Abstract
Recent studies have revealed numerous examples in which oxidation and reduction of cysteines in proteins are integrated into specific cascades of biological regulatory systems. In general, these reactions proceed as thiol-disulfide exchange events. However, it is not exactly understood how a disulfide bond is created de novo. DsbB, an Escherichia coli plasma membrane protein, is one of the enzymes that create a new disulfide bond within itself and in DsbA, the direct catalyst of protein disulfide bond formation in the periplasmic space. DsbB is associated with a cofactor, either ubiquinone or menaquinone, as a source of an oxidizing equivalent. The DsbB-bound quinone undergoes transition to a pink (λmax, ≈500 nm, ubiquinone) or violet (λmax, ≈550 nm, menaquinone)-colored state during the course of the DsbB enzymatic reaction. Here we show that not only the thiolate form of Cys-44 previously suggested but also Arg-48 in the α-helical arrangement is essential for the quinone transition. Quantum chemical simulations indicate that proper positioning of thiolate anion and ubiquinone in conjunction with positively charged guanidinium moiety of arginine allows the formation of a thiolate-ubiquinone charge transfer complex with absorption peaks at ≈500 nm as well as a cysteinylquinone covalent adduct. We propose that the charge transfer state leads to the transition state adduct that accepts a nucleophilic attack from another cysteine to generate a disulfide bond de novo. A similar mechanism is conceivable for a class of eukaryotic dithiol oxidases having a FAD cofactor.
Keywords: FAD, ubiquinone, DsbA
Cysteines in primary translation products are in the reduced thiol form, certain pairs of which are subsequently oxidized to form disulfide bridges. Thus, all of the disulfide bonds in cellular proteins must have once been created by net oxidation processes at the expense of stoichiometric amounts of electrons, whether or not they are subsequently transferred to other proteins by redox-neutral dithiol-disulfide exchange processes. The primary compartment for the disulfide bond creation is the periplasmic space in prokaryotes and the endoplasmic reticulum in eukaryotes, where dedicated factors participate to facilitate disulfide bond formation. Although in Escherichia coli a periplasmic protein DsbA donates its own disulfide (Cys-30-Cys-33) to client proteins, it is an integral plasma membrane protein, DsbB, that plays a pivotal role in the de novo creation of disulfides (for reviews, see refs. 1 and 2). In essence, DsbB acts as a molecular machine that transforms an oxidizing equivalent of bound quinone into a protein disulfide.
The primary role of DsbB is to reoxidize reduced active-site cysteines of DsbA in a manner dependent on cellular respiratory components (3). DsbB is associated with a respiration-coupled cofactor, either aerobic ubiquinone (UQ8) or anaerobic menaquinone (MK8) (4-6). Each of its two periplasmic domains carries a pair of essential cysteine residues, Cys-41-Cys-44 and Cys-104-Cys-130, respectively (7). While there is general consensus that the former cysteine pair interacts with UQ (8) and that the latter is engaged in the thiol-disulfide exchange with DsbA (9-11), different views have been put forward about the details of DsbB's reaction mechanisms (11-15).
We have characterized reactivities and properties of DsbB that is free from any bound quinones (11), of DsbB that bears UQ (16), and of DsbB that bears menaquinone (6). These analyses taken together revealed that there are two pathways of DsbB/quinone-mediated oxidation of DsbA. In the “rapid” pathway, direct exchange of the DsbA thiols with DsbB Cys-104-Cys-130 disulfide is coupled with rapid intramolecular thiol-disulfide equilibration in DsbB and quinone-dependent oxidation of the Cys-41-Cys-44 pair. In the “slow” pathway, an intermediate disulfide is formed between the reactive Cys-30 residue of DsbA and Cys-104 of DsbB. This complex is then resolved by a quinone-dependent reaction that produces oxidized forms of both DsbA and DsbB (see Fig. 5, which is published as supporting information on the PNAS web site, for easier grasp). It is thought that reduced quinones produced in these reactions will be recycled back to oxidized quinones by terminal oxidases of the respiratory chain.
A remarkable feature of the DsbB-bound quinones is that they undergo a striking transition during the reaction to exhibit strong absorption at ≈500 nm (with pink color in the case of UQ; ref. 16) or at ≈550 nm (with violet color in the case of menaquinone; ref. 6). Whereas the spectral transition of quinone can be observed only transiently and briefly during the reactions with wild-type DsbB and wild-type DsbA, it can be seen evidently and stably when DsbB is forced to form a stable disulfide-bonded complex with DsbA(C33S) lacking one of the active-site cysteines. In this complex that mimics the slow pathway intermediate, disulfide bond rearrangements occur in DsbB such that Cys-130 is now disulfide-bonded with Cys-41 and Cys-44 is unpaired and reduced (11, 12, 16). Indeed, a series of our mutational analyses indicated that the quinone transition takes place whenever Cys-44 is unpaired. Consistent with the involvement of Cys-44, the transition is sensitive to a thiol modification agent and to pH changes, giving an apparent pKa of ≈6.6 expected for thiolate deprotonation (6, 16).
Regeimbal et al. (17) reported that their DsbB preparation was purple-colored and discussed that it was due to the formation of quinhydrone-like charge transfer (CT) complex between UQ and hydroquinone that was also bound to DsbB. However, the lack of supporting evidence other than the similarity of the absorbance curve with that of quinhydrone made us question the likelihood of the involvement of the stacked two quinone rings for the spectroscopic behavior of DsbB. Because our experimental findings that suggest the importance of the unpaired and deprotonated Cys-44 residue fit very well with accumulating information on the DsbB-DsbA reaction mechanisms, we explored further the involvement of Cys-44 and nearby residues in the UQ transition and DsbB reactivities by both mutagenesis experiments and theoretical considerations.
Materials and Methods
Preparation of DsbB and DsbA and Their Derivatives. DsbB and its variants contained a C-terminal His-6 tag and the Cys8Ala and Cys49Val silent mutations. Additional amino acid alterations were introduced by QuikChange (Stratagene) mutagenesis using appropriate sets of primers. DsbB and DsbA were overproduced and purified as described in ref. 16. The DsbB preparations generally contained 0.5-1 molar equivalent of endogenous UQ8 (5). To estimate relative UQ8 contents of different DsbB preparations, they were extracted with organic solvent (6) and subjected to reverse-phase column chromatography (11).
Spectroscopic Analyses. DsbB preparations were scanned for absorbance at 340-700 nm by using a Hitachi U-3310 spectrophotometer in 50 mM Na-phosphate buffer (pH 8.0) containing 0.1 M NaCl, 0.1% dodecyl-β-d-maltoside, and 0.1 mg/ml E. coli phospholipids (Avanti Polar Lipids) (18).
Quantum Chemistry Calculations. Optimal geometry and absorption maxima (excitation energies) of CT complexes composed of benzoquinone (BQ), thiolate, and guanidinium groups were assessed for the general arrangement of these components shown in Fig. 3 by quantum chemistry calculations using the program package gamess (19), without taking dielectric environment into account. Geometry optimizations for the complex were carried out by the density functional calculations with the B3LYP exchange-correlation functional. We used 6-31G** basis sets with additional diffuse functions on BQ and thiolate moieties. The geometry was optimized with a constraint that keeps an angle of C2 of BQ, S of thiolate, and Cζ of guanidinium groups to be 120°. The constraint virtually maintains orientations of those groups in a manner consistent with the experimentally supported inference that Cys-44 and Arg-48 are positioned with mutual proximity in the α-helical secondary structure. The angle constraint also furnishes the CT complex with a tetrahedral coordination centered at the thiolate anion, which gives rise to chemically stable interaction involving this thiolate anion.
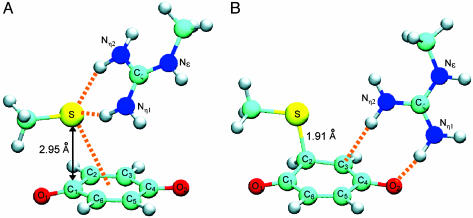
Fig. 3.
Quantum chemical simulation models for a CT complex and an adduct between thiolate and quinone in conjunction with guanidinium. Molecular structures of CT complex (A) and adduct (B), both consisting of BQ (bottom), thiolate (upper left), and guanidinium (upper right) moieties, were modeled by density functional theory calculations. No dielectric environment was taken into account throughout the calculations. Orange broken lines in A indicate putative hydrogen bonds between thiolate and guanidinium as well as CT interaction between thiolate and BQ, whereas those in B indicate hydrogen bonds between BQ and guanidinium. Atoms are color-coded: light blue, carbon; dark blue, nitrogen; red, oxygen; yellow, sulfur; white, hydrogen. The CT complex in A exhibits strong CT photo-absorptions at 510 and 487 nm, with oscillator strengths of 0.15 and 0.04, respectively. The adduct complex in B forms through a bond formation between the S atom of thiolate and the C2 atom of BQ accompanied by negative CT from the thiolate anion to BQ. The guanidinium group accordingly moves and stabilizes the transferred negative charge distributing in a region around the O2 atom of BQ.
The excitation energies were computed by the multiconfigurational quasidegenerate perturbation theory (MCQDPT) method following complete active space self-consistent field (CASSCF) calculations. The basis functions used were the same as those for the geometry optimization. The complete active space is constructed with 12 electrons in 10 active orbitals including valence π and π* ones of BQ and two lone pair ones of thiolate. The ground state and two nearly degenerate CT excited states characterized by excitations from lone pairs of thiolate to π* of BQ were averaged with weights of 0.5, 0.25, and 0.25, respectively. The oscillator strengths of the CT excitations were evaluated at the CASSCF level with corrections using the MCQDPT excitation energies.
The adduct complex shown in Fig. 3B was found by approaching thiolate to BQ. During the geometry optimization of the adduct complex, an oxygen atom of the reduced BQ moiety was suggested to withdraw a nearby proton of the guanidinium group. The occurrence of this proton transfer in DsbB was questionable because the computation did not take protein environments surrounding the complex into account; in the actual protein, several polar groups stabilize the positive charge of the guanidinium group, keeping its pKa high. We therefore imposed a constraint on the Nη1—H bond length, a factor important for such hypothetical proton transfer, such that it is to be 1.01 Å, the Nη1—H bond length in isolation. Otherwise, the geometry optimization was carried out without any constraints.
Results and Discussion
Single Quinone Undergoes Spectral Transition on DsbB. Previously, we prepared quinone-free DsbB [DsbB(ΔQ)] by expressing DsbB in the ubiA- menA- mutant cells defective in the biosynthesis of UQ and menaquinone (11, 16). The absence of any detectable quinone species was shown by HPLC elution profiles of the organic solvent extracts of the preparation (11). The quinone-free DsbB supported the development of ≈500-nm absorbance when supplemented with UQ1 and DsbA(C33S) (16). The absorbance intensity was UQ1 concentration-dependent and saturable at an equimolar UQ1:DsbB ratio (16). Because this reaction took place in the absence of any previously bound quinone species, involvement of a quinhydrone-like CT complex (17) composed of two quinone rings is highly unlikely. Instead, a CT complex of quinone could involve a different partner, the Cys-44 thiolate anion (16).
In Addition to Cys-44, Arg-48 Is Essential for the Quinone Transition. A segment immediately C-terminal to Cys-44 is important for both DsbB functionality (20) and quinone transition (16). The particular importance of Arg-48, an evolutionarily conserved residue (18) located at the distal end of this segment and around the membrane-periplasm interface, has been suggested by deleterious alanine (20) and histidine (18) substitutions, the latter of which increasing DsbB's Km for UQ ≈7-fold (18).
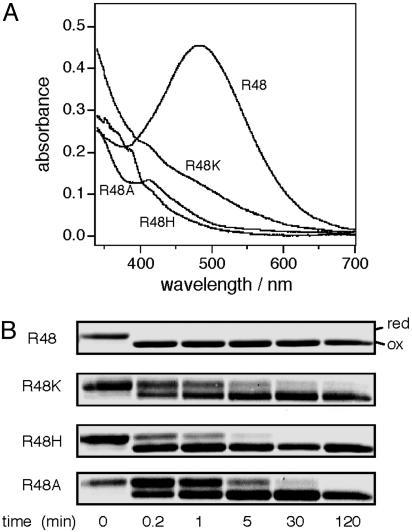
We examined effects of different amino acid substitutions at this position on UQ transition, using the SCSS form of DsbB, in which the UQ red shift is induced constitutively (16). Substitutions of His and Ala abolished the 500-nm absorbance peak, and that of Lys decreased it to form a shoulder around this wavelength (Fig. 1A). All of the mutant forms of DsbB[SCSS] contained somewhat reduced but substantial levels of bound UQ (Fig. 6, which is published as supporting information on the PNAS web site). The complete disappearance of the 500-nm peak observed with the Arg48His and Arg48Ala mutants (Fig. 1 A) is difficult to explain solely in terms of the moderate reduction in the bound UQ. Furthermore, they did not develop any appreciable 500-nm absorbance peak even when supplemented with excess UQ1 (K. Inaba, unpublished result). We suggest that chemical properties of Arg-48 together with the Cys-44 thiolate anion contribute to the electronic transition of UQ on DsbB.
Fig. 1.
Importance of Arg-48 of DsbB in spectral transition of bound UQ and UQ-dependent oxidation of Cys-41 and Cys-44. (A) Arg-48 mutational effects on DsbB[SCSS] absorbance spectrum. Absorption spectra at pH 8.0 were recorded for 100 μM solutions of DsbB[SCSS] and its variants having the indicated amino acid substitution. (B) Mutational effects on UQ-dependent formation of the Cys-41-Cys-44 disulfide bond. Reduced forms of DsbB[CCSS] and its variants (10 μM), purified as quinone-free forms (11), were mixed with UQ1 (100 μM). The mixtures in 50 mM Na-Pi (pH 8.0) containing 0.3 M NaCl, 0.1% dodecyl-β-d-maltoside, and 0.1 mg/ml E. coli phospholipids were incubated at 25°C. At specified time points, samples were withdrawn into an equal volume of 10% trichloroacetic acid and then processed for 4-acetamide-4′-maleimidylstilbene-2-2′-disulfonate modification, SDS/PAGE (12%), and Coomassie brilliant blue staining (6, 16).
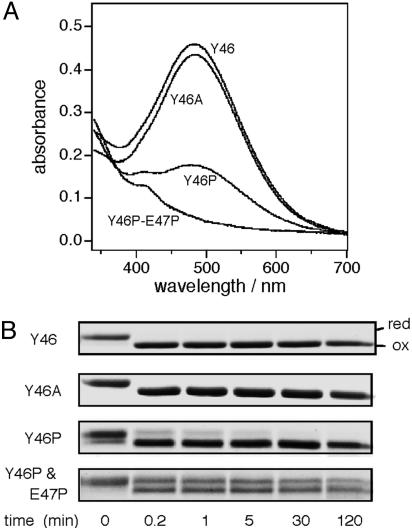
Importance of an α-Helical Arrangement That Coordinates the Positioning of Cys-44 and Arg-48 Side Chains in UQ Transition. The psipred program suggested that the Leu-43-to-Ile-63 segment assumes an α-helix (21). Thus, Cys-44 and its fourth neighbor, Arg-48, will have their side chains oriented to the same side of the helix axis. To examine the importance of the helical configuration of this region, we substituted proline, a helix breaker, for position 46 and for both positions 46 and 47 in DsbB[SCSS]. The Tyr46Pro variant exhibited greatly reduced A500 peak, and the Tyr46Pro-Glu47Pro double mutant protein lacked the peak entirely (Fig. 2A). In contrast, Ala substitution for Tyr-46 did not significantly affect the absorption spectrum. The decreased A500 observed with the proline mutants was not due to a decreased UQ content, because they proved to retain nearly normal amounts of UQ (Fig. 7, which is published as supporting information on the PNAS web site). Thus, destabilization of the putative helical structure around position 46 impairs the DsbB[SCSS]'s ability to induce electronic transition of UQ. These results point to the importance of Arg-48 that is placed at the fourth position from Cys-44 in a context of α-helix.
Fig. 2.
UQ transition and UQ-dependent Cys-41-Cys-44 disulfide bond formation are sensitive to helix-breaking alterations between Cys-44 and Arg-48. (A) Position 46 and 47 mutational effects on DsbB[SCSS] absorbance spectrum. Absorption spectra at pH 8.0 were recorded for 100 μM solutions of DsbB[SCSS] and its variants having the indicated amino acid substitution(s). (B) Mutational effects on UQ-dependent formation of the Cys-41-Cys-44 disulfide bond. DsbB[CCSS] variants as indicated were subjected to the UQ-dependent oxidation assay as described in the legend to Fig. 1B.
The importance of the proper positioning of Cys-44 and Arg-48 can be reinforced by our previous observations, in which insertion of a single alanine after Cys-44, Ile-45, Tyr-46, or Glu-47 impaired the in vivo function of DsbB (20) as well as the in vitro induction of UQ transition shown for the Ala insertion after Tyr-46 (16). These insertions will rotate the Cys-44 and Arg-48 side chains by ≈100°. In contrast, neither Ile45Ala, Tyr46Ala, or Glu47Ala substitution nor Ala insertion after Arg-48 compromised the DsbB function (20). Although Kobayashi et al. (20) discussed that the distance between Cys-44 and the membrane surface may be important for the respiratory chain coupling of DsbB, we now consider that what is actually important may be the length of the helix that separates Cys-44 and Arg-48.
Correlations Among UQ Transition, DsbB Functionality, and UQ-Dependent de Novo Formation of the Cys-41-Cys-44 Disulfide. UQ selectively oxidizes the Cys-41 and Cys-44 pair of DsbB cysteines (8, 14), and this can be regarded as a main reaction of disulfide bond generation (see below). Thus, we used DsbB[CCSS] and the 4-acetamide-4′-maleimidylstilbene-2-2′-disulfonate modification assay to follow this important reaction in vitro. At 25°C, it was completed very rapidly (within 12 sec, the earliest possible sampling point) (R48 and Y46A in Figs. 1B and 2B). We then studied the effects of the mutations at positions 46-48 by introducing them into DsbB[CCSS]. All of the mutations that affected the CT transition of UQ retarded the oxidation reaction as well (Figs. 1B and 2B; mutants other than Tyr46Ala). Among them, Arg48Ala and the double proline substitution showed the most pronounced effects. Such a correlation supports the notion that the CT state of UQ is important for the generation of the Cys-41-Cys-44 disulfide bond. Its importance for the enzymatic activity of the intact DsbB is shown in Table 1, in which the in vitro DsbA oxidation activities of different variants correlate well with their CT propensities.
Table 1. DsbA oxidation activities and absorbance at 500 nm for DsbB variants constructed in this study.
| DsbB | Rate of DsbA oxidation, nM DsbA/nM DsbB per s | A500* |
|---|---|---|
| Wild type | 5.3 | 0.45 |
| DsbB (R48K) | 1.4 × 10-1 | 0.10 |
| DsbB (R48H) | 1.0 × 10-1 | 0.02 |
| DsbB (R48A) | <1.0 × 10-2 | 0.04 |
| DsbB (Y46A) | 1.0 | 0.42 |
| DsbB (Y46P) | 2.1 × 10-2 | 0.17 |
| DsbB (Y46P and E47P) | <1.0 × 10-2 | 0.05 |
Simulated Chromophore Generation for a Thiolate-Quinone Complex in Association with a Positively Charged Guanidinium Group. Our biochemical results suggest strongly that the spectral transition of quinone represents the formation of a CT complex between Cys-44 thiolate anion and quinone. The role of Arg-48 could be to stabilize the CT state by electrostatically counteracting the thiolate. These observations and considerations prompted us to analyze CT excitation of a thiolate-quinone complex by means of quantum chemistry simulation.
First, density functional theory calculations (see Materials and Methods) were used to model structure of the thiolate-BQ CT complex that is further associated with a guanidinium group. Fig. 3A depicts the modeled CT complex structure. The thiolate anion is coordinated to the π orbitals of BQ by a strong CT interaction indicated by the short distance (2.95 Å) between the thiolate S atom and the proximal C1 atom of BQ. The guanidinium group of Arg-48 interacts with the thiolate anion in a tetrahedral manner and stabilizes the negative charge of the thiolate.
Absorption maxima of the CT excitations of the complex were then computed by using the multiconfigurational quasidegenerate perturbation theory (22) (see Materials and Methods). There are two nearly degenerate and low-lying CT excited states characterized by the excitations from each of two lone pairs of thiolate to a π * orbital of BQ. Excitation energies of the CT states were calculated to be 510 and 487 nm and are in good agreement with the experimentally observed absorption maximum, ≈500 nm. Accordingly, the relatively broad peak observed at 500 nm, as shown in Fig. 1 A, should have contained the two CT excitation components.
Our calculations predicted a substantial absorbance intensity for the CT excitations; oscillator strengths were estimated to be 0.15 and 0.04, respectively (Fig. 3A), and the overall oscillator strength of the CT band (0.19) is ≈50% of the allowed π-π* excitation (0.44) of BQ (23). In fact, the experimentally observed absorbance of the CT band of UQ on DsbB was about a half of the strong π-π* band at 280 nm (K. Inaba, unpublished results), consistent with the estimation by calculations based on Fig. 3A.
It should be noted that the present simulation procedures provide a molecular model of the CT complex in only qualitative terms, because of the lack of structural information for the surrounding protein environments. Nevertheless, our simple model is consistent with available experimental evidence in a manner to extract basic molecular origin of the anomalous spectral transition. An arginine residue can fulfill the role of stabilizing the UQ-cysteine CT complex, because of its extremely high pKa value and long side chain. The high pKa and the flexible side chain will allow a firm interaction of arginine with the cysteine thiolate, keeping the anionic state of the latter, an acid of relatively high pKa value. The anionic character of thiolate will enhance its ability to transfer negative charges to quinone and to stabilize the CT complex. Although lysine could also act as a counter ion group, its lower pKa that originates from less pronounced intramolecular delocalization of the positive charge may induce an extensive intermolecular transfer of its own charges or a proton to the thiolate, thus counteracting the thiolate-to-quinone CT and resulting in the lowered absorbance at 500 nm, as seen in Fig. 1 A.
It is also noteworthy that the UQ-cysteine CT interaction could itself lower the pKa of the electron donor, as suggested for the interaction between flavin and acyl-CoA thioester in acyl-CoA dehydrogenase (24). Thus, the arginine group may play a catalytic role in the synergistic formation of the thiolate anion and the cysteine-UQ CT complex.
Possible Formation of a Cysteinyl-Quinone Adduct Having a Covalent S—C Linkage. Interestingly, computer simulations predicted further that a quinone-cysteinyl covalent adduct can be formed if further approach of the thiolate anion to quinone is possible (see Materials and Methods). Fig. 3B illustrates the molecular structure of this complex, in which a chemical bond of 1.91 Å has been formed between the thiolate S atom and the quinone C2 atom. In this complex, charge redistributions occur such that a negative charge originally located on the thiolate anion is transferred to a region around the O2 atom of quinone, and the guanidinium group accordingly undergoes a conformational shift to stabilize the translocated negative charge. This molecular state could mean that the positively charged guanidinium group adjacent to the UQ ring increases the electrophilicity of BQ. Our calculation showed that the potential energy of the covalent adduct (Fig. 3B) is 5.3 kcal/mol lower than that of the CT complex (Fig. 3A), indicative of inherent stability of the adduct. However, we believe that this difference that accounts for the adduct in virtual isolation does not necessarily hold exactly for the situations in the actual protein environments. Thus, relative stability of the CT complex and the thiolate-BQ adduct may be modulated in DsbB such that the former can persist under certain conditions and the latter can only exist transiently as a transition state of the reaction (see below).
Geometry of Cys-44, Arg-48, and UQ Responsible for the 500-nm Absorption. The 500-nm absorption is abolished by protein denaturants such as guanidine-HCl and SDS (K. Inaba, unpublished results), showing the importance of native DsbB conformation for the CT transition. We have already discussed that Cys-44 and Arg-48 should be placed on an α-helix and separated by three residues in between. This feature is conserved among the DsbB/DsbI family of proteins (25). Arg-48 is immediately followed by the second transmembrane sequence and is likely to be “snorkeling” from the lipid phase (26). Thus, its guanidinium group will be available for the electrostatic interaction with the Cys-44 thiolate in the aqueous milieu of the periplasm. At the same time, the membrane anchoring will restrict the movement of this segment and could provide a platform on which the thiolate-UQ CT reaction can take place efficiently. Such an arrangement will be particularly advantageous in this case, because UQ is also anchored to the membrane by means of its hydrophobic isoprenyl tail.
Essential information that is still lacking is the primary site of UQ binding in DsbB. Photo-crosslinking experiments using a derivatized quinone analogue revealed that a N-terminal region of the second periplasmic domain was in the vicinity of quinone (27). Although functional importance of Gln-95 and His-91 in this region was suggested (21, 28), a His91Ala variant of DsbB was able to develop A500 in the presence of DsbA(C33S), albeit with somewhat reduced efficiency (K. Inaba, unpublished results). In any case, UQ might interact with both of the two periplasmic domains of DsbB near their membrane-embedded regions in a specific orientation that enables CT from the Cys-44 thiolate.
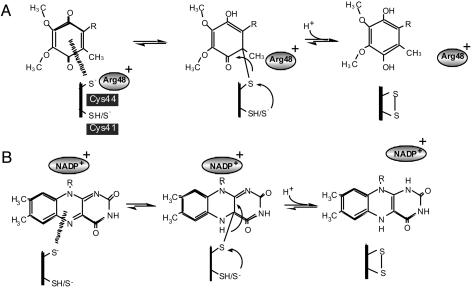
Thiolate-UQ Adduct Could Generate a Transition State in the de Novo Formation of a Disulfide Bond in DsbB/DsbA. Both the rapid and slow pathways of DsbA oxidation can proceed without any involvement of quinone up to significant points, after which quinone becomes essential (ref. 11 and Fig. 5). Remarkably, both pathways involve the quinone spectral transition (11, 16). In the predominant rapid pathway, direct disulfide exchange between Cys-104-Cys-130 (DsbB) and the reduced active site of DsbA is followed by rapid thiol-disulfide equilibration between the Cys-104/Cys-130 and the Cys-41/Cys-44 pairs within DsbB. The role of UQ is to oxidize the latter pair of cysteines; in other words, UQ creates this disulfide bond. We propose that the formation of the Cys-44-UQ adduct with the C—S bond is a crucial event in the de novo disulfide bond formation, in which a nucleophilic attack of the C—S bond by Cys-41 is induced to form the Cys-41-Cys-44 disulfide and to reduce the BQ ring (Fig. 4A). In agreement with this model, the DsbB mutations that impaired the quinone transition significantly retarded the UQ-dependent formation of the Cys-41-Cys-44 disulfide bond.
Fig. 4.
A critical step of disulfide bond generation that is coupled with Cys-44 to UQ electron transfer. (A) Proposed reaction scheme for DsbB. When Cys-44 is reduced, it induces a Cys-44-UQ CT complex that is in equilibrium with its adduct form having the S—C covalent linkage. At a moment of the latter, Cys-41 of DsbB attacks the Cys-44-UQ covalent bond to gain the Cys-41-Cys-44 disulfide at the expense of UQ reduction to UQH2. Arg-48 that is positively charged plays a crucial role in stabilization of the Cys-44-UQ complexes by increasing either nucleophilicity of Cys-44 or electrophilicity of UQ. (B) A conceptually similar electron transfer mechanism proposed for FAD-containing enzymes, lipoamide dehydrogenase, and mercuric ion reductase (29, 31). In these cases, FAD and NADP+ may assume the role assigned above for UQ and Arg-48, respectively.
In the slow pathway, the thiolate-UQ adduct will induce a cascade of concerted nucleophilic attacks, in which Cys-33 attacks Cys-30, Cys-104 attacks Cys-130, and Cys-41 attacks the C—S bond of the adduct, resulting in a net gain of one disulfide bond. Taken together, we conclude that the transient formation of the Cys-44-UQ CT and adduct complexes acts as a key molecular device that enables the DsbB-DsbA system to serve as the source of protein disulfide bonds in the periplasm.
Do Quinone- and Flavin-Dependent Disulfide Oxidoreductases Share a Convergent Reaction Strategy? Cysteine-based complexes of FAD were described for lipoamide dehydrogenase and mercuric ion reductase, in which a cysteine-FAD CT complex with absorption of ≈500∼600 nm is transiently generated during the catalytic reaction of these enzymes (29-32). CT and adduct complexes between cysteine and FAD are significantly stabilized by NAD+/NADP+, another cofactor of the system, the effect of which can be explained in terms of the positive charge enhancement of the electrophilicity of the adjacent FAD isoalloxazine ring. Thus, we can find a convergent strategy of electron transfer in some thiol-based oxidoreductases, in which both common and divergent tripartite elements, thiolate, the BQ/isoalloxazine ring, and a positively charged molecule participate (Fig. 4).
It is possible that this strategy is adopted also by Ero1p and Erv2p, FAD-containing sulfhydryl oxidases from eukaryotic cells. Sevier et al. (21) proposed that these enzymes and DsbB share similar architecture in their catalytic domains, in which a four-helix bundle scaffold structure embraces a cofactor inside, near one of active site cysteines. Although crystal structure of the oxidized form of the Ero1p soluble domain did not reveal the presence of a positive charge placed close to the isoalloxazine ring (33), it does possess highly conserved cationic amino acids on both sides of Cys-355 adjacent to the ring. Erv2p has Lys-78 and FAD nearby Cys-124 (34); the Lys-78 Nζ—H2 is only 2.96 Å away from a carbonyl O2 atom of the isoalloxazine ring. The equivalent residue is Arg in Erv-like domains of human and rat augmenter-of-liver-regeneration proteins (35, 36). Furthermore, some flavoenzymes that contain an ERV/ALR domain exhibit prominent thiolate-to-flavin CT bands (37, 38). It is of prime interest to ask whether Erv2p/Ero1 undergoes any spectroscopic transition in the course of its catalytic reactions.
Conclusion
DsbB provides the first example of biological oxidoreduction mechanism that utilizes the cysteine-UQ CT complex. It was found that three essential elements, a thiolate anion, a counteracting positive-charge moiety, and a quinone ring, allow a thiolate-to-quinone CT and covalent bonding that facilitate the system to create one S—S bond de novo. Information obtained in this system by combined genetic, biochemical, spectroscopic, and quantum chemical approaches should be important for our understanding not only of cellular oxidative protein folding but also of the expanding roles of UQ-dependent disulfide bond generation in cellular control (39). Atomic-level structural analysis of these systems is most eagerly awaited, because such information should also contribute to our understanding of similar processes involving FAD as a cofactor, which is widely conserved in eukaryotic cells.
Supplementary Material
Acknowledgments
We thank Robert Gennis and Colin Thorpe for drawing our attention to the flavin-cysteinate complexes. We are also grateful to Yoshinori Akiyama for useful suggestions and Junko Moriwaki-Hattori and Kiyoko Mochizuki for technical support. This work was supported by PRESTO (K. Inaba and S.H.) and Core Research for Evolutional Science and Technology (to K. Ito), Japan Science and Technology Agency, grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and by its National Project on Protein Structural and Functional Analyses (K. Ito).
Author contributions: K. Inaba, K. Ito, and S.H. designed research; K. Inaba, Y.-h.T., and S.H. performed research; K. Inaba, Y.-h.T., and S.H. analyzed data; and K. Inaba, K. Ito, and S.H. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: UQ, ubiquinone; BQ, benzoquinone; CT, charge transfer.
References
- 1.Collet, J.-F. & Bardwell, J. C. A. (2002) Mol. Microbiol. 44, 1-8. [DOI] [PubMed] [Google Scholar]
- 2.Kadokura, H., Katzen, F. & Beckwith, J. (2003) Annu. Rev. Biochem. 72, 111-135. [DOI] [PubMed] [Google Scholar]
- 3.Kobayashi, T., Kishigami, S., Sone, M., Inokuchi, H., Mogi, T. & Ito, K. (1997) Proc. Natl. Acad. Sci. USA 94, 11857-11862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bader, M., Muse, W., Ballou, D. P., Gassner, C. & Bardwell, J. C. A. (1999) Cell 98, 217-221. [DOI] [PubMed] [Google Scholar]
- 5.Bader, M., Xie, T., Yu, C.-A. & Bardwell, J. C. A. (2000) J. Biol. Chem. 275, 26082-26088. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi, Y.-h., Inaba, K. & Ito, K. (2004) J. Biol. Chem. 279, 47057-47065. [DOI] [PubMed] [Google Scholar]
- 7.Jander, G., Martin, N. L. & Beckwith, J. (1994) EMBO J. 13, 5121-5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kobayashi, T. & Ito, K. (1999) EMBO J. 18, 1192-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guilhot, C., Jander, G., Martin, N. L. & Beckwith, J. (1995) Proc. Natl. Acad. Sci. USA 92, 9895-9899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kishigami, S., Kanaya, E., Kikuchi, M. & Ito, K. (1995) J. Biol. Chem. 270, 17072-17074. [DOI] [PubMed] [Google Scholar]
- 11.Inaba, K., Takahashi, Y.-h. & Ito, K. (2005) J. Biol. Chem. 280, 33035-33044. [DOI] [PubMed] [Google Scholar]
- 12.Kadokura, H. & Beckwith, J. (2002) EMBO J. 21, 2354-2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inaba, K. & Ito, K. (2002) EMBO J. 21, 2646-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Regeimbal, J. & Bardwell, J. C. A. (2002) J. Biol. Chem. 277, 32706-32713. [DOI] [PubMed] [Google Scholar]
- 15.Grauschopf, U., Fritz, A. & Glockshuber, R. (2003) EMBO J. 22, 3503-3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inaba, K., Takahashi, Y.-h., Fujieda, N., Kano, K., Miyoshi, H. & Ito, K. (2004) J. Biol. Chem. 279, 6761-6768. [DOI] [PubMed] [Google Scholar]
- 17.Regeimbal, J., Gleiter, S., Trumpower, B., Yu, C.-A., Diwaker, M., Ballou, D. P. & Bardwell, J. C. A. (2003) Proc. Natl. Acad. Sci. USA 100, 13779-13784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kadokura, H., Bader, M., Tian, H., Bardwell, J. C. A. & Beckwith, J. (2000) Proc. Natl. Acad. Sci. USA 97, 10884-10889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt, M. W., Baldridge, K. K., Boatz, J. A., Elbert, S. T., Gordon, M. S., Jensen, J. H., Koseki, S., Matsunaga, N., Nguyen, K. A., Su, S., et al. (1993) J. Comput. Chem. 14, 1347-1363. [Google Scholar]
- 20.Kobayashi, T., Takahashi, Y. & Ito, K. (2001) Mol. Microbiol. 39, 158-165. [DOI] [PubMed] [Google Scholar]
- 21.Sevier, C. S., Kadokura, H., Tam, V. C., Beckwith, J., Fass, D. & Kaiser, C. A. (2005) Protein Sci. 14, 1630-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakano, H. (1993) J. Chem. Phys. 99, 7983-7992. [Google Scholar]
- 23.Brint, P., Connerade, J.-P., Tsekeris, P., Bolovinos, A. & Baig, A. (1986) J. Chem. Soc. Fraday Trans. 2 82, 367-375. [Google Scholar]
- 24.Dmitrenko, O., Thorpe, C. & Bach, D. (2003) J. Phys. Chem. B 107, 13229-13236. [Google Scholar]
- 25.Raczko, A. M., Bujnicki, J. M., Pawlowski, M., Godlewska, R., Lewandowska, M. & Jagusztyn-Krynicka, E. (2005) Microbiology 151, 219-231. [DOI] [PubMed] [Google Scholar]
- 26.Liang, J., Adamian, L. & Jackups, R., Jr. (2005) Trends Biochem. Sci. 30, 355-357. [DOI] [PubMed] [Google Scholar]
- 27.Xie, T., Yu, L., Bader, M., Bardwell, J. C. A. & Yu, C.-A. (2002) J. Biol. Chem. 277, 1649-1652. [DOI] [PubMed] [Google Scholar]
- 28.Tan, J., Lu, Y. & Bardwell, J. C. A. (2005) J. Bacteriol. 187, 1504-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thorpe, C. & Williams, C. H., Jr. (1976) J. Biol. Chem. 251, 7726-7728. [PubMed] [Google Scholar]
- 30.Thorpe, C. & Williams, C. H., Jr. (1981) Biochemistry 20, 1507-1513. [DOI] [PubMed] [Google Scholar]
- 31.Miller, S., Massey, V., Ballou, D., Williams, C. H., Distefano, M. D., Moore, M. J. & Walsh, C. T. (1990) Biochemistry 29, 2831-2841. [DOI] [PubMed] [Google Scholar]
- 32.Williams, C. H., Jr. (1992) Chemistry and Biochemistry of Flavoenzymes, ed. Müller, F., III (CRC, Boca Raton, FL), pp. 121-211.
- 33.Gross, E., Kastner, D. B., Kaiser, C. A. & Fass, D. A. (2004) Cell 117, 601-610. [DOI] [PubMed] [Google Scholar]
- 34.Gross, E., Sevier, C. S., Vala, A., Kaiser, C. A. & Fass, D. A. (2002) Nat. Struct. Biol. 9, 61-67. [DOI] [PubMed] [Google Scholar]
- 35.Wu, C. K., Dailey, T. A., Dailey, H. A., Wang, B. C. & Rose, J. P. (2003) Protein Sci. 12, 1109-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farrell, S. R. & Thorpe, C. (2005) Biochemistry 44, 1532-1541. [DOI] [PubMed] [Google Scholar]
- 37.Raje, S. & Thorpe, C. (2003) Biochemistry 42, 4560-4568. [DOI] [PubMed] [Google Scholar]
- 38.Hofhaus, G., Lee, J. E., Tews, I., Rosenberg, B. & Lisowsky, T. (2003) Eur. J. Biochem. 270, 1528-1535. [DOI] [PubMed] [Google Scholar]
- 39.Malpica, R., Franco, B., Rodriguez, C., Kwon, O. & Georgellis, D. (2004) Proc. Natl. Acad. Sci. USA 101, 13318-13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.