast cancer.
This table contains the same 69 entries as in Figure 5 plus additional details on the identified peptides (listed as m/z values), MS ion intensities, and signatures. The significance levels of 3 different Mann-Whitney U tests (columns 6–8) and of a multiclass Kruskal-Wallis test (column 9) are given. The actual signatures (blue, green, or red) are composed of entries that showed clear peptide ion marker potential (adjusted P < 0.0002) for at least 1 type of cancer. Adjusted P value is the overriding criterion, leading to final signatures of 26 (prostate), 50 (bladder), and 25 (breast) peptide ions (identical to those shown in Figure 5). The second column lists median intensities of each m/z peak in the control samples. Peak intensity ratios (columns 3–5) were calculated by dividing the median values of each m/z peak in each cancer group by the median value of the corresponding peak in the control samples. Ratios (r) for those peptides that are part of 1 or more signatures are shaded dark grey when the median signal is of higher intensity in a particular cancer (r ≥ 1.4) and lighter gray when it is lower (r ≤ 0.75). Norm., normalized.
Sequence clusters within the marker signatures derive from abundant serum peptides and protein precursors.
Three sequence clusters are derived from naturally occurring serum peptides, fibrinopeptide A (FPA), complement C3f, and bradykinin, which are each generated at an earlier stage from various plasma proteins through endoproteolytic cleavage, either at the initiation of the ex vivo intrinsic pathway (bradykinin, cleaved from high molecular weight–kininogen [HMW-kininogen] by plasma kallikrein) or during serum preparation (FPA, N terminally cleaved from fibrinogen by thrombin to form fibrin; C3f, released by factors I and H after prior conversion of C3 to C3b) (37, 38). The full-length founder peptides end with Arg or Lys preceded by a hydrophobic amino acid (Val, Leu, or Phe). Arg is partially removed from C3f and bradykinin (to form desArg-bradykinin [bradykinin that has the Arg removed]). Similar trypsin-like cleavages (Arg/Lys–Xaa) underlie formation of all other peptide clusters as well (see below). The C terminal basic amino acid is preceded by a hydrophobic amino acid (F, L, V, I, W, A) in 21 and by S, Q, or N in 15 out of the 39 observed cleavage sites (see Supplemental Table 4). Arg/Lys is typically removed (fully or in part) by a carboxypeptidase, except when preceded by Pro (3 out of 3 cases) or sometimes when preceded by Val (2 out 4). Further exoprotease degradation then proceeds at the N terminal or C terminal ends either to completion or until it stalls; many or all of the intermediates are typically represented (Figure 5 and Supplemental Table 3). This will be a recurring theme with most other clusters (see below).
Diagnostic MALDI-TOF spectral patterns consisting of N terminal FPA and C3f truncations have previously been found in sera of myocardial infarction patients (30). In contrast, we detected almost all these peptides (19 total) in control sera and showed that their presence is either consistently lower (all FPA fragments in all cancers; 3 C3f fragments in breast cancer) and/or higher (several C3f fragments in bladder and prostate cancer; 1 FPA fragment in breast cancer) in patient sera (Figures 5 and 7). Full-length C3f was present in all samples at equally high levels; full-length FPA was virtually absent in sera from bladder cancer patients. No fibrinopeptide B or fragments thereof were found in any of the samples. Decreased levels of FPA (fragments) in prostate, bladder, and breast cancer patients, as shown here, also contrast with earlier findings of elevated phospho-FPA levels in sera of ovarian cancer patients (measured by electrospray ionization–MS; ref. 31) and of FPA levels in gastrointestinal and breast cancers (measured immunochemically; ref. 39, 40).
Figure 7. Median ion intensities of serum peptides of selected sequence clusters relative to the corresponding values in the control group.
Median intensity for each peptide in each of the 3 cancer groups is plotted as the ratio versus the median intensity of the counterpart in the control group (r = patient/control). Ratios are plotted on a log scale ranging from 0.1 to 10. Bars pointing to the left (r < 1) or right (r > 1) indicate, respectively, lower or higher median intensities in a cancer group as compared with the control group. Peptides that didn’t show much difference in median ion intensity between patient and control groups map closely to or onto the center line (r = 1).
Bradykinin is believed to be a cancer growth factor, and various antagonists have therefore been tested as anticancer agents (41). We now find that bradykinin and desArg-bradykinin levels are higher in sera of breast cancer patients and lower in bladder cancer patients (Figure 5). The prohydroxylated forms (42) of each peptide also followed that trend (data not shown). The bradykinin and FPA parent proteins, fibrinogen α and HMW-kininogen, each contributed 1 additional sequence cluster, located in a different section of the precursor sequence, to the cancer serum peptide signatures (Figures 5 and 7 and Supplemental Tables 3 and 4). Interestingly, the bradykinin and other kininogen-derived peptides have opposite marker properties. For example, whereas bradykinin and desArg-bradykinin were of lower ion intensity in bladder cancer than in control sera, the other peptides (1944 and 2209) showed higher relative intensities (Figures 5 and 6). This observation provides a decisive argument against the most straightforward explanation of why some peptide ion intensities are higher or lower as compared with a control group, namely because the parent protein is up- or downregulated. As the concentration of HMW-kininogen can’t be up and down at the same time, this is clearly not the case.
One of the peptides (2724; Figure 5) in a cluster derived from the inter-α-trypsin inhibitor heavy chain H4 (ITIH4) precursor (43) covers amino acids 662–687 (Supplemental Tables 3 and 4) and is bracketed by 2 kallikrein cleavage sites (Phe-Arg–Xaa). Residues 662–688 likely represent a propeptide of unknown function (44). Like bradykinin, it ends with Pro-Phe-Arg. Several longer ITIH4 precursor fragments span the first kallikrein cleavage site, including a peptide (3272; at 658–687) reported to be a biomarker for early stage ovarian cancer (32). It further appears that variations in N terminal truncation in the ITIH4 cluster by just a few amino acids can produce fairly selective ion markers for different cancers. Median ion intensities of peptides 3971 and 3273, for instance, were clearly highest in bladder cancer samples, peptides 2358 and 2184 were highest in breast cancer, and 2271 was highest in prostate cancer. Also of note, peptide 2115 matches the sequence of an ITIH4 splice variant (PRO1851; Supplemental Table 4) and appears to have biomarker capacity for each cancer type, particularly for bladder and breast (Figure 6).
Another cluster consisting of 2 × 4 peptides located on either site of a single Ile-Arg—Xaa cleavage site is derived from the complement C4a precursor (45) (Figure 5 and Supplemental Tables 3 and 4). This C4a cluster has the highest incidence of ion markers for breast cancer, more than in any other cluster and also more than C4a-derived bladder cancer markers (Figure 6). Only a single ion (peptide 1763) of this cluster is a marker for prostate cancer and is shared in that capacity with the other 2 cancer types. On the other hand, all but 1 ion marker derived from apoA-I, apoA-IV, and apoE are bladder cancer specific, all with appreciably higher ion intensities; the exception (apoA-IV, peptide 1971) is actually highly selective and statistically the most significant (P = 5.5 × 10–13) ion marker for breast cancer (Figures 5 and 6).
Upregulation of clusterin (i.e., apoJ) has been correlated by immunohistochemistry with progression of both prostate and bladder cancer (46–48). The 10–amino acid clusterin fragment that we detected at elevated concentrations in sera of bladder and prostate cancer patients is located at the C terminus of the β chain (Supplemental Tables 2 and 3). A single cut is sufficient to release this peptide, following separation of the clusterin β (N-t) and α (C-t) chains by cleavage of a Val-Arg–Xaa bond. A 6–amino acid subfragment thereof has in turn statistically significant marker potential for bladder cancer (Figures 5 and 6), which is in keeping with the trend for most other peptides from apoA-I, apoA-IV, and apoE.
Finally, 2 ions (peptides 2602 and 2451), each with higher median intensities in breast cancer samples than in controls, corresponded to peptides derived from Factor XIIIa and thransthyretin (Figures 5 and 6). Peptide 2602 corresponded to the C terminal 25 amino acids of the factor XIIIa propeptide (37 residues long) (Supplemental Tables 3 and 4). Interestingly, Factor XIII itself has been found downregulated in breast tumors compared with normal mammary tissues (49). While we don’t know whether this was also the case in the patients from whom the blood samples in our study were obtained, it would contrast with our observations, further arguing against a model that higher ion intensities (i.e., peptide concentrations) are the simple consequence of upregulated precursors.
Cancer type–specific peptide signatures contain selected members from several different sequence clusters.
In all, 69 serum peptides are listed in Figure 5 (with matching information provided in Figure 6). Of those, 61 have clear MALDI-TOF MS ion marker potential (adjusted P < 0.0002) for at least 1 type of cancer and are color coded in blue (prostate cancer), green (bladder cancer) or red (breast cancer). The resulting signatures for the 3 cancer types consist of 26 (prostate), 50 (bladder), and 25 (breast) peptides, several of which occur in 2 or all 3 cancer groups. Compared with healthy control samples, median intensities of ion markers can be higher (Figure 5) or lower in any particular cancer group: 16 higher and 10 lower (16+/10–) in prostate cancer; 31+/19– in bladder cancer; and 19+/6– in breast cancer. Only 3 peptides in each of the up or down categories were shared by all cancer groups. One peptide from the C4a and 2 from the ITIH4 cluster had consistently higher ion intensities in all cancers than in healthy controls; 3 FPA fragments were lower in all cancers. The rest of the ion markers were either in common between 2 groups or, more often, unique to a single patient cohort (Figure 5). Of note are 9 apo peptides (apoA-I, apoA-IV, apoE, and apoJ) and 3 C3f peptides of selectively higher ion intensities in bladder cancer and 4 C4a, 2 bradykinin, and 1 transthyretin peptides higher in breast cancer. All 3 peptide ions that were of uniquely lower intensity in breast cancer derived from C3f. Interestingly, some of the shared marker ions had higher median intensities compared with controls in 1 type of cancer but lower in another (Figures 5 and 6). For instance, 5 peptide ions had higher than control median intensities in breast cancer samples, lower than control intensities in bladder cancer samples, and no appreciable marker value for prostate cancer. A single ITIH4 peptide (842; HAAYPF) was relatively higher in prostate cancer patients but virtually absent in bladder cancer.
It appeared there were no clear rules or trends in what clusters and in particular what rungs in the peptide sequence ladders may have ion marker value for one or another type of cancer, if any. In an attempt to find such trends or to at least better visualize any global differences that might exist, we plotted the ratios of the median ion intensities for each of the peptides in 4 major clusters between each cancer group and the healthy controls (i.e., r = patient/control). The center line in the panels of Figure 7 represents no difference (r = 1); bars pointing to the left (r < 1) or right (r > 1) indicate, respectively, lower or higher median. Even in the case of the FPA ladder where nearly all peptides in cancer sera produced ion signals of lower intensities than in controls, the actual ratios vary for each rung and for each cancer type. Of note is the seemingly total absence (r = 0) of full-length FPA in sera of bladder cancer patients. The 3 other clusters exhibited a pronounced internal variability with median intensity ratios that were mostly over but also equal to or under 1. Visual inspection of the 4 color-coded graphs (33 × 3 data points) in Figure 7 readily distinguishes the 3 cancer types. There is a trend for peptides in bladder cancer sera to exhibit relatively high ion intensities in the C3f cluster and rather variable intensities in the C4a and ITIH4 clusters and for some peptides in the C3f cluster to be of lower intensity and others in the C4a cluster to be of higher intensity in breast cancer sera. Ion intensities of peptides in prostate cancer sera don’t seem to follow those trends but are selectively more pronounced in some of the smaller peptides of the ITIH4 cluster. Interestingly, there is 1 rung in each of the C3f, C4a, and ITIH4 ladders (Figure 7) for which median ion intensities in the control samples were virtually zero yet were much higher in all 3 cancer types, resulting in very high ratios for each.
Taken together, the data in Figure 7, based in equal parts on statistical analysis (Figure 6), visual inspection of spectra overlays (Figure 3), peptide sequencing (Figures 4 and 5), and relative ion intensity analysis, indicate that the human serum peptidome holds information in the form of signatures consisting of a few dozen peptides each that can distinguish 3 different cancers from controls as well as from each other.
Peptide ion signatures provide accurate class prediction for an external validation set of prostate cancer samples.
To evaluate the robustness of the identified groups of markers, we tested the peptide signatures on a set of 41 independent serum samples from patients with advanced prostate cancer (prostate 2 [PR2]) (Figures 8 and 9A). The assignment of the prostate cancer samples into the training set (prostate 1 [PR1]) or the test set (PR2) was random but preserved the same demographic/pathological parameters (e.g., age, PSA levels, Gleason score, and survival time). None of the samples in the test set had been previously included in the supervised analysis, which therefore allowed for the estimation of true predictive accuracy. The 41-member test set was analyzed following standard protocol and a new spreadsheet generated that also included all data from the original 106 training samples. Peptide ions from feature list 2 (68 peptides; see Figures 2A and 8) and from the prostate cancer signature (26 sequenced peptides; Figures 5 and 6) were then selectively used for comparison of the control, PR1, and PR2 groups by hierarchical clustering (Figure 9B) and principal component analysis (Figure 9C). Samples from PR1 and PR2 were for the most part separated from the controls. Individual comparisons of each of these 26 peptide ions among the 3 sample groups indicated that the intensities of 26 out of 26 were statistically different (adjusted P < 0.0002, i.e., the P value to create the signature; see Figure 6) between PR1 and control, 23 out of 26 between PR2 and control, and only 1 out of 26 between PR1 and PR2. Finally, support vector machine–based (SVM-based) class predictions in either binary or multiclass formats were then carried out using all 651 or the 68 or 26 selected (see above) peptide ions. We obtained similar sensitivities in 3 instances, namely 100% (41/41) and 97.5% (40/41) accuracy for, respectively, binary and multigroup class predictions (Table 1).
Figure 8. Study overview.
The diagram shows the approach used for development and validation of the 68-peptide ion signature and the prostate cancer signature consisting of 26 serum peptides with known sequence (blue in Figure 5). Numbers that are circled indicate total number of selected peptides at that stage of the study.
Figure 9. Independent set of prostate cancer serum samples for validation of established peptide signature biomarkers.
(A) Study design. See Figure 8 and Results. (B) Hierarchical cluster analysis of all spectra from PR1, PR2, and control groups. Either the 68 peptide ions with statistically significant intensity differences for the 3 binary comparisons (Figure 2) or 26 of the sequenced peptides that constitute the prostate cancer signature (blue in Figure 5) were used; the rest of the approximately 650 peptide ions were ignored. The heat map scale of normalized ion intensities ranges from 0 (green) to 2,000 (red) with the midpoint at 1,000 (yellow). (C) Principal component analysis of the PR1 and PR2 groups plus controls, based on the same peptide ions as in B. The first 3 principal components, accounting for most of the variance in the original data set, are shown.
Table 1 .
Class prediction of a prostate cancer validation set (PR2) using SVM (linear kernel) and the 651-, 68-, and 26-feature sets
Aminoprotease activities in plasma generate a sequence ladder from synthetic C3f.
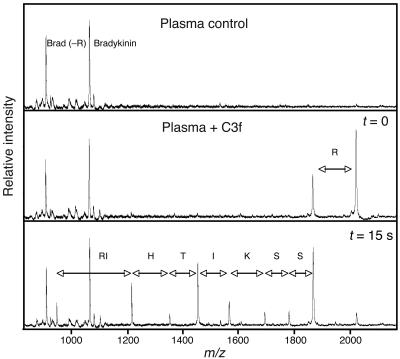
It appears that the serum peptidome is largely the product of resident substrates, more specifically their proteolytic breakdown products (ref. 17; this study), and therefore represents a read-out of the repertoire of proteases that exist in plasma and/or become activated during clotting. With the exception of bradykinin, we have consistently observed much higher peptide concentrations in serum than in plasma (Figure 10 and data not shown), which makes sense as ex vivo coagulation and complement activation underlie generation of the founder peptides of nearly every cluster. Peptides from plasma prepared in heparin-containing blood collection tubes are likely the result of low-level clotting and heparin-induced complement activation (ref. 17; J. Villanueva and P. Tempst, unpublished results). Apparently, the inducible plasma and serum peptidome is then amplified by exoprotease activities, which may also account for many or all of the observed differences. The data presented in this study suggest that cancer cells may contribute unique proteases, perhaps exoproteases, which result in subtle but signature alterations of the complex equation of hundreds of peptides that can be resolved from human serum. In an effort to begin to understand the presence and roles of exoproteases, synthetic C3f was added to fresh plasma at a concentration close to that observed in serum. As shown in Figure 10, degradation was very fast. C terminal Arg was removed within seconds, and the N terminal truncations occurred in 10–15 minutes. The resulting pattern was similar to the endogenous one observed in serum and also illustrated the disparate ion intensities for different rungs in the ladder. However, most of the C3f ladder, except its smallest rung, disappeared upon prolonged incubation (data not shown). Exoproteolytic degradation of synthetic FPA in plasma followed a similar time course, but fibrinopeptide B (FPB) was completely degraded in just a few minutes (data not shown), which may explain why the endogenous form was never observed in our serum profiling analyses. The results suggest that the operative exoprotease concentrations and activities are roughly equivalent in plasma and serum and therefore not the consequence of coagulation.
Figure 10. Plasma exoproteases degrade synthetic C3f in a manner similar to proteolysis of the endogenous peptide (derived from C3 precursor) in serum.
A MALDI-TOF MS read-out of fresh plasma (top panel) indicates very low levels of small peptides except for bradykinin and desArg-bradykinin. After addition of synthetic C3f (1 pmol/μl plasma), an aliquot was immediately (i.e., after ~15–20 seconds) withdrawn, and another was withdrawn after 15 minutes. The sample was kept at room temperature at all times. The middle panel indicates removal of the C terminal Arg by a carboxypeptidase in a matter of seconds. C3f is then further degraded by the activity of aminopeptidases to result in a type of sequence ladder as endogenously present in serum. Brad (–R), bradykinin minus C-terminal Arg; R, Arg; RI, Arg-Ile; H, His; T, Thr; I, Ile; K, Lys; S, Ser.
Discussion
In the search for clinically relevant biomarkers, the low mass range of the serum proteome, particularly peptides with a molecular mass below 3,000 Da, has not received the same attention as higher molecular weight peptides and proteins. Small, preexisting peptides are not readily picked up by high-throughput liquid chromatography/liquid chromatography–MS/MS (LC/LC-MS/MS) analyses of whole-proteome tryptic digests and have also been underrepresented in surface-enhanced laser desorption/ionization–TOF (SELDI-TOF) MS-based screens that seem to favor polypeptides in the 5- to 15-kDa mass range (19–24). The current study and a recent analysis by Koomen et al. (17) provide the first details on the composition of the peptide pool in serum and plasma. Overall, it appears that a large part of the human serum peptidome as detected by MALDI-TOF MS is produced ex vivo by degradation of endogenous substrates by endogenous proteases. As illustrated in Figure 11, peptides are generated during the proteolytic cascades that occur in the intrinsic pathway of coagulation and complement activation (50). Some of these are known bioactive molecules, others represent cleaved propeptides, and still others are seemingly random internal fragments of the precursor proteins. However, the observed cleavage sites are generally consistent with trypsin- and chymotrypsin-like activities of known serine proteases (kallikreins, plasmin, thrombin, factor I, etc.). Once generated, the founder peptides are trimmed down by exoproteases into ladder-like clusters.
Figure 11. Activity of serum proteases.
Many serum peptides are generated by a 2-step proteolytic process. When used in the proper combinations, 1 or more selected members of 6–12 different clusters create diagnostic signatures in the form of ion intensities measured by direct MALDI-TOF MS that can predict cancer and cancer type. Amino acids are color coded to represent sequence clusters of C3f (left) or FPA (right), which are just 2 examples of all the observed clusters.
Exoproteases form a heterogeneous group of enzymes that play a role in the regulation of biologically active peptides (51–53). For instance, leucine aminopeptidase (LAP), aminopeptidase A (AP-A), aminopeptidase N (AP-N), carboxypeptidase N (CP-N), and the kininase I family of carboxypeptidases are involved in the production of angiotensin, bradykinin, and vasopressin (53), and TAFI (a carboxypeptidase B enzyme) in the regulation of fibrinolysis (54). Several exoproteases are transmembrane proteins, anchored in the plasma membrane of vascular endothelial cells. Heterogeneous distribution results in the production of a wide variety of proteolytic peptides in different tissues and contexts (51). In addition, some exoproteases like AP-N and placental LAP (P-LAP) are shed from cells through the action of ADAM family proteases (55) and end up in the bloodstream in soluble form (55, 56), thereby degrading resident polypeptides in the blood, plasma, and serum.
Depending on the analytical approach and the objectives of a diagnostic marker search, there are opposing views on the presence of a vast peptide pool (degradome) in plasma or serum generated from blood proteins as described above (Figure 11). It can be considered background noise in peptide marker discovery efforts, making it all but impossible to find any naturally occurring, true biomarkers in the peptidome or to obtain mechanistic insights in specific activities of tumor-associated proteases. Those who subscribe to this view believe that exoprotease activity, or all protease activity for that matter, should be blocked at the time of sample collection. However, it has been correctly pointed out (17) that the protein degradome is the only segment of the serum peptidome that can be readily interrogated by direct MALDI-TOF MS. Fragments of bona fide marker proteins (for example, PSA in sera of prostate cancer patients), if present, are currently undetectable because of sensitivity, ion suppression, and mass resolution issues inherent in the technology. It can therefore be argued that precisely this degradome offers the best opportunity at this point for biomarker or surrogate biomarker discovery.
Whereas the only comprehensive, high-resolution MS analysis of the plasma/serum peptides to date aimed at providing an inventory (17), we undertook to find peptides and patterns with marker potential for specific types of solid tumor cancers. In the discovery phase of our studies, we sorted through hundreds of features to identify several that were most predictive of outcome and showed that reduction in the number of key peptides to a few (i.e., the signatures) that were easily recognized between samples did not adversely affect class predictions. We then demonstrated that this signature could be used to discriminate between cancer and control in an independent validation set comprised of serum samples obtained from patients with advanced prostate cancer. Strikingly, all 46 sequence-identified peptides from the initial set of 68 rigorously selected discriminant peptide signals were part of the serum degradome. With two-thirds of the initial marker group now characterized, we trust that these findings can be generalized.
The small number of blood proteins that are the source of nearly all the peptides in prostate, bladder, and breast cancer signatures are naturally not biomarkers but simply serve as an endogenous substrate pool for the real biomarkers, i.e., proteases. There is no actual relationship between the substrate concentrations and the MS-ion intensities of many of the degradation products. Highly abundant serum proteins such as albumin and immunoglobulins were not represented, and fragments of proteins with a more than 10-fold difference in concentration had comparable ion intensities. On the other hand, whereas full-length C3f produced nearly identical ion intensities in all cancer groups and controls, several of its truncated forms did not. In fact, 2 or more patient sera peptides (say, x and y) that derived from the same protein had often opposite relative ion intensities (i.e., the ion intensity divided by that of the corresponding peptide in the control group); for instance, the signal of peptide x was higher and that of peptide y lower than that of their counterparts in control sera. Finally, several of the protein degradome peptides that we observed and that had high surrogate marker value were virtually absent from the controls (e.g., several entries in Figure 6 that list a median normalized intensity value of 1 for the control). In fact, 7 such peptides (Figures 5 and 6; m/z = 998, 1278, 2053, 2409, 2565, 2704, and 3971), each unique to 1 or more types of cancer, were not reported in the high-resolution blanket analyses of plasma peptides, possibly because that blood sample was obtained from a healthy individual (17).
The 2-step proteolytic process depicted in Figure 11 that generates the most abundant layer of the serum peptidome is subject to changes in enzyme panels, cofactors, inhibitors, and various other controlling elements and conditions, which make for a virtually unlimited combinatorial variability to produce peptides of different sizes and composition. Direct MALDI-TOF MS–based serum peptide profiling is thus a form of activity-based proteomics, monitoring surrogate biomarkers in the form of proteome metabolomic products. This can be exploited for diagnostic and predictive purposes as a phenotypic read-out of catalytic and other metabolic activities in body fluids or tissues, utilizing endogenous (or exogenous) substrates and quantitative product analysis. It also makes this approach particularly well suited for detection of cancer, as proteases are well-established components of cancer progression and invasiveness (57–60). We provide evidence here that exoprotease activities superimposed on the ex vivo coagulation and complement-degradation pathways contribute to generation of not only cancer-specific but also cancer type–specific serum peptides.
Exoproteases have been previously implicated in cancer (58). For instance, AP-N/CD13 is highly expressed in bladder, gastric, thyroid, and hepatic carcinomas (61–64), and the concentration of its soluble form is also increased in cancer patients (56). Similarly, increased concentration of a lysosomal dipeptidyl-aminopeptidase (DAP II) has been observed in sera of tumor-bearing animals and cancer patients (65). LAP, aminopeptidase P (AP-P), and enkephalin-degrading tyrosyl aminopeptidase (EDA) have been associated with breast cancer (57, 66–68) and AP-A, methionine aminopeptidase 2 (Met-AP2), and glycylproline dipeptidyl aminopeptidase (GPDA) with various other types of cancers (69–71). Increased activity and expression of AP-N and Met-AP2 have been functionally correlated with metastasis of cancer cells by promotion of angiogenesis (72–75). As for carboxypeptidases, carboxypeptidase D (CP-D) is selectively more highly expressed in hematopoietic tumor cells (76), and PSMA is overexpressed in prostate cancer and has been implicated in tumor invasion (14, 77).
How all the above and other, currently unidentified enzymes may contribute mechanistically to the observed differences in serum peptide patterns among the 3 different cancers remains unexplained and may require a great deal of future study to understand. Nonetheless, the differences are statistically significant. It is also important to note some of the overlaps between the groups. Despite the sex difference, the breast and bladder cancer signatures overlapped by 8 peptide ions that deviated in median intensities from the corresponding control ions in a similar manner; only 1 peptide ion (1865) showed diametrically up- or downregulated intensities. Breast and bladder (85% males in the study cohort; see Supplemental Table 1) cancer shared 7 peptide ions with similarly up- or downregulated intensities; 7 others were either higher in breast cancer but lower in bladder cancer or vice versa, relative to the control. Finally, 23 out of the 26 prostate cancer marker peptides were also part of the larger bladder cancer signature. However, 19 of these 23 had markedly better P values for bladder cancer, and 4 were better for prostate cancer, relative to the controls. We think it unlikely that the overlaps or differences are sex related, as a preliminary comparison of serum peptide profiles from healthy men and women indicated only statistically insignificant differences (J. Villanueva and P. Tempst, unpublished observations). Furthermore, most peptide ion markers for each cancer type were equally well separated from both male and female subsets of the control group (Supplemental Figure 1). A more likely explanation for the bladder/prostate cancer overlap is that the prostate gland and bladder (partially) are derived embryologically from endodermal tissues in the urogenital sinus and likely share biological features not seen in tissues from outside the genitourinary tract. For instance, tissue recombination studies have shown that urogenital mesenchyme can actually induce differentiation of bladder epithelium toward a prostatic epithelial–differentiated phenotype, but this property is restricted to endodermal epithelia (as in the bladder) with similar embryonic origin to the prostate (78). Overall, the prostate cancer signature was sufficiently robust to predict the class of members of an independent validation set with 97.5% sensitivity in multiclass SVM analysis (Table 1).
In conclusion, it is our view that proteolytic degradative patterns in the serum peptidome hold important information that may have direct clinical utility as a surrogate marker for detection and classification of cancer. Our findings also suggest that future work to optimize serum peptidomics for clinical practice should be carried out with the recognition that endogenous proteolytic activities contribute important cancer type–specific information. Use of protease inhibitors and, as we have previously cautioned (29), even the slightest deviation from standard protocol for specimen collection, storage and handling, analytical chemistry, and MS signal processing are particularly ill advised. We anticipate that as we scale up these efforts using the same general methodology, we will expand and refine our definition of key discriminatory peptides for prediction of each cancer type. The patterns may also have diagnostic value for identifying cancer subtype and stage or may mark a given clinical outcome of interest or may reliably distinguish clinically insignificant from significant cancer. Such a blood test could, for example, identify patients with newly diagnosed prostate cancer who might safely avoid surgery or radiation. Focused MS quantitation of key peptides derived from either endogenous or custom synthetic substrate and utilizing isotopically labeled standards should then facilitate introduction of this technology into clinical practice.
Methods
Serum samples.
Blood samples from healthy volunteers (mixed sexes; ages 23 to 56; see Supplemental Table 1) with no known malignancies and from patients diagnosed with either prostate cancer, bladder cancer, or breast cancer were all collected at Memorial Sloan-Kettering Cancer Center (MSKCC) following a standard clinical protocol (29). Details on patient age, sex, and pathologic diagnosis are given in Supplemental Table 1. All collections were approved by the MSKCC Institutional Review and Privacy Board. Informed consent was obtained from all patients. Blood samples were obtained in 8.5-ml, BD Vacutainer, glass red-top tubes (BD; 366430), allowed to clot at room temperature for 1 hour, and centrifuged at 1,400–2,000 g for 10 minutes at room temperature. Sera (upper phase) were transferred to four 4-ml cryovials (Fischer Scientific International, 0566966) with approximately 1 ml serum in each and stored frozen at –80°C until further use (29). A similar procedure was followed for preparation of plasma in heparin-containing green-top tubes (BD, 366480), except that centrifugation was done immediately after blood collection. Upon delivery at the MS lab, the cryovials (source vials) were barcoded. One cryovial of each sample was thawed on ice and used to generate 9 smaller aliquots (50 μl each) in barcoded microeppendorf tubes and stored at –80°C in barcoded freezer boxes. In this study, all serum samples were always frozen and thawed twice, the second thawing step immediately before peptide extraction and MS analysis. We have made a concerted effort to instruct nurses, phlebotomists, messenger service staff, and clinical technicians about the importance of strict adherence to the standard protocol.
Analytical chemistry.
Automated, solid-phase peptide extraction, MALDI-TOF MS profiling, signal processing and spectral alignments, and the use of custom mass spectral viewing tools were all performed as previously developed in the authors’ laboratory (18, 29). Additional details and a description of tandem MS identification of selected serum peptides are given in Supplemental Methods.
Statistics.
The binned spreadsheet containing data from spectra obtained for all samples of cancer patients or healthy subjects (106 samples total; 651 m/z values, with normalized intensities for each sample; > 70,000 data points) as well as the test set for prostate cancer (PR2; 41 samples; ~27,000 data points) were imported into the GeneSpring program (version 7; Agilent Technologies) and analyzed using various statistical algorithms such as 1-way ANOVA, principal component analysis, hierarchical clustering, k-nearest neighbor (k-NN), and SVM. Different experiments were created in GeneSpring to represent the masses. No normalizations were applied to the experiment since the masses were normalized by the database that binned them. In the parameter section of the experiments, a parameter called cancertype was created to label samples as prostate cancer, breast cancer, bladder cancer, or control. In the experiment interpretation section, the analysis mode was set to ratio (signal/control), and all measurements were used. No cross-gene error model was used for either.
ANOVA.
Once the experiments were created, the m/z values (peaks) were filtered by using nonparametric tests: the Mann-Whitney U test (for binary comparisons) and the Kruskal-Wallis test (for multiclass comparisons). The Benjamini and Hochberg method was used to adjust P values for multiple comparisons (79). The threshold for significance was an expected false discovery rate of less than 1 × 10–5. These tests are meant to find peaks that show statistically significant differences between the clinical groups studied.
Hierarchical clustering.
The 651 m/z values were subjected to average-linkage hierarchical clustering, using standard correlation (also known as Pearson correlation around zero) as a distance metrics (GeneSpring program). The peaks were organized by creating mock-phylogenetic trees (dendrograms) termed gene trees and experiment trees in the software. The trees were displayed with the samples along the x axis and the masses along the y axis.
Class prediction.
SVM and k-NN analyses were done by using the class prediction tool in GeneSpring. The training groups were either a binary comparison (PR1 and control) or a multiclass comparison (PR1, breast cancer, bladder cancer, and control). The test set was PR2. The parameter to predict was set to cancertype. The gene selection was set to use different groups of masses previously selected (e.g., 651, 68, 26). In k-NN the number of neighbors was set to 5 with a P value decision cutoff of 1. The SVM was done with the same training sets and parameters and set to predict the PR2 test set. The kernel used was polynomial dot product (order 1) with a diagonal scaling of 0.
Supplementary Material
Acknowledgments
This work was supported by NIH grants 1-R21-CA1119425, 5-P30-CA08748, and 5-P50-CA92629 and awards from the Prostate Cancer Foundation, the Vakil Research Fund, and Accelerate Brain Cancer Cure. We thank Larry Norton and Mark Kris for support; Richard Robbins, Mark Robson, and Chris Sander for helpful discussions; San San Yi for peptide synthesis; Lynne Lacomis for help with the artwork; and all volunteers for generous donation of blood samples.
Footnotes
Nonstandard abbreviations used: AP-A, aminopeptidase A; desArg-bradykinin, bradykinin that has the Arg removed; FPA, fibrinopeptide A; HMW, high molecular weight; ITIH4, inter-α-trypsin inhibitor heavy chain H4; k-NN, k-nearest neighbor; LAP, leucine aminopeptidase; MALDI-TOF, matrix-assisted laser desorption/ionization–time-of-flight; MS, mass spectrometric, mass spectrometry; PR1, prostate 1 (group); PSA, prostate-specific antigen; PSMA, prostate-specific membrane antigen; SVM, support vector machine.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 116:271–284 (2006). doi:10.1172/JCI26022
See the related Commentary beginning on page 26.
References
- 1.Lander E.S., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Hood L. Leroy Hood expounds the principles, practice and future of systems biology. Drug Discov. Today. 2003;8:436–438. doi: 10.1016/S1359-6446(03)02710-7. [DOI] [PubMed] [Google Scholar]
- 3.Etzioni R., et al. The case for early detection. Nat. Rev. Cancer. 2003;3:243–252. doi: 10.1038/nrc1041. [DOI] [PubMed] [Google Scholar]
- 4.Chung C.H., Bernard P.S., Perou C.M. Molecular portraits and the family tree of cancer. Nat. Genet. 2002;32(Suppl.):533–540. doi: 10.1038/ng1038. [DOI] [PubMed] [Google Scholar]
- 5.Staudt L.M. Gene expression profiling of lymphoid malignancies. Annu. Rev. Med. 2002;53:303–318. doi: 10.1146/annurev.med.53.082901.103941. [DOI] [PubMed] [Google Scholar]
- 6.Anderson N.L., Anderson N.G. The human plasma proteome: history, character, and diagnostic prospects. Mol. Cell. Proteomics. 2002;1:845–867. doi: 10.1074/mcp.R200007-MCP200. [DOI] [PubMed] [Google Scholar]
- 7.Adkins J.N., et al. Toward a human blood serum proteome: analysis by multidimensional separation coupled with mass spectrometry. Mol. Cell. Proteomics. 2002;1:947–955. doi: 10.1074/mcp.M200066-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Sidransky D. Emerging molecular markers of cancer. Nat. Rev. Cancer. 2002;2:210–219. doi: 10.1038/nrc755. [DOI] [PubMed] [Google Scholar]
- 9.Bidart J.M., et al. Kinetics of serum tumor marker concentrations and usefulness in clinical monitoring. Clin. Chem. 1999;45:1695–1707. [PubMed] [Google Scholar]
- 10.Jortani S.A., Prabhu S.D., Valdes R., Jr. Strategies for developing biomarkers of heart failure. . Clin. Chem. 2004;50:265–278. doi: 10.1373/clinchem.2003.027557. [DOI] [PubMed] [Google Scholar]
- 11.Watts N.B. Clinical utility of biochemical markers of bone remodeling. Clin. Chem. 1999;45:1359–1368. [PubMed] [Google Scholar]
- 12.Gillette M.A., Mani D.R., Carr S.A. Place of pattern in proteomic biomarker discovery. J. Proteome Res. 2005;4:1143–1154. doi: 10.1021/pr0500962. [DOI] [PubMed] [Google Scholar]
- 13.Hugosson J., et al. Prostate specific antigen based biennial screening is sufficient to detect almost all prostate cancers while still curable. . J. Urol. 2003;169:1720–1723. doi: 10.1097/01.ju.0000061183.43229.2e. [DOI] [PubMed] [Google Scholar]
- 14.Ghosh A., Wang X., Klein E., Heston W.D. Novel role of prostate-specific membrane antigen in suppressing prostate cancer invasiveness. . Cancer Res. 2005;65:727–731. [PubMed] [Google Scholar]
- 15.Richter R., et al. Composition of the peptide fraction in human blood plasma: database of circulating human peptides. J. Chromatogr. B Biomed. Sci. Appl. 1999;726:25–35. doi: 10.1016/S0378-4347(99)00012-2. [DOI] [PubMed] [Google Scholar]
- 16.Tirumalai R.S., et al. Characterization of the low molecular weight human serum proteome. Mol. Cell. Proteomics. . 2003;1:1096–1103. doi: 10.1074/mcp.M300031-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Koomen J.M., et al. Direct tandem mass spectrometry reveals limitations in protein profiling experiments for plasma biomarker discovery. . J. Proteome Res. 2005;4:972–981. doi: 10.1021/pr050046x. [DOI] [PubMed] [Google Scholar]
- 18.Villanueva J., et al. Serum peptide profiling by magnetic particle-assisted, automated sample processing and MALDI-TOF mass spectrometry. Anal. Chem. 2004;76:1560–1570. doi: 10.1021/ac0352171. [DOI] [PubMed] [Google Scholar]
- 19.Petricoin E.F., et al. Use of proteomic patterns in serum to identify ovarian cancer. Lancet. 2002;359:572–577. doi: 10.1016/S0140-6736(02)07746-2. [DOI] [PubMed] [Google Scholar]
- 20.Adam B.L., et al. Serum protein fingerprinting coupled with a pattern-matching algorithm distinguishes prostate cancer from benign prostate hyperplasia and healthy men. Cancer Res. 2002;62:3609–3614. [PubMed] [Google Scholar]
- 21.Li J., Zhang Z., Rosenzweig J., Wang Y.Y., Chan D.W. Proteomics and bioinformatics approaches for identification of serum biomarkers to detect breast cancer. Clin. Chem. 2002;48:1296–1304. [PubMed] [Google Scholar]
- 22.Ebert M.P., et al. Identification of gastric cancer patients by serum protein profiling. . J. Proteome Res. 2004;3:1261–1266. doi: 10.1021/pr049865s. [DOI] [PubMed] [Google Scholar]
- 23.Ornstein D.K., et al. Serum proteomic profiling can discriminate prostate cancer from benign prostates in men with total prostate specific antigen levels between 2.5 and 15.0 ng/ml. J. Urol. 2004;172:1302–1305. doi: 10.1097/01.ju.0000139572.88463.39. [DOI] [PubMed] [Google Scholar]
- 24.Conrads T.P., et al. High-resolution serum proteomic features for ovarian cancer detection. Endocr. Relat. Cancer. 2004;11:163–178. doi: 10.1677/erc.0.0110163. [DOI] [PubMed] [Google Scholar]
- 25.Coombes K.R., Morris J.S., Hu J., Edmonson S.R., Baggerly K.A. Serum proteomics profiling-a young technology begins to mature. Nat. Biotechnol. 2005;23:291–292. doi: 10.1038/nbt0305-291. [DOI] [PubMed] [Google Scholar]
- 26.Diamandis E.P. Mass spectrometry as a diagnostic and a cancer biomarker discovery tool: opportunities and potential limitations. Mol. Cell. Proteomics. 2004;3:367–378. doi: 10.1074/mcp.R400007-MCP200. [DOI] [PubMed] [Google Scholar]
- 27.Check E. Proteomics and cancer: running before we can walk? Nature. 2004;429:496–497. doi: 10.1038/429496a. [DOI] [PubMed] [Google Scholar]
- 28.Ransohoff D.F. Opinion: bias as a threat to the validity of cancer molecular-marker research. Nat. Rev. Cancer. 2005;5:142–149. doi: 10.1038/nrc1550. [DOI] [PubMed] [Google Scholar]
- 29.Villanueva J., et al. Correcting common errors in identifying cancer-specific serum peptide signatures. J. Proteome Res. 2005;4:1060–1072. doi: 10.1021/pr050034b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marshall J., et al. Processing of serum proteins underlies the mass spectral fingerprinting of myocardial infarction. J. Proteome Res. 2003;2:361–372. doi: 10.1021/pr030003l. [DOI] [PubMed] [Google Scholar]
- 31.Bergen H.R., 3rd, et al. Discovery of ovarian cancer biomarkers in serum using NanoLC electrospray ionization TOF and FT-ICR mass spectrometry. . Dis. Markers. 2003;19:239–249. doi: 10.1155/2004/797204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Z., et al. Three biomarkers identified from serum proteomic analysis for the detection of early stage ovarian cancer. Cancer Res. 2004;64:5882–5890. doi: 10.1158/0008-5472.CAN-04-0746. [DOI] [PubMed] [Google Scholar]
- 33.Weinberger S.R., Dalmasso E.A., Fung E.T. Current achievements using ProteinChip Array technology. Curr. Opin. Chem. Biol. 2002;6:86–91. doi: 10.1016/S1367-5931(01)00282-4. [DOI] [PubMed] [Google Scholar]
- 34.Kapp E.A., et al. Mining a tandem mass spectrometry database to determine the trends and global factors influencing peptide fragmentation. Anal. Chem. 2003;75:6251–6264. doi: 10.1021/ac034616t. [DOI] [PubMed] [Google Scholar]
- 35.Gao J., Opiteck G.J., Friedrichs M.S., Dongre A.R., Hefta S.A. Changes in the protein expression of yeast as a function of carbon source. J. Proteome Res. 2003;2:643–649. doi: 10.1021/pr034038x. [DOI] [PubMed] [Google Scholar]
- 36.Fach E.M., et al. In vitro biomarker discovery for atherosclerosis by proteomics. Mol. Cell. Proteomics. 2004;3:1200–1210. doi: 10.1074/mcp.M400160-MCP200. [DOI] [PubMed] [Google Scholar]
- 37. Jandl, J.H. 1996. Blood: textbook of hematology. Little, Brown and Co. New York, New York, USA. 1510 pp. [Google Scholar]
- 38.Sahu A., Lambris J.D. Structure and biology of complement protein C3, a connecting link between innate and acquired immunity. Immunol. Rev. 2001;180:35–48. doi: 10.1034/j.1600-065X.2001.1800103.x. [DOI] [PubMed] [Google Scholar]
- 39.Abbasciano V., Levato F., Zavagli G. Specificity of fibrinopeptide A (FpA) as a marker for gastrointestinal cancers before and after surgery. . Med. Oncol. Tumor Pharmacother. 1987;4:75–79. doi: 10.1007/BF02934943. [DOI] [PubMed] [Google Scholar]
- 40.Auger M.J., Galloway M.J., Leinster S.J., McVerry B.A., Mackie M.J. Elevated fibrinopeptide A levels in patients with clinically localised breast carcinoma. Haemostasis. 1987;17:336–339. doi: 10.1159/000215766. [DOI] [PubMed] [Google Scholar]
- 41.Stewart J.M. Bradykinin antagonists as anti-cancer agents. Curr. Pharm. Des. 2003;9:2036–2042. doi: 10.2174/1381612033454171. [DOI] [PubMed] [Google Scholar]
- 42.Kato H., Matsumura Y., Maeda H. Isolation and identification of hydroxyproline analogues of bradykinin in human urine. FEBS Lett. 1988;232:252–254. doi: 10.1016/0014-5793(88)80427-7. [DOI] [PubMed] [Google Scholar]
- 43.Salier J.P., Rouet P., Raguenez G., Daveau M. The inter-alpha-inhibitor family: from structure to regulation. Biochem. J. 1996;315:1–9. doi: 10.1042/bj3150001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nishimura H., et al. cDNA and deduced amino acid sequence of human PK-120, a plasma kallikrein-sensitive glycoprotein. FEBS Lett. 1995;357:207–211. doi: 10.1016/0014-5793(94)01364-7. [DOI] [PubMed] [Google Scholar]
- 45.Belt K.T., Carroll M.C., Porter R.R. The structural basis of the multiple forms of human complement component C4. Cell. 1984;36:907–914. doi: 10.1016/0092-8674(84)90040-0. [DOI] [PubMed] [Google Scholar]
- 46.July L.V., et al. Clusterin expression is significantly enhanced in prostate cancer cells following androgen withdrawal therapy. Prostate. 2002;50:179–188. doi: 10.1002/pros.10047. [DOI] [PubMed] [Google Scholar]
- 47.Scaltriti M., et al. Clusterin (SGP-2, ApoJ) expression is downregulated in low- and high-grade human prostate cancer. Int. J. Cancer. 2004;108:23–30. doi: 10.1002/ijc.11496. [DOI] [PubMed] [Google Scholar]
- 48.Miyake H., Gleave M., Kamidono S., Hara I. Overexpression of clusterin in transitional cell carcinoma of the bladder is related to disease progression and recurrence. Urology. 2002;59:150–154. doi: 10.1016/S0090-4295(01)01484-4. [DOI] [PubMed] [Google Scholar]
- 49.Jiang W.G., Ablin R., Douglas-Jones A., Mansel R.E. Expression of transglutaminases in human breast cancer and their possible clinical significance. Oncol. Rep. 2003;10:2039–2044. [PubMed] [Google Scholar]
- 50. Tietz, N.W. 1995. Clinical guide to laboratory tests. Philadelphia, Pennsylvania, USA. W.B. Saunders Co. 1096 pp. [Google Scholar]
- 51.Sanderink G.J., Artur Y., Siest G. Human aminopeptidases: a review of the literature. J. Clin. Chem. Clin. Biochem. 1988;26:795–807. doi: 10.1515/cclm.1988.26.12.795. [DOI] [PubMed] [Google Scholar]
- 52.Silveira P.F., Gil J., Casis L., Irazusta J. Peptide metabolism and the control of body fluid homeostasis. Curr. Med. Chem. Cardiovasc. Hematol. Agents. 2004;2:219–238. doi: 10.2174/1568016043356264. [DOI] [PubMed] [Google Scholar]
- 53.Mitsui T., Nomura S., Itakura A., Mizutani S. Role of aminopeptidases in the blood pressure regulation. Biol. Pharm. Bull. 2004;27:768–771. doi: 10.1248/bpb.27.768. [DOI] [PubMed] [Google Scholar]
- 54.Nesheim M., et al. Thrombin, thrombomodulin and TAFI in the molecular link between coagulation and fibrinolysis. Thromb. Haemost. 1997;78:386–391. [PubMed] [Google Scholar]
- 55.Ito N., et al. ADAMs, a disintegrin and metalloproteinases, mediate shedding of oxytocinase. Biochem. Biophys. Res. Commun. 2004;314:1008–1013. doi: 10.1016/j.bbrc.2003.12.183. [DOI] [PubMed] [Google Scholar]
- 56.van Hensbergen Y., et al. Soluble aminopeptidase N/CD13 in malignant and nonmalignant effusions and intratumoral fluid. Clin. Cancer Res. 2002;8:3747–3754. [PubMed] [Google Scholar]
- 57.Martinez J.M., et al. Aminopeptidase activities in breast cancer tissue. Clin. Chem. 1999;45:1797–1802. [PubMed] [Google Scholar]
- 58.Matrisian L.M., Sledge G.W., Jr., Mohla S. Extracellular proteolysis and cancer: meeting summary and future directions. Cancer Res. 2003;63:6105–6109. [PubMed] [Google Scholar]
- 59.Egeblad M., Werb Z. New functions for the matrix metalloproteinases in cancer progression. . Nat. Rev. Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 60.Rao J.S. Molecular mechanisms of glioma invasiveness: the role of proteases. Nat. Rev. Cancer. 2003;3:489–501. doi: 10.1038/nrc1121. [DOI] [PubMed] [Google Scholar]
- 61.Moffatt S., Wiehle S., Cristiano R.J. Tumor-specific gene delivery mediated by a novel peptide-polyethylenimine-DNA polyplex targeting aminopeptidase N/CD13. Hum. Gene Ther. 2005;16:57–67. doi: 10.1089/hum.2005.16.57. [DOI] [PubMed] [Google Scholar]
- 62.Kehlen A., Lendeckel U., Dralle H., Langner J., Hoang-Vu C. Biological significance of aminopeptidase N/CD13 in thyroid carcinomas. Cancer Res. 2003;63:8500–8506. [PubMed] [Google Scholar]
- 63.Rocken C., et al. Ectopeptidases are differentially expressed in hepatocellular carcinomas. Int. J. Oncol. 2004;24:487–495. [PubMed] [Google Scholar]
- 64.Carl-McGrath S., et al. The ectopeptidases CD10, CD13, CD26, and CD143 are upregulated in gastric cancer. Int. J. Oncol. 2004;25:1223–1232. [PubMed] [Google Scholar]
- 65.Kojima K., et al. Serum activities of dipeptidyl-aminopeptidase II and dipeptidyl-aminopeptidase IV in tumor-bearing animals and in cancer patients. Biochem. Med. Metab. Biol. 1987;37:35–41. doi: 10.1016/0885-4505(87)90007-7. [DOI] [PubMed] [Google Scholar]
- 66.Essler M., Ruoslahti E. Molecular specialization of breast vasculature: a breast-homing phage-displayed peptide binds to aminopeptidase P in breast vasculature. Proc. Natl. Acad. Sci. U. S. A. 2002;99:2252–2257. doi: 10.1073/pnas.251687998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Carrera M.P., et al. Serum enkephalin-degrading aminopeptidase activity in N-methyl nitrosourea-induced rat breast cancer. Anticancer Res. 2005;25:193–196. [PubMed] [Google Scholar]
- 68.Pulido-Cejudo G., et al. A monoclonal antibody driven biodiagnostic system for the quantitative screening of breast cancer. Biotechnol. Lett. 2004;26:1335–1339. doi: 10.1023/B:BILE.0000045629.57791.5a. [DOI] [PubMed] [Google Scholar]
- 69.Suganuma T., et al. Regulation of aminopeptidase A expression in cervical carcinoma: role of tumor-stromal interaction and vascular endothelial growth factor. Lab. Invest. 2004;84:639–648. doi: 10.1038/labinvest.3700072. [DOI] [PubMed] [Google Scholar]
- 70.Selvakumar P., et al. High expression of methionine aminopeptidase 2 in human colorectal adenocarcinomas. Clin. Cancer Res. 2004;10:2771–2775. doi: 10.1158/1078-0432.CCR-03-0218. [DOI] [PubMed] [Google Scholar]
- 71.Ni R.Z., Huang J.F., Xiao M.B., Li M., Meng X.Y. Glycylproline dipeptidyl aminopeptidase isoenzyme in diagnosis of primary hepatocellular carcinoma. World J. Gastroenterol. 2003;9:710–713. doi: 10.3748/wjg.v9.i4.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sheppard G.S., et al. 3-Amino-2-hydroxyamides and related compounds as inhibitors of methionine aminopeptidase-2. Bioorg. Med. Chem. Lett. 2004;14:865–868. doi: 10.1016/j.bmcl.2003.12.031. [DOI] [PubMed] [Google Scholar]
- 73.Griffith E.C., et al. Molecular recognition of angiogenesis inhibitors fumagillin and ovalicin by methionine aminopeptidase 2. Proc. Natl. Acad. Sci. U. S. A. 1998;95:15183–15188. doi: 10.1073/pnas.95.26.15183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pasqualini R., et al. Aminopeptidase N is a receptor for tumor-homing peptides and a target for inhibiting angiogenesis. Cancer Res. 2000;60:722–727. [PMC free article] [PubMed] [Google Scholar]
- 75.Petrovic N., Bhagwat S.V., Ratzan W.J., Ostrowski M.C., Shapiro L.H. CD13/APN transcription is induced by RAS/MAPK-mediated phosphorylation of Ets-2 in activated endothelial cells. . J. Biol. Chem. 2003;278:49358–49368. doi: 10.1074/jbc.M308071200. [DOI] [PubMed] [Google Scholar]
- 76.O’Malley P.G., Sangster S.M., Abdelmagid S.A., Bearne S.L., Too C.K. Characterization of a novel, cytokine-inducible carboxypeptidase-D isoform in hematopoietic tumor cells. Biochem. J. 2005;390:665–673. doi: 10.1042/BJ20050025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fair W.R., Israeli R.S., Heston W.D. Prostate-specific membrane antigen. Prostate. 1997;32:140–148. doi: 10.1002/(SICI)1097-0045(19970701)32:2<140::AID-PROS9>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 78.Marker P.C., Donjacour A.A., Dahiya R., Cunha G.R. Hormonal, cellular, and molecular control of prostatic development. Dev. Biol. 2003;253:165–174. doi: 10.1016/S0012-1606(02)00031-3. [DOI] [PubMed] [Google Scholar]
- 79.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. (Ser. B.) 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.